Abstract
Kinase-associated protein phosphatase interacts specifically with plant receptor-like protein kinases. This interaction is thought to be a key step in signal perception and transduction. The minimal kinase interaction (KI) domain of kinase-associated protein phosphatase was mapped to a 119-aa segment spanning residues 180 to 298. A forkhead-associated (FHA) homology region resides in this minimal KI domain. Site-directed mutagenesis of four highly conserved sites in this FHA homology region abolishes the KI domain’s interaction with receptor-like protein kinases, indicating that the FHA region is essential for binding. Serial deletion analysis indicates that 30 aa on each side of the FHA region are also needed for binding; this minimal functional unit is designated as the KI domain. Kinetic studies using surface plasmon resonance indicate that the binding between the KI domain and receptor-like protein kinases has a dissociation constant (KD) of about 25–100 nM, which is similar to the binding affinity of two other well characterized phosphorylation-dependent protein-binding domains (14-3-3 and Src homology 2) and their high-affinity phosphopeptide ligands.
Kinase-associated protein phosphatase (KAPP) is a downstream regulatory component in receptor-like protein kinase (RLK) signal transduction pathways that recognizes the phosphorylated form of an RLK protein kinase domain. Arabidopsis KAPP was cloned through its interaction with the cytoplasmic domain of RLK5, a serine/threonine protein kinase (1). KAPP contains three functional domains: a type I membrane anchor, a kinase interaction (KI) domain, and a type 2C protein phosphatase. KAPP has been identified in Arabidopsis and maize (1, 2). Northern blots and in situ hybridization analyses showed that KAPP mRNA is expressed in many plant tissues, such as roots, leaves, and apical and floral meristems (1, 3). In vitro binding studies revealed that KAPP interacts with a subset of plant RLKs (2). Therefore, KAPP is considered a downstream regulatory protein in many RLK signal transduction pathways. The role of KAPP in the Arabidopsis CLAVATA1 (CLV1) signal transduction pathway has recently been elucidated by both biochemical and genetic approaches (3–5). CLV1 is a leucine-rich repeat RLK that plays an important role in the proper balance between cell proliferation and differentiation in the Arabidopsis shoot apical and floral meristems (6). In vitro, KAPP binds to CLV1 protein kinase in a phosphorylation-dependent manner (3, 4). The in vivo interaction between the KI domain and CLV1 in plant cells was confirmed by coimmunoprecipitation analysis (4, 5). Introduction of a KAPP overexpression transgene into wild-type Arabidopsis resulted in the production of plants with a weak clv1 phenotype (3). Conversely, reduction of KAPP mRNA levels in an intermediate clv1 allele background (clv1–1) recovered a wild-type Arabidopsis phenotype (4). These results suggest that KAPP binds CLV1 and negatively regulates CLV1 signaling.
The physical interaction between KAPP and RLKs is thought to be important for their roles in signal transduction. The KI domain binds RLKs in vitro in a phosphorylation-dependent manner and does not bind autophosphorylation-deficient mutants of RLKs, such as RLK5, CLV1, and KIK1 (KI domain interacting kinase 1, a RLK from maize with an unknown biological function) (1–3). Despite its importance, the KI domain has not been fully defined. Sequence alignment showed that the KI domain has no sequence similarity with several well characterized phosphopeptide-binding domains, such as Src homology 2 (SH2) domains (7–10), 14-3-3 proteins (11, 12), protein tyrosine-binding domains (8, 13), cAMP-responsive element binding protein-binding protein (14–17), and WW domains (18, 19). However, within the KI domain there exists a short stretch (52 aa) homologous to the forkhead-associated (FHA) domain, a conserved region in the forkhead protein family (20). This homology was discovered by using a generalized sequence profile method. Forkhead proteins are putative transcription factors with a highly conserved DNA-binding region of about 110 aa named the forkhead domain (21). At least 40 members of the family have been identified. The FHA domain is located outside the DNA-binding domain, indicating a function different from that of the forkhead domain. The FHA domain contains 55 to 75 aa and has three highly conserved blocks (20). FHA domains have been identified in various signal transduction proteins such as protein kinases, protein phosphatases, transcription factors, and other proteins (20). The observation that the FHA domain was found within the region of KAPP, known to be important for the interaction with phosphorylated RLKs, led to the suggestion that the FHA domain is involved in binding phosphorylated proteins (20). This hypothesis has recently been supported by the observation that a FHA-domain-containing protein from yeast interacts with another phosphorylated protein (22). Using the yeast two-hybrid system with a loss-of-function RAD53 mutant bait, Sun et al. identified a protein, RAD9, which interacts with RAD53 when RAD9 is phosphorylated (22). Deletion analysis revealed that one of the two FHAs in RAD53, FHA2, is involved in its interaction with RAD9 in response to DNA damage. These data show that the FHA homology region is involved in protein–protein interactions and supports the idea that the FHA domain may be part of a new class of phosphoprotein-binding modules (23).
