Abstract
Intact influenza A virions were bombarded with thermally activated tritium atoms, and the intramolecular distribution of the label in the matrix protein M1 was analyzed to determine the in situ accessibility of its tryptic fragments. These data were combined with the previously reported x-ray crystal structure of the M1 fragment 2–158 [Sha, B. & Luo, M. (1997) Nat. Struct. Biol. 4, 239–244] and the predicted topology of the C domain (159–252) to propose a model of M1 arrangement in the virus particle.
Keywords: tritium planigraphy, three-dimensional modeling, topology, secondary structure prediction
The influenza virus (Flu) is a representative of intricately organized enveloped viruses. Notwithstanding the vast material accumulated on the molecular design of the Flu virion, its structure is far from fully established. The four main Flu structural proteins differ markedly in their localization and functions (1). X-ray data are available only for the hemagglutinin (2) and neuraminidase (3) ectodomains and for the N-proximal moiety of the M1 protein (4) crystallized in an aqueous milieu. The central role of M1 in sustaining the virion structure and virus replication has brought it under quite intense study (for a recent review, see ref. 5). A hypothetical model of M1 packing in the virion has been advanced on the basis of the x-ray data on the protein fragment in crystal (4) and the electron cryomicroscopy data on M1 localization in the virus (6).
To assess the in situ spatial organization of the M1 protein, we subjected intact FluA particles to tritium bombardment (7, 8). In a number of studies over the last decade, this approach has been used successfully to probe directly the accessible surface of the protein in biological macromolecular entities such as plant viruses (9, 10), the ribosome (11), membrane structures (12), and the erythrocyte (13). In essence, the method exposes a specially prepared target maintained at the temperature of liquid nitrogen to a beam of “hot” tritium atoms (generated through catalytic dissociation of molecular tritium at the surface of a tungsten filament heated to 2,000 K). The initial energy of incident particles is just sufficient for abstraction of hydrogen atoms from the target molecule and substitution of tritium for hydrogen. Even a single unreactive collision instantly reduces the energy of the tritium atom below the threshold required for displacing hydrogen. Thus, the label is incorporated at the first impact or not incorporated at all. In further analysis, the label distribution along the polypeptide chain characterizes the steric accessibility for the bombarding tritium atoms; the most exposed parts of the macromolecule are the most intensely labeled ones.
Here, we present quantitative data on the in situ labeling topography of the FluA matrix protein M1. On the strength of these and other relevant results, we take steps toward elucidating the spatial organization of M1 in the virion.
METHODS
Influenza virus A/Aichi/2/68 (H3N2) was propagated in 10-day embryonated chicken eggs (14) and purified by centrifugation through 20% (vol/vol) sucrose in STE buffer (100 mM NaCl/10 mM Tris⋅HCl/1 mM EDTA, pH 7.4) at 21,000 rpm for 90 min at 8°C in an SW 27.1 rotor of a Beckman–Spinco L5-75 centrifuge. Purified virus was resuspended in the same buffer and stored in liquid nitrogen if not used immediately.
Tritium bombardment was carried out as described (15) by using 1.5 ml of virus suspension (1–2 mg/ml) in STE buffer to prepare the target and by heating the tungsten wire twice for 15 s in each of eight injections of molecular tritium into the vacuum reactor flask, with the target frozen on its walls (a total of 4 min of bombardment). These conditions yielded virus preparations with specific radioactivity of 1 × 105 to 20 × 105 dpm/μg protein. After labeling, the samples were purified by centrifugation through 20% (vol/vol) sucrose in STE buffer (SW 40.1 rotor; 30,000 rpm; 8°C; 60 min).
Isolation and purification of M1 protein has been described in detail (16). Covalent attachment of the M1 protein to thiopropyl Sepharose 6B, tryptic digestion, high-performance chromatography equipment, and conditions of separation of tryptic peptides have been described (17). Tryptic peptides were hydrolyzed, and amino acid analysis was run on a Hitachi-835 (Tokyo) analyzer in the standard mode for protein hydrolysate analysis with cation-exchange separation and ninhydrin postcolumn derivatization. The radioactivity of tryptic peptides was determined in a toluene liquid scintillator on a Delta-3000 counter (Tracor Analytic Instruments, Austin, TX). The specific activities of the peptides were obtained by relating the radioactivity to the peptide yield according to the amino acid analysis data.
