Abstract
The ligand hepatocyte growth factor/scatter factor (HGF) and its receptor tyrosine kinase, c-Met, are highly expressed in most human malignant mesotheliomas (MMs) and may contribute to their increased growth and viability. Based upon our observation that RNA silencing of fos-related antigen 1 (Fra-1) inhibited c-met expression in rat mesotheliomas (1), we hypothesized that Fra-1 was a key player in HGF-induced proliferation in human MMs. In three of seven human MM lines evaluated, HGF increased Fra-1 levels and phosphorylation of both extracellular signal–regulated kinase 5 (ERK5) and AKT that were inhibited by the phosphatidylinositol 3-kinase (PI3K) inhibitor, LY290042. HGF-dependent phosphorylation and Fra-1 expression were decreased after knockdown of Fra-1, whereas overexpression of Fra-1 blocked the expression of mitogen/extracellular signal–regulated kinase kinases (MEK)5 at the mRNA and protein levels. Stable MM cell lines using a dnMEK5 showed that basal Fra-1 levels were increased in comparison to empty vector control lines. HGF also caused increased MM cell viability and proliferating cell nuclear antigen (PCNA) expression that were abolished by knockdown of MEK5 or Fra-1. Data suggest that HGF-induced effects in some MM cells are mediated via activation of a novel PI3K/ERK5/Fra-1 feedback pathway that might explain tumor-specific effects of c-Met inhibitors on MM and other tumors.
Keywords: hepatocyte growth factor/scatter factor, fos-related antigen 1, phosphatidylinositol 3-kinase, MEK5, mesothelioma
CLINICAL RELEVANCE
Data here suggest that hepatocyte growth factor/scatter factor–-induced effects in some malignant mesothelioma (MM) cells are mediated via activation of a novel phosphatidylinositol 3-kinase/mitogen/extracellular signal–regulated kinase kinases 5/fos-related antigen 1 feedback pathway that might explain differential effects of c-Met inhibitors on MM and other tumor types.
Malignant mesothelioma (MM) is an insidious tumor associated historically with occupational exposure to asbestos (2, 3). The average survival of patients with MM is less than 1 year after initial diagnosis, and no successful treatment options exist. Although the mechanisms of development of MM are obscure, the initiation of signaling events after interaction with asbestos fibers may govern transactivation of genes governing cell proliferation and transformation (4). Cell survival, proliferation, and progression of MMs are complex and may be related to overexpression of several autocrine growth factors, including insulin-like growth factor (5, 6), platelet-derived growth factor (7), fibroblast growth factor (8), transforming growth factor β (8), and hepatocyte growth factor/scatter factor (HGF) (9).
Increased expression of Fos/Jun family members and activator protein-1 (AP-1) transactivation are observed in mesothelial cells after exposure to asbestos and erionite fibers in contrast to other nonpathogenic fibers and particles (10–12). In comparison to other fos/jun mRNAs, fos-related antigen 1 (fra-1) expression is more protracted and critical to expession of c-met and cd44 in a rat model of mesothelial cell transformation (1).
Although HGF and its receptor, c-Met, are known to be involved in chemotaxis, growth, and invasion of a number of tumor types including MMs (13–15), the mechanisms of HGF/Met signaling and their functional ramifications in MMs remain unclear. We previously reported that HGF stimulates AKT phosphorylation in human MMs (16). Moreover, we have shown that inhibition of extracellular regulated kinases 1 and 2 (ERK1/2) activation results in decreases in Fra-1 expression and inhibition of morphologic transformation of rat MMs in vitro (1, 17). Although we have reported that ERK1/2 and ERK5 cooperate in asbestos-induced lung epithelial cell proliferation (18), the role of ERK5 in cell signaling and proliferation in human mesotheliomas is unclear. Here we hypothesized that HGF might phosphorylate ERK5 through a phosphatidylinositol 3-kinase (PI3K)-dependent pathway linked to Fra-1 expression in human MMs. In support of our hypothesis, we report that modulating the PI3K/mitogen/extracellular signal–regulated kinase kinases (MEK)5 pathway regulates Fra-1 expression in some MMs that is linked causally to HGF-dependent viability and proliferation, as measured by expression of proliferating cell nuclear antigen (PCNA). These results suggest a novel PI3K/MEK5/Fra-1 pathway as a possible target for therapy of MMs. Moreover, we document a negative feedback loop whereby overexpression of Fra-1 blocks expression of MEK5. Results here may also explain disparate effects of Fra-1 overexpression and alterations in cell proliferation in other tumors (19).