Both the demonstration of KAPP’s key role in the Arabidopsis CLAVATA1 signal transduction pathway and the observation that the FHA domain of RAD53 is important for interaction with RAD9 have prompted us to characterize the FHA-containing KI domain. Serial deletion mutants have been generated to map the KI domain of KAPP to a minimum 119-residue segment of the protein. Furthermore, alteration of four conserved residues of FHA abolished binding, implicating FHA in RLK interaction. The kinetics of binding between KI domain and RLKs have been measured by surface plasmon resonance (SPR). This information provides new insights into this domain and allows the comparison of the KI domain with other phosphopeptide-interaction domains.
MATERIALS AND METHODS
Construction of Glutathione S-Transferase (GST) and MBP Fusion Protein Expression Vectors.
All KAPP recombinant vectors were constructed by PCR and cloned into the EcoRI and HindIII sites of a GST fusion protein vector, GTK+, derived from pGEX-2T (Amersham Pharmacia) (2). The modified GST fusion construct contains a Flag sequence and a protein kinase A site just before EcoRI site. The end points of the KAPP deletion mutants are shown in Fig. 1A. Four site-directed missense mutants, G211E (amino acid 211 glycine to glutamic acid), S226A, H229L, and N250S, were generated on GST-KID239 (1) by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). In-frame cloning and point mutations were confirmed by DNA sequencing. The maltose-binding protein (MBP)-RLKs used in this paper have been reported (1–3).
Figure 1.
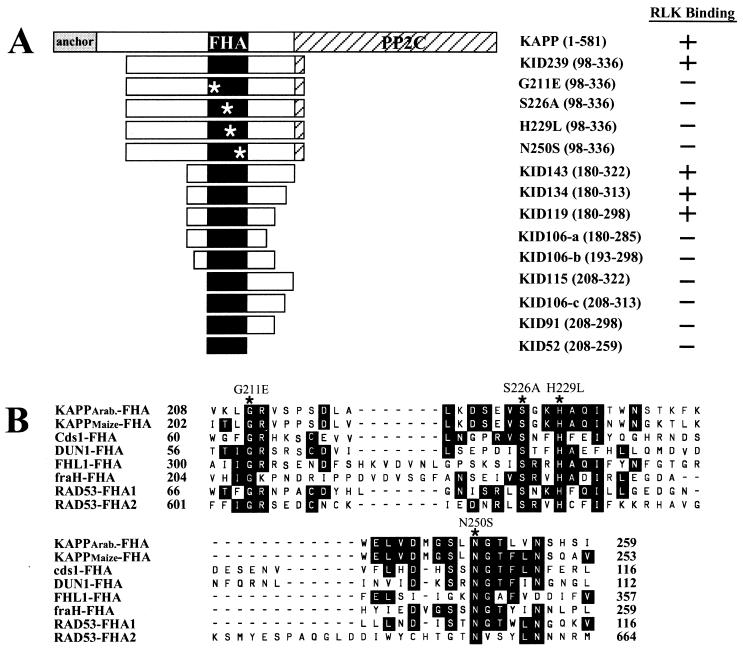
Schematic diagram of KAPP mutants and FHA alignments. (A) KAPP mutants including nine deletion mutants and four site-directed mutants. The largest KAPP fragment, 239 aa in length, was initially identified by interaction cloning (1). The smallest fragment is a 52-aa region with homology to the FHA sequence. (Right) Summary of binding between these KAPP mutants and RLKs from three different binding analyses. (B) Amino acid sequence alignment of the FHA domains from eight representative proteins. The FHA homology region has been identified in many eukaryotic proteins, including transcription factors [FHL1 (26)], kinases [Cds1 (27); DUN1 (28); and RAD53 (29)], phosphatases [KAPPs (1, 2)] and other proteins (fraH, GenBank accession no. U14553). Four highly conserved amino acid residues were chosen for site-directed mutagenesis (∗). Amino acid positions of KAPP were based on the posted sequence from GenBank (accession no. U09505).
Purification of GST and MBP Fusion Proteins.