RESULTS AND DISCUSSION
Procedure and Adequacy Checks.
The goal of the work was to evaluate the extent of labeling of the matrix protein M1 within the FluA virion and to examine the label distribution along the polypeptide chain to envisage its spatial organization in the viral membrane. The target for bombardment with atomic tritium was a freshly prepared and rapidly frozen vitrified aqueous suspension of virus particles, which were then isolated by mild sedimentation through a sucrose solution in saline (14). Virion integrity was checked by functional tests, dynamic light scattering, and an electron-microscopic dye-exclusion assay, and virion integrity proved to be the same in the specimens before and after bombardment.
The very first experiments on FluA identified labeling of the spike glycoprotein hemagglutinin and the matrix protein M1 but virtually no labeling of the nucleocapsid protein in the virion interior. Although M1 is believed to be embedded in the inner leaflet of the membrane (6) and hence to have no regions exposed on the membrane outer surface, we did expect tritium incorporation in this protein. Indeed, the earlier control experiments with liposomes showed that their bilayer, withstanding the target preparation and irradiation procedures, is partly penetrable for the tritium flux (18), acting as a semitransparent screen. Therefore, we had grounds for supposing that M1 labeling was not an artifact caused by rupture of the viral membrane bilayer. The label was truly incorporated into the amino acid residues of the protein, not just residual lipids or unsaturated fatty acids that acylate a portion of M1 molecules. On protein elution from gel, acid hydrolysis, and amino acid analysis with radioactivity counting in the eluate fractions, the sum radioactivity of the amino acids proved to be 70–75% of the total M1 radioactivity, a proportion observed for various protein preparations and merely indicative of incomplete removal of the water-exchangeable label; complete removal can be achieved only by protein hydrolysis to amino acids (19).
Comparing the specific radioactivities of the M1 protein labeled in situ and in isolation and taking the flux attenuation coefficient obtained with liposomes (18) that relates the screening effect of the lipid bilayer to its thickness, we could make a rough estimate that the distance from the outer virion surface to the “center” of this protein does not exceed the membrane span, which is quite consistent with the data available (6). Thus, it can be concluded that our approach is adequate to the system under study.
Label Distribution in M1.
Analysis of the intramolecular distribution of the tritium label in M1 proved to be quite complicated. The high hydrophobicity of the protein hindered its dissolution in aqueous buffers and enzymatic hydrolysis. Therefore, we resorted to a special tactics detailed elsewhere (17): the protein was “stretched” by covalent binding to an activated carrier thiopropyl Sepharose, which rendered M1 susceptible to trypsin and allowed fractionation of the resulting fragments. The tryptic peptides that could be isolated and assessed for specific labeling covered 93% of the M1 sequence (excluding the shortest T11, T17, the C-terminal T23 and T24, as well as arginines 77 and 78; Fig. 1).
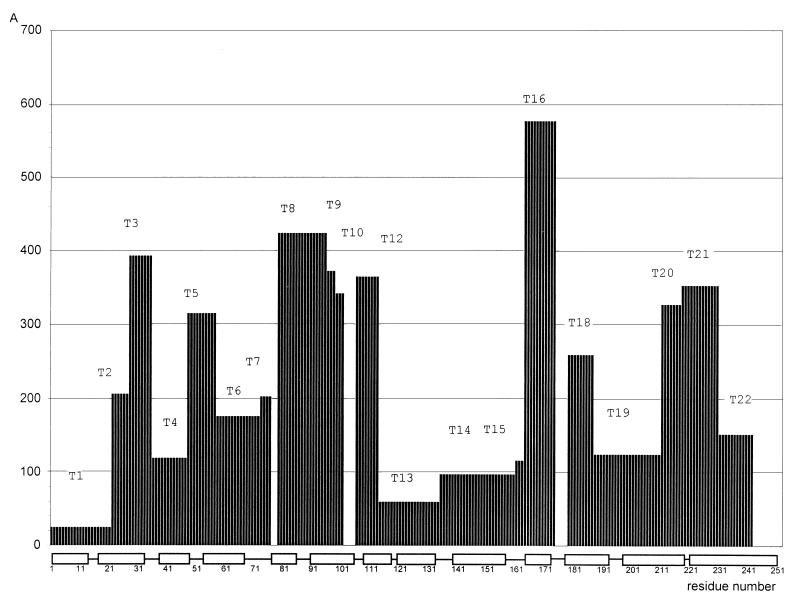
Figure 1.