MATERIALS AND METHODS
Human Mesothelial and Mesothelioma Cell Lines
The human mesothelial LP9/TERT-1, an hTERT-immortalized line was obtained from Dr. James Rheinwald (20). Human pleural mesothelioma cell lines were isolated from patients at autopsy or after surgical dissection of MMs and were kindly provided by Dr. Michele Carbone (Loyola University, Maywood, IL) (MC1, MC2, MC3, and MC4), and Dr. Harvey Pass (New York University, New York, NY) (MP2, MP5). The MM cell line MA1 (#CRL-2081) was obtained from the ATCC (Manassas, VA). All MM cell lines were tested for the mRNA expression of large and small Simian Virus 40 (SV40) T/t-antigen by PCR before each experiment. The MC3, MC4, MA1, and MP2 lines were negative for SV40 large T/t-antigen (SV40−), whereas MC1, MC2, and MP5 were SV40 positive (SV40+). Cells were propagated in DMEM/F12 medium containing 10% fetal bovine serum (FBS) and hydrocortisone (100 ng/ml), insulin (2.5 μg/ml), transferrin (25 μg/ml), and selenium (2.5 ng/ml) (Sigma, St. Louis, MO). For all experiments, cells were grown to confluence, and maintenance medium containing 0.5% FBS was added 24 hours before addition of HGF at 10 ng/ml medium. This concentration of HGF was selected from preliminary data showing that among other concentrations (1, 5, 10, 20, 50, 100 ng/ml medium), 10 ng/ml gives the maximum AKT activation with no toxic effects (data not shown). Stock solutions of the PI3K inhibitor, LY294002 (Calbiochem, La Jolla, CA) was diluted in dimethyl sulfoxide (DMSO) and used at the effective nontoxic concentrations, 10 or 20 μM, as reported previously (21). Control cells received DMSO in medium (< 0.1%).
Western Blot Analyses
Nearly confluent MM cells were washed three times with cold PBS, scraped from culture plates, and collected by centrifugation at 14,000 rpm for 1 minute. The pellet was resuspended in lysis buffer (20 mM Tris [pH 7.4], 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 1 mM Na3VO4, 2 mM pyrophosphate, 1 mM PMSF, 10 μg/ml leupeptin, 1 mM DTT, 10 mM NaF, 1% aprotinin), incubated at 4°C for 15 minutes, and centrifuged at 14,000 rpm for 20 minutes. Protein concentrations were determined using a Bio-Rad assay (Bio-Rad, Hercules, CA). Twenty micrograms of protein in sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 50 mM dithiothreitol, 0.1% wt/vol bromophenol blue) was resolved by electrophoresis in 10% SDS-polyacrylamide gels, and transferred to nitrocellulose using a semi-dry transfer apparatus (Ellard Instrumentation, Ltd., Seattle, WA). Blots were incubated in blocking buffer (Tris-buffered saline [TBS] containing 5% nonfat dry milk plus 0.1% Tween-20 [Sigma]) for 1 hour, washed three times for 5 minutes each in TBS/0.1% Tween-20, and incubated at 4°C overnight with anti-rabbit antibodies specific to phospho-AKT or total AKT, phospho-ERK5 or total ERK5, all at a 1:1,000 dilution (Cell Signaling Technology, Beverly, MA), Fra-1 (R-20) at a dilution of 1:500 (Santa Cruz Biotechnology Inc., Santa Cruz, CA), MEK5 antibody (ab24828) at a dilution of 2 μg/ml (Abcam, Inc, Cambridge, MA), or PCNA at a dilution of 1:10,000 (Bethyl Laboratories Inc., Montgomery, TX). Blots were then washed three times with TBS/0.1% Tween-20 and incubated with a specific peroxidase-conjugated secondary antibody (anti-rabbit) at a dilution of 1:3,000 (Amersham Pharmacia Biotech, Piscataway, NJ) for 1 hour. Blots were washed three times in TBS/0.1% Tween-20, and protein bands were visualized with the LumiGlo enhanced chemiluminescence detection system (Kirkgaard and Perry Laboratories, Gaithersburg, MD) and quantitated by densitometry (18). Blots were reprobed with an antibody to α-Tubulin at a dilution 1:1,000 (Santa Cruz Biotechnology, Santa Cruz, CA) to validate equal protein loading between lanes (17).