All fusion constructs (KAPP fragment fused to GST, RLK fragment fused to MBP) were introduced into Escherichia coli. The expression levels were examined by running an SDS-polyacrylamide gel of the total bacterial proteins followed by Coomassie blue staining. The highest expressing colony for each particular construct was picked and used for further protein purification as described by Horn and Walker (24). The GST-KAPP fusion proteins were purified by affinity chromatography on glutathione agarose resin (Sigma). MBP-RLK fusion proteins were purified by affinity chromatography on amylose agarose (Sigma).
Dot-Blot Analysis.
The MBP-RLK (RLK catalytic domain) fusion proteins were immobilized on a nitrocellulose membrane by hydrophobic interaction (1 μg per spot). The filter was blocked in PBS-T buffer (10 mM NaH2PO4,/150 mM NaCl/0.2% Tween 20, pH 7.5) with 5% nonfat dry milk for at least 30 min at room temperature and cut into pieces to react with different GST fusion proteins. GST fusion proteins (truncated KAPP and KAPP site-directed mutants) were diluted to 1 μg/ml in PBS-T buffer and incubated with the membrane pieces overnight at 4°C. The membrane was washed three times (10 min each time) in PBS-T at room temperature, incubated with 1:3,000 PBS-T diluted anti-Flag monoclonal antibody for 1 hr, washed three times with PBS-T at room temperature, incubated with 1:10,000 PBS-T-diluted peroxidase labeled anti-mouse antibody for 1 hr, and washed three times with PBS-T. The signal was detected with an enhanced chemiluminescence kit (Amersham Pharmacia).
Biotinylation of Fusion Proteins.
The purified GST-KAPP or MBP-RLK fusion proteins were diluted to 1 mg/ml in HBS buffer [10 mM Hepes, pH 7.4/150 mM NaCl/3 mM EDTA/0.005% (vol/vol) Surfactant P20]. To 40 μl protein solution, 5 μl 0.9 mM biotinylated reagent NHS-LC-Biotin (Pierce; dissolved in 2 mM NaOAc, pH adjusted to 6.0 with NaOH) was added to give a final concentration of 100 μM. After 4 hr at 4°C, 500 μl 1 M Tris⋅HCl, pH 7.8, was added and reacted for at least 2 hr at 4°C to quench unreacted biotinylating reagent. After 250 μg BSA was added as a carrier, the solution was concentrated and washed four times with HBS buffer on a Centricon 10 ultrafilter (Amicon). The solution was filtered through a micropure separator 0.45 μM filter (Millipore) to remove aggregates before use.
ELISA.
Purified MBP-RLK fusion proteins were coated on a 96-well ELISA plate (100 ng/well). Biotinylated GST-KAPP fusion proteins at three concentrations were incubated with the coated protein overnight at 4°C in TBS-T buffer (10 mM Tris⋅HCl/150 mM NaCl, pH 7.5/0.5% Tween 20), after which the plates were washed and developed as described previously (25).
SPR Analysis.
SPR analysis was performed on a Pharmacia BIAcore 2000 instrument (Pharmacia Biosensor, Uppsala, Sweden). Biotinylated MBP-RLK proteins were immobilized at various levels on streptavidin chips. The immobilized or bound proteins can be quantified as response units (RUs), which correlate with the accumulated protein mass on the chip matrix. The ratio of interaction between a ligand (an immobilized RLK) and an analyte (a soluble KAPP mutant) can be directly estimated by the SPR signal. Two chips were prepared for the kinetic studies of binding to KIK1 and to CLV1, each chip with four flow cells (FC1 to FC4). On the KIK1 chip, FC1 was left blank as a negative control, and 132 RU, 258 RU, and 482 RU of MBP-KIK1 were immobilized on FC2, FC3, and FC4, respectively. On the CLV1 chip, 0 RU, 640 RU, 1160 RU, and 2493 RU of MBP-CLV1 were coated on flow cells FC1–FC4, respectively.
Preliminary studies showed that both KIK1 and CLV1 chips could be regenerated to 98% capacity with a 2-min exposure to a solution containing 0.1 M glycine (adjusted pH to 12 with NaOH) and 0.3% Triton X-100. This solution was used as the stripping buffer for the remaining experiments. Analytes at three concentrations (25 μg/ml, 50 μg/ml, and 100 μg/ml) in HBS buffer were passed through flow cells FC1–FC4 in series at 10 μl/min at 25°C for 10 min, and association of the analytes with the immobilized RLK was continuously monitored by SPR. To study dissociation of the binding, the HBS buffer alone was passed through the flow cells, with dissociation continuously monitored by SPR. After each 20-min analysis, the chip was stripped for 2 min with stripping buffer and equilibrated with HBS before the next analyte sample was analyzed.