Distribution of the tritium label in the influenza virus matrix protein M1 on bombardment of intact virions. The ordinate is the specific radioactivity (cpm/nmol) of the residue averaged for each tryptic peptide isolated (Tn). The string at the bottom indicates the helices established by x-ray analysis (4) and predicted in this work.
The maximally labeled peptide T16 was taken as a reference point to calculate the relative accessibility of every peptide: T16 (1.0), T8 (0.73), T20 (0.68)… T1 (0.04). As an initial discrimination, we considered the peptides as exposed to tritium bombardment when relative accessibility was no less than 0.45 (i.e., T3, T5, T8–T10, T12, T16, T18, T20, and T21; these parts are marked in Fig. 2).
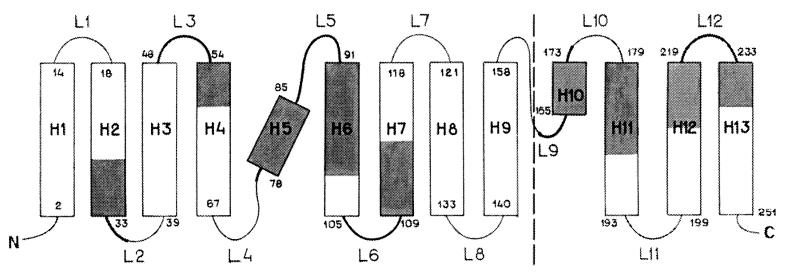
Figure 2.
Regions most exposed to tritium bombardment, mapped on the topology of the M1 protein. The dashed line separates the x-ray data (ref. 4; on the left) and the prediction for the C domain (on the right). α-Helices are represented by boxes (with residue numbers at both ends) and marked as H1–H13; loops are marked as L1—L12; the N and C termini are indicated. Segments accessible for tritium labeling are shaded in the helices and shown with thicker stretches in the loops, in roughly the actual size proportion within each element.
Spatial Reconstruction.
There is an essential difference in considering the tritium bombardment data for a free globular protein (which is randomly oriented in the target and can thus be viewed as uniformly irradiated in all directions) and for a membrane protein, especially in an intact virion (which is effectively irradiated only “from the outside” in the direction not deviating greatly from the normal to the membrane surface). In other words, this difference is roughly the same as the difference between omnidirectional and unidirectional exposure.
At this stage of research, we deliberately restrict ourselves to a “rough resolution” of the label distribution (tryptic peptides) and a single cutoff (exposed/not exposed); hence, we do not have to distinguish between attenuation of the incident labeling beam by the lipid layer above the protein, and shielding of parts of the polypeptide chain by other overlying parts (which has a far greater effect, as the penetration of hot tritium atoms into protein does not exceed 5 Å; ref. 15). In any case, the labeling intensity first and foremost reflects the closeness of the corresponding protein region to the membrane outer surface.
Thus the maximally labeled peptide T16 was taken to be “uppermost” (i.e., representing the part of M1 most embedded in the viral membrane from the interior and closest to its outer surface), and, conversely, the least labeled peptides were taken to belong to the lower or hidden M1 regions.
In other words, the tritium bombardment approach, which has been called tritium planigraphy (8, 20) as applied to isolated proteins, may here be regarded as “tritium stratigraphy.”
According to the x-ray data (4) on the crystal structure of the M1 2–158, the fragment comprises nine α-helices (H1—H9) and eight loops (L1–L8) and has no β-strands; H1–H4 form the first, amino terminal domain N (2–67), and H6–H9 form the second, middle domain M (91–158). If we assume that the helices reported for the crystal are indeed present in the protein in situ and map our tritium bombardment data for tryptic peptides on these helices and loops, we get a picture displayed in the left part of Fig. 2. There is no contradiction with the secondary structure elements established by x-ray analysis; for instance, there is no case when both ends of an α-helix are labeled when its middle is not, an arrangement that would require bending or breaking to fit into the scheme. This model also indirectly supports the essential postulate of a uniform disposition of the M1 molecules in every membrane and in the virion population, at least in terms of overwhelming prevalence.