The kinetic parameters were determined at three density levels of each RLK (ligand) and the two lowest concentrations of each analyte, kon and koff, rate constants were calculated with biaevaluation software (Pharmacia Biosensor, Uppsala, Sweden); the dissociation constant KD was calculated as KD = koff/kon.
RESULTS
Production of KAPP Deletion and Amino Acid Replacement Mutants Fused to GST.
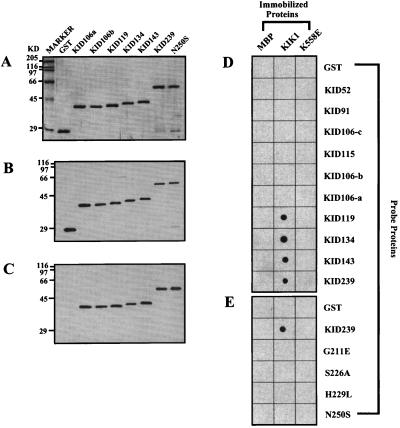
As shown in Fig. 1A, a series of deletion and point mutants of KID239, a 239-residue fragment spanning amino acids 97 to 336 of the Arabidopsis KAPP shown previously to bind the catalytic domain of RLK5 in a phosphorylation-dependent manner, were generated as fusions to GST, with a Flag epitope placed between GST and the KAPP fragment. Deletion end points were chosen on the basis of conserved amino acid sequence blocks between Arabidopsis and maize KAPP proteins, which showed almost identical binding characteristics with a panel of RLKs in a previous study (2). The highly conserved sites in the FHA homology region chosen for mutagenesis are shown in Fig. 1B, an alignment modified from Hofmann and Bucher (20). The quality of the proteins was checked on a Coomassie blue-stained SDS/PAGE gel. All purified proteins had a major band of the predicted size (Fig. 2A).
Figure 2.
Representative binding results from dot-blot immuno analysis. A modified dot-blot immuno analysis was performed to test the binding between GST fusions (KAPP mutants) and MBP fusions (MBP, KIK1 and K558E). Equal amounts (1 μg) of MBP fusion proteins were spotted on the nitrocellulose membrane. The membrane was then cut into many small strips. Each strip, containing three MBP fusion protein spots (MBP, KIK1, and K558E), was incubated with each of the KAPP mutants. The Flag epitope was used to detect the bound GST fusion proteins by chemiluminescent immuno analysis. (A) Coomassie blue-stained SDS/PAGE gel showing the representative KAPP mutant proteins used in the binding experiment. Each lane contained 0.75 μg purified recombinant protein. GST fusion proteins used in these experiments contains a Flag epitope. (B) Immunoblotting results with anti-Flag antibody as the primary antibody and anti-mouse horseradish peroxidase-conjugated IgG as the secondary antibody. Proteins were loaded in the same order as in A. Each lane contains 75 ng of purified protein. (C) Same blot as B. The blot was stripped off and reused for the immunoblot with anti-KAPP antibody as the primary antibody and anti-rabbit peroxydase-labeled IgG as the secondary antibody. (D) Binding results between truncated GST-KAPP fusion proteins and MBP fusions. (E) Binding results between site-directed GST-KAPP mutant proteins and MBP fusions.
Production of RLK Fusion Proteins to Test as Ligands for KAPP Mutants.
Representative RLK catalytic domains were produced as fusions to MBP (2, 4, 24). KIK1, a maize RLK that had previously been shown to bind KAPP particularly well (2) but whose function is not yet known, served as a model RLK. CLV1 is an Arabidopsis RLK that has been shown by genetic and biochemical approaches to interact with KAPP to regulate the development of floral and meristematic tissues. RLK5 is another Arabidopsis RLK; its function has not been reported, but its binding to KAPP was the route through which the KAPP clone was originally identified by interaction cloning. A nonautophosphorylating KIK1 amino acid replacement mutant, K558E, and the MBP fusion partner served as negative controls.
Assessing Binding of KAPP to RLKs by Dot Blotting, ELISA, and SPR.
Three different methods were used to evaluate binding between the KAPP mutants and the various RLKs. In dot-blot analysis, various RLKs and controls were spotted on nitrocellulose membranes, which were then incubated with soluble KAPP mutant proteins. After the membrane was washed to remove unbound KAPP mutant protein, bound KAPP mutant protein was detected via its Flag epitope. The detection sensitivity and reliability of Flag and KAPP antibodies were compared for all the GST fusion proteins involved in the experiment. Both antibodies recognized the same purified fusion protein bands with identical quality (Fig. 2 B and C). The monoclonal anti-Flag antibody was then used for the binding experiments. Representative binding results are shown in Fig. 2 D and E.
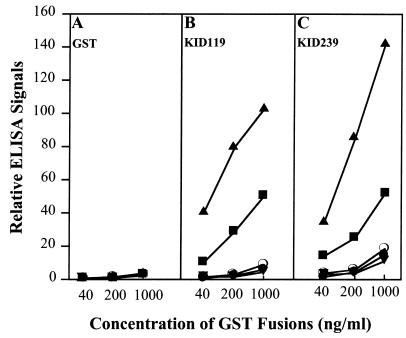
Wells of ELISA dishes were coated with various RLKs and reacted with KAPP mutant proteins at graded concentrations. After the wells were washed to remove unbound KAPP mutant protein, bound KAPP mutant protein was quantified as detailed in Materials and Methods. Representative ELISA results are presented in Fig. 3.
Figure 3.
Representative binding results between GST fusion proteins (GST, KID119, and KID239) and MBP fusion proteins [MBP (●), RLK5 (○), CLV1 (■), KIK1 (▴), and K558E (▾)] by ELISA. MBP fusion proteins were coated on a 96-well ELISA dish (100 ng/well). Biotinylated GST fusion proteins (40 ng/ml, 200 ng/ml, and 1,000 ng/ml) were incubated with the MBP fusion protein-coated wells overnight. Streptavidin-conjugated alkaline phosphatase and its substrate were used for ELISA color development. (A) Binding results of GST with all five MBP fusion proteins. (B) Binding results of KID119 with all five MBP fusion proteins. (C) Binding of KID 239 with MBP fusion proteins.
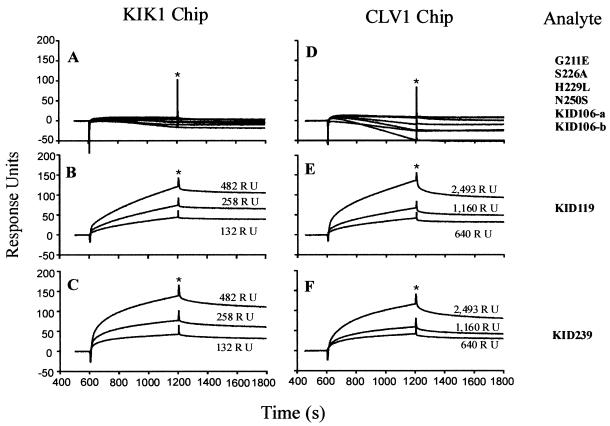
For kinetic analysis by SPR (details in Materials and Methods), various RLK ligands were immobilized at graded levels in the flow cells of the sensor chip. The various KAPP mutant proteins served as analytes; solutions of these proteins at graded concentrations were passed through the flow cells in series, with binding detected in real time as an increase in the SPR signal. After 10 min, blank buffer was allowed to flow through the same flow cells, and dissociation of the KAPP mutant analyte from the immobilized RLK ligand into the bulk solution stream was detected in real time as a decrease in SPR signal. Representative association/dissociation curves are shown in Fig. 4.
Figure 4.
Representative SPR sensorgrams of the binding between analytes, GST-KAPP mutants, and ligands, MBP-KIK1 and MBP-CLV1, from BIAcore 2000. The sensorgrams shown here represent the binding between two immobilized MBP fusions (KIK1 and CLV1) at three density levels (illustrated as RU above each curve) and various soluble analytes (all at 25 μg/ml). (A–C Left) Binding results on a KIK1 chip. (D–F) Binding results on a CLV1 chip. Right-most column shows the corresponding analytes for each corresponding row. All the analytes (KAPP mutants) incapable of binding with the two ligands are summarized in A and D. The binding results of KID119 (mapped KI domain) and KID239 are shown in B, C, E, and F, respectively. The corresponding ligand densities in A and D are 258 RU and 1160 RU, respectively. The binding signal from the blank surface of chips has been subtracted from the original results. All curves have been adjusted to the same baseline (zero) to make comparison of sensorgrams easier. Phases before asterisks (∗) represent the association sensorgrams; phases after asterisks represent the dissociation sensorgrams.
The results of all these binding experiments were remarkably consistent and support the conclusions detailed in the next four subsections. Numerous negative controls were included in the experiments: GST, the fusion partner of the KAPP mutant constructs, was used in place of KAPP; MBP, the fusion partner of the RLKs, was used in place of RLK; and K558E, a nonautophosphorylating mutant of KIK1, was used in place of KIK1. Little or no binding was observed in any of these negative controls.