A separate problem was to predict the topology of the C-proximal moiety of the M1 protein: there are no x-ray data, and Sha and Luo (4) simply assert that it should form at least one other domain C. In contrast to globular proteins, for which the methods for predicting the secondary structure elements rest on more than a hundred x-ray structures, for membrane proteins there is no reliable prediction tool even regarding their transmembrane elements. Still, if we suppose that α-helices, as in the N and M domains, are formed by hydrophobic amino acid clusters and represent the sequence of residues 159–252 as a continuous helix, we can discern four extensive hydrophobic regions (165–170, 173–193, 199–219, and 223–251) and can break this helix further at hydrophobic stretches (21) into four α-helices (H10–H13) and four loops (L9–L12). In addition, four helical regions are predicted in the C domain with the Ptitsyn–Finkelstein algorithm (22) for integral membrane proteins (not shown). Applying the same accessibility criterion as above (0.45 threshold) and mapping the in situ tritium labeling data on the four-helix model of the C-proximal part of M1, we see two most exposed regions 164–187 (T16 and T18) and 211–230 (T20 and T21) and without much ambiguity arrive at a fold depicted in the right part of Fig. 2.
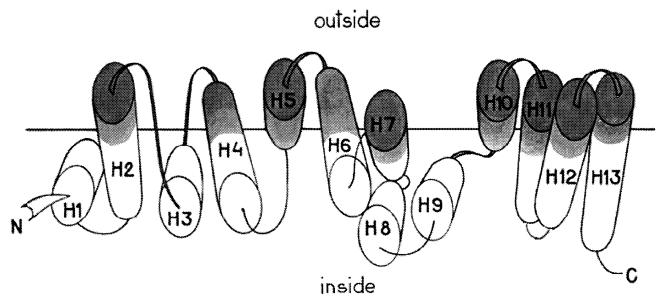
As we proceed from the general topology to the helix packing, we immediately see that the most straightforward version, an array of antiparallel helices perpendicular to the membrane surface, is inconsistent with the labeling data, especially for the N and M domains. This result is neither surprising nor discouraging, as M1 is not a transmembrane protein in the strict sense (see above); even for classical representatives of membrane-spanning proteins such as cytochrome oxidase or bacteriorhodopsin, the transmembrane helices are known to be tilted with respect to the bilayer normal by 21° on average and sometimes twice that (23), and of course no a priori expectations should be placed on the orientation of helices in the M1 regions at or beyond the membrane inner surface. A plausible arrangement of the 13 helices (without claiming particular helix–helix angles or contact zones) is shown as a “spread” in Fig. 3; one can see that some helices or parts thereof and adjacent loops are not labeled because they lie farther from the membrane outer surface and/or are shielded by other helices.
Figure 3.
Schematic disposition of the secondary structure elements of the M1 protein in the influenza virus membrane. The elements are spread for clarity; the horizontal line corresponds to the labeling threshold (see Results and Discussion) and does not represent any part of the membrane.
The finer details of M1 organization in the membrane await more sophisticated analysis (including examination of individual residue labeling to identify the contacts of the secondary structure elements).
Model Comparison.
We wish to refrain from discussing here such issues as the monomer/dimer state of M1 or the nucleoprotein–RNA interaction site(s) considered by Sha and Luo (4). The former does not influence the protein topology essentially, and the RNA-binding region would be much the same, because we have assumed their x-ray secondary structure elements for the N and M domains in our modeling. Apart from the orientation and packing of helices in the N and M domains, our model defies the purely speculative disposition of the C domain proposed by Sha and Luo. Indeed, its constituent T16 is the segment most heavily labeled after tritium bombardment of intact virions and, hence, is the part of M1 closest to the outer membrane surface, testifying to substantial embedding of the C domain. However, Sha and Luo view T16 as an appendage hanging into the virion interior. Furthermore, there are no firm grounds for believing that such a protein would have the same structure in crystal (aqueous milieu) and in the viral membrane (lipid milieu); on the contrary, the lipid environment is known to influence the folding of membrane proteins essentially (see, e.g., ref. 24 for review). Sha and Luo (4) admit that binding of M1 to the membrane would require a large-scale rearrangement (“flipping out”) to expose the highly hydrophobic surface of the N domain, which in the crystal at pH 4.0 is hidden at the N–M domain interface. With the helix arrangement proposed by us, the bifunctional properties of M1 (see ref. 4 and references therein) can be realized easily without additional structural alterations.
Acknowledgments
We thank Prof. T. A. Egorov for helpful suggestions and comments, M. B. Viryasov for technical assistance, and V. A. Shishkov for computer help in preparing the figures.
References
- 1.Schulze I T. Adv Virus Res. 1973;18:1–56. [Google Scholar]
- 2.Wilson I A, Skehel J J, Wiley D C. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 3.Varganese J N, Lover W G, Colman P M. Nature (London) 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 4.Sha B, Luo M. Nat Struct Biol. 1997;4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 5.Garoff H, Hewson R, Opstelten D-J E. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujiyoshi Y, Kume N P, Sakata K, Sato S. EMBO J. 1994;13:318–326. doi: 10.1002/j.1460-2075.1994.tb06264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shishkov A V, Filatov E S, Simonov E F, Unukovich M S, Goldanskii V I, Nesmeyanov A N. Dokl Akad Nauk SSSR. 1976;228:1237–1239. [PubMed] [Google Scholar]
- 8.Bogacheva E N, Goldanskii V I, Shishkov A V, Galkin A V, Baratova L A. Proc Natl Acad Sci USA. 1998;95:2790–2794. doi: 10.1073/pnas.95.6.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldanskii V I, Kashirin I A, Shishkov A V, Baratova L A, Grebenshchikov N I. J Mol Biol. 1988;201:567–574. doi: 10.1016/0022-2836(88)90638-9. [DOI] [PubMed] [Google Scholar]
- 10.Baratova L A, Grebenschikov N I, Dobrov E N, Gedrovitch A V, Kashirin I A, Shishkov A V, Efimov A V, Jarvekulg L, Radavsky Y L, Saarma M. Virology. 1992;188:175–180. doi: 10.1016/0042-6822(92)90747-d. [DOI] [PubMed] [Google Scholar]
- 11.Agafonov D E, Kolb V A, Spirin A S. Proc Natl Acad Sci USA. 1997;94:12892–12897. doi: 10.1073/pnas.94.24.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsetlin V I, Alyonycheva T N, Shemyakin V V, Neiman L A, Ivanov V T. Eur J Biochem. 1988;178:123–129. doi: 10.1111/j.1432-1033.1988.tb14437.x. [DOI] [PubMed] [Google Scholar]
- 13.Gordeeva L V, Baratova L A, Margolis L B, Shishkov A V. Biofizika. 1989;34:970–975. [PubMed] [Google Scholar]
- 14.Zhirnov O P, Ovcharenko A V, Bukrinskaya A G. J Gen Virol. 1985;66:1633–1638. doi: 10.1099/0022-1317-66-7-1633. [DOI] [PubMed] [Google Scholar]
- 15.Gedrovich A V, Goldanskii V I, Rumyantsev Y M, Unukovitch M S, Shishkov A V. Radiokhimiya. 1984;4:483–494. [Google Scholar]
- 16.Zhirnov O P. Virology. 1992;186:324–330. doi: 10.1016/0042-6822(92)90090-c. [DOI] [PubMed] [Google Scholar]
- 17.Fedorova N V, Ksenofontov A L, Viryasov M B, Baratova L A, Timofeeva T A, Zhirnov O P. J Chromatogr. 1998;706:83–89. doi: 10.1016/s0378-4347(97)00558-6. [DOI] [PubMed] [Google Scholar]
- 18.Glushakova S E, Ksenofontov A L, Fedorova N V, Mazhul L A, Ageeva O N, Margolis L B, Baratova L A, Shishkov A V. Biosci Rep. 1991;11:131–137. doi: 10.1007/BF01182481. [DOI] [PubMed] [Google Scholar]
- 19.Shishkov A V, Baratova L A. Russian Chem Rev. 1994;63:781–796. [Google Scholar]
- 20.Shishkov A V. Soviet Chem Phys. 1991;10:878–890. [Google Scholar]
- 21.Lim V I. J Mol Biol. 1974;88:872–894. doi: 10.1016/0022-2836(74)90404-5. [DOI] [PubMed] [Google Scholar]
- 22.Ptitsyn O B, Finkelstein A V. Biopolymers. 1983;22:15–25. doi: 10.1002/bip.360220105. [DOI] [PubMed] [Google Scholar]
- 23.Bowie J. J Mol Biol. 1997;272:780–789. doi: 10.1006/jmbi.1997.1279. [DOI] [PubMed] [Google Scholar]
- 24.Booth P J. Fold Des. 1997;2:R85–R92. doi: 10.1016/s1359-0278(97)00045-x. [DOI] [PubMed] [Google Scholar]