KID119 Is a Minimal KI Domain.
KAPP deletion proteins containing 119 residues with wild-type sequence in the FHA domain were the only proteins capable of binding. Deletion of only 13 amino acids from either end of the KID119 segment (KID106-a and KID106-b) abolished binding (Figs. 2–4; results summarized in Fig. 1A Right). KID119 includes the FHA homology region, but that domain by itself (corresponding to KID52) is not sufficient for binding.
The FHA Homology Region Is Necessary for Binding RLK Ligands.
None of the four KAPP mutant proteins with amino acid replacements at conserved positions in the FHA homology region (KAPP positions 211, 226, 229, and 250) were found to bind RLKs (Figs. 2E and 4 A and D; summarized in Fig. 1A Right). Thus this domain, although not sufficient for binding, is evidently necessary.
Differences in the Binding of Various RLKs.
KIK1 consistently showed higher binding to the functional KAPP proteins than did CLV1 (Figs. 3 B and C, 4 B, C, E and F; dot-blot data not shown). The ratio of binding, which could be assessed by SPR, corresponded to about 0.9 and 0.2 molecules of KAPP bound per molecule of KIK1 and CLV1, respectively. Furthermore, binding curves for KIK1 and CLV1 ligands were very similar when the flow cells had about five times as many immobilized CLV1 as KIK1 molecules (in Fig. 4, compare B with E and C with F). Although the extent of phosphorylation of the recombinant RLKs is not known, the ratio of binding supports the hypothesis that greater than 90% of KIK1 molecules, but only 20% of the CLV1 molecules, have a functional epitope for KAPP binding. The results also show that KAPP binds these two sites with equivalent kinetics. RLK5 showed little binding above background by any of the methods used (Figs. 3 B and C; dot-blot data not shown). Previous studies used radiolabeled KAPP to detect binding to RLK5. The dot-blot and ELISA methods are indirect measures of binding and may be less sensitive than the binding assay using radiolabeled KAPP. It may also be that only a small portion of the recombinant RLK5 proteins have functional binding epitopes, which limits the sensitivity of these assays.
Both Association and Dissociation Between KAPP and RLKs Are Slow.
KIK1 and CLV1 ligands were immobilized at graded surface densities in the flow cells of two sensor chips. Because of the higher binding capacity of KIK1, it was immobilized at densities about five times lower than CLV1. KAPP mutant proteins at concentrations of 25, 50, and 100 μg/ml were used as analytes, yielding nine association/dissociation curves for each RLK/KAPP construct pair. For the two lowest analyte concentrations, the six curves for each pair were reasonably consistent with a simple reversible binding model Ligand + Analyte ↔ Complex governed by a single pair of kon and koff rate constants; these rate constants are reported in Table 1, along with the corresponding dissociation equilibrium constant KD = koff/kon. Results with the highest analyte concentration, 100 μg/ml, did not fit a simple binding model; the reason for this discrepancy is unknown, but one possibility is that the KAPP proteins aggregate at the highest concentration. The estimated kon and koff rate constants are very similar for all eight RLK-KAPP pairs studied, varying over only 3- and 4-fold ranges, respectively. In particular, the rate constants for reaction of a particular KAPP analyte with KIK1 are very similar to the constants for its reaction with CLV1. These data suggest that the differing binding capacities of KIK1 and CLV1 are not primarily caused by differences in affinity.
Table 1.
Apparent binding kinetics of KIK1 and CLV1 with different KAPP deletion mutants
| Ligand | Analyte | kon, M−1s−1 × 103 | koff, s−1 × 10−4 | KD, nM |
|---|---|---|---|---|
| MBP-KIK1 | KID239 | 5.38 ± 1.85 | 3.90 ± 0.63 | 80 ± 24 |
| KID143 | 2.00 ± 0.55 | 1.13 ± 0.49 | 56 ± 19 | |
| KID134 | 3.92 ± 1.32 | 1.02 ± 0.18 | 27 ± 5 | |
| KID119 | 2.11 ± 0.65 | 1.86 ± 0.57 | 88 ± 16 | |
| MBP-CLV1 | KID239 | 6.81 ± 1.43 | 4.98 ± 0.45 | 80 ± 29 |
| KID143 | 4.07 ± 2.31 | 3.40 ± 1.73 | 91 ± 27 | |
| KID134 | 3.49 ± 0.99 | 1.82 ± 0.32 | 55 ± 13 | |
| KID119 | 2.35 ± 1.19 | 2.40 ± 0.91 | 103 ± 31 |
Each parameter was the mean and standard deviation from six sets of data collected from the interaction between three density levels of each ligand (KIK1 or CLV1) and two lowest concentrations of each analyte (KAPP deletion mutants).
DISCUSSION
We have used a simplified dot-blot immuno analysis to test binding of RLKs with various KAPP deletions and site-directed mutants. ELISA and SPR have been used semiquantitatively and quantitatively, respectively, to confirm the binding results obtained by dot-blot analysis. The results from all three experiments are consistent, mapping the minimal functional KI domain to a 119-residue segment of KAPP. Removal of an additional 13-aa polypeptide at either side of this segment completely eliminated binding with RLKs. The binding results are summarized in Fig. 1A Right.
A 52-aa region with homology to the FHA region exists in the middle of the mapped KI domain. Binding analysis with a fusion protein of this FHA homology region indicates that it alone is not sufficient for interaction with RLKs. Mutational analysis at the four most conserved residues of the FHA homology region, G211, S226, H229, and N250, resulted in a KI domain incapable of binding. This result strongly suggests that the FHA homology region is necessary for RLK binding. These results closely parallel studies of the yeast protein RAD53 (22), which contains two FHA domains, FHA1 and FHA2, and which binds its ligand, RAD9, only when the latter is phosphorylated. Amino acid replacements at FHA2 positions 622 (homologous to KAPP position 229) and 655–657 (655 is homologous to KAPP position 250) abolished binding, suggesting that the FHA of KAPP and FHA2 of RAD53—and, by plausible extension, all or most FHAs—share similar protein–protein interaction determinants. Deletion analysis indicates that flanking sequences of the FHA homology region are also needed in protein–protein interaction. The functional KI domain consists, therefore, of both the FHA homology region and about 30 aa at each side. Determining the precise function of the flanking regions will require a three-dimensional structure analysis of the KI domain bound to a ligand. We hypothesize that the flanking sequences might stabilize the three-dimensional structure required for protein–protein interaction.
The FHA domain was discovered on the basis of its homology to a conserved region in the forkhead protein family (20). Unlike forkhead domains, which are DNA- binding domains, FHA domains are not restricted to a specific protein or protein group. Functions of FHA domains have not been reported until very recently. Sun et al. (22) have shown that in RAD53 of Saccharomyces cerevisiae, an FHA2 is involved in protein–protein interaction with RAD9 and that it functions in a DNA damage checkpoint. Sun et al. also showed that the flanking sequences of the FHA homology region are important for the interaction of RAD53 with RAD9 (22). According to their results, the minimal FHA-containing functional unit in RAD53 is 182 aa long and is located between positions 549 to 730. The FHA2 homology region in RAD53 extends from position 601 to 664, as shown in Fig. 1B. In this paper, we are reporting the involvement of the FHA homology region in the KI domain. It will be interesting to test the possible interaction between the KI domain and RAD9 or RAD53 with RLKs because it may reveal whether flanking regions of FHA control the binding specificity. Our results and the results from Sun et al. (22) suggest that the FHA homology region may represent a new phosphopeptide interaction motif used in various signal transduction pathways of different organisms. To determine whether this FHA domain possesses other biological functions will require the investigation of other FHA homology-containing proteins.
We have indirectly shown that the phosphorylation level of RLKs can alter the binding capacity to the KI domain, although kinetic analysis indicates that it does not change the binding affinity with the apparent dissociation constant (KD) between 25 and 100 nM. For example, the observed weaker binding signal of the CLV1 to the KI domain in comparison to the interaction of KIK1 and the KI domain is likely the result of less phosphorylation on KI domain-binding sites of CLV1 than KIK1. The difference of autophosphorylation levels may be caused by using the E. coli expression system. Alternatively, expression of CLV1 in E. coli might result in a majority of the protein being inactive. We observed only very weak binding signals for the interaction between RLK5 and the KI domain and did not determine the binding kinetics.
In comparison to the kinetic data of other phosphoprotein binding domains, the interaction between the KI domain and RLKs has a relatively slow association rate (≈103 M−1 s−1). For example, the association of 14-3-3 proteins and high-affinity phosphoserine-containing peptide ligands is about 104 M−1 s−1 (12), and the association of SH2 domains and high-affinity phosphotyrosine-containing peptide ligands is about 105–106 M−1 s−1 (10). The dissociation rate of the KI domain and RLKs (≈10−4 s−1) is also slower than that of both 14-3-3 proteins and high-affinity peptide ligands (≈10−3 s−1) (12) and SH2 domains and high-affinity peptide ligands (≈10−1 s−1) (10). The dissociation constant of the KI domain and RLKs (25–100 nM) is similar to both SH2 and high-affinity peptide ligands (50–150 nM) (10) and 14-3-3 proteins and high-affinity peptide ligands (50–200 nM) (12). The kinetics indicate that the KI domain binds to RLKs slowly. Once bound, the reaction is very stable. It may reflect its physiological function in the plants. In vivo, KAPP may bind phosphorylated RLKs until the dephosphorylation process (negative regulation) by the protein phosphatase is completed. This binding property may have important consequences in determining the extent of activity of a signaling pathway initiated by a RLK during plant growth and development.
Acknowledgments
These studies were supported by a National Science Foundation grant (MCB9809884) to J.C.W. We are grateful to Kevin Lease, Erika Ingham, and Dr. Tsung-Luo Jinn for their critical comments on the manuscript. We also thank Drs. Julie M. Stone and David Braun for their helpful discussions throughout this research.
ABBREVIATIONS
- SH2
Src homology 2
- KAPP
kinase-associated protein phosphatase
- KI
kinase interaction
- FHA
forkhead associated
- RLK
receptor-like protein kinase
- MBP
maltose-binding protein
- GST
glutathione S-transferase
- SPR
surface plasmon resonance
- RU
response unit
- KIK1
KI domain interacting kinase 1
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Stone J M, Collinge M A, Smith R D, Horn M A, Walker J C. Science. 1994;266:793–796. doi: 10.1126/science.7973632. [DOI] [PubMed] [Google Scholar]
- 2.Braun D M, Stone J M, Walker J C. Plant J. 1997;12:83–95. doi: 10.1046/j.1365-313x.1997.12010083.x. [DOI] [PubMed] [Google Scholar]
- 3.Williams R W, Wilson J M, Meyerowitz E M. Proc Natl Acad Sci USA. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stone J M, Trotochaud A E, Walker J C, Clark S E. Plant Physiol. 1998;117:1217–1225. doi: 10.1104/pp.117.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trotochaud A E, Hao T, Wu G, Yang Z, Clark S E. Plant Cell. 1999;11:393–405. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark S E, Williams R W, Meyerowitz E M. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 7.Waksman G, Kominos D, Robertson S C, Pant N, Baltimore D, Birge R B, Cowburn D, Hanafusa H, Mayer B J, Overduin M, et al. Nature (London) 1992;358:646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 8.Cohen G B, Ren R. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 9.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 10.Marengere L E M, Songyang Z, Gish G D, Schaller M D, Parsons J T, Stern M J, Cantley L C, Pawson T. Nature (London) 1994;369:502–505. doi: 10.1038/369502a0. [DOI] [PubMed] [Google Scholar]
- 11.Muslin A J, Tanner J W, Allen P M, Shaw A S. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 12.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin S J, Cantley L C. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 13.Kavanaugh W M, Turck C W, Williams L T. Science. 1995;268:1177–1179. doi: 10.1126/science.7539155. [DOI] [PubMed] [Google Scholar]
- 14.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 15.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bächinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan I, Pérez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 17.Parker D, Ferreri K, Nakajima T, Lamorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu P-J, Zhou X Z, Shen M, Lu K P. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 19.Barinaga M. Science. 1999;283:1247–1249. doi: 10.1126/science.283.5406.1247. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann K, Bucher P. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 21.Kaestner K H, Lee K-H, Schlöndorff J, Hiemisch H, Monaghan A P, Schütz G. Proc Natl Acad Sci USA. 1993;99:7628–7631. doi: 10.1073/pnas.90.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Z, Hsiao J, Fay D S, Stern D F. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 23.Walworth N C. Science. 1998;281:185–186. doi: 10.1126/science.281.5374.185. [DOI] [PubMed] [Google Scholar]
- 24.Horn M A, Walker J C. Biochim Biophys Acta. 1994;1208:65–74. doi: 10.1016/0167-4838(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 25.Yu J, Smith G P. Methods Enzymol. 1996;267:3–27. doi: 10.1016/s0076-6879(96)67003-7. [DOI] [PubMed] [Google Scholar]
- 26.Hermann-Le Denmat S, Werner M, Sentenac A, Thuriaux P. Mol Cell Biol. 1994;14:2905–2913. doi: 10.1128/mcb.14.5.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsay H D, Griffiths D J, Edwards R J, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, Elledge S J. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- 29.Stern D F, Zheng P, Beidler D R, Zerillo C. Mol Cell Biol. 1991;11:987–1001. doi: 10.1128/mcb.11.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]