Chromatin Binding Analysis
The chromatin binding assay was performed as described previously (22). Briefly, approximately 4 × 106 cells per sample were rinsed with PBS and then with ice-cold CSK buffer (10 mM HEPES [pH 7.4], 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2). Cells were scraped from the plate, pelleted by centrifugation at 500 × g, and lysed in CSK-Triton buffer (CSK buffer containing 0.5% Triton X-100, 1 μg of leupeptin/ml, 1 μg of aprotinin/ml, 1 mM NaF, 1 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride) at 107 cells/ml for 10 minutes on ice. Nuclei were pelleted (designated fraction P1) by centrifugation at 1,500 × g for 5 minutes at 4°C. Supernatants (fraction S1), containing cytoplasmic and unbound nuclear proteins, were removed and further clarified by centrifugation at 16,000 × g for 10 minutes at 4°C. The pelleted nuclei then were washed with 1 ml of CSK-Triton buffer, pelleted by centrifugation, and suspended in CSK-Triton buffer at 107 nuclei/ml. The washed nuclei were then either used for Western blotting analysis directly or treated with nuclease to release chromatin-bound proteins. For nuclease treatments, washed nuclei were resuspended at 107 nuclei/ml in CSK-Triton buffer containing 160 U of DNase I/ml and 50 mM MgCl2 and incubated on ice for 10 minutes. Nuclear remnants were then pelleted by centrifugation as before, and the proteins released into the supernatant by the nuclease treatment were separated from the proteins remaining in the pellet. For total-cell lysates, cells were rinsed twice with PBS and then lysed with E1A lysis buffer (50 mM HEPES [pH 7.0], 250 mM NaCl, 5 mM EDTA, 0.1% NP-40, 1 mM dithiothreitol, 1 μg of leupeptin/ml, 1 μg of aprotinin/ml, 1 mM NaF, 1 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride) on ice for 30 minutes, and insoluble debris was removed by centrifugation. Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad Laboratories). Equivalent amounts of lysate were mixed with sodium dodecyl sulfate (SDS) sample buffer and heated to 95°C for 5 minutes. Western blots were performed as described above.
SYBR Green Real-Time Quantitative PCR
Total RNA (1 μg) was reverse-transcribed with random primers using the Promega AMV Reverse Transcriptase kit (Promega, Madison, WI) according to the recommendations of the manufacturer. PCR amplifications were performed using the ABI PRISM 7,700 Sequence Detection System (Perkin Elmer Applied Biosystems, Foster, CA). Reactions were performed in a 50 μl reaction mixture that included 25 μL SYBR Green JumpStart Taq ReadyMix (Sigma), distilled H2O, DNA template, and 0.2 μM each primer from QuantiteTect primer assays (Qiagen, Valencia, CA). Amplification was performed by initial denaturation at 94°C for 2 minutes, and 40 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 1 minute, and extension for 1 minute at 72°C. Then followed a dissociation cycle of 95°C for 15 seconds, 60°C for 15 seconds, and 95°C for 15 seconds. CT (threshold cycles) for both the mRNAs of interest and the 18S rRNA control were determined. Original input RNA amounts were calculated using the Comparative CT Method (2dd^CT) to analyze changes in gene expression in the samples relative to the untreated control sample. Duplicate assays were performed with RNA samples isolated from at least two independent experiments. The values obtained from cDNAs and 18S controls provided relative gene expression levels for the gene locus investigated.
Constructs and Transfection Techniques
A dominant-negative MEK5 pcDNA3 dnMEK5(A) was obtained from Dr. Lee Jiing-Dwan (The Scripps Research Institute, La Jolla, CA). The dominant-negative construct was created by PCR-based mutagenesis of the dual phosphorylation site (ser311 and thr315 with alanine) (23) and cloned into pcDNA3 vector (Invitrogen, San Diego, CA). A wild type Fra-1 of human origin was constructed and cloned into pcDNA3.1 (Invitrogen). For creation of permanent cell lines, cells were grown to 80 to 90% confluence, trypsinized, counted, and resuspended at 3 × 106 cells/ml at room temperature. An aliquot of the cell suspension (400 μl) was mixed with 10 μg of plasmid DNA (expression or control plasmids) and electroporated at 400 V and 330 μF capacitance. Cells were immediately plated in fresh growth medium in 35-mm culture dishes and allowed to recover overnight. After an overnight recovery, cells were selected for neo resistance using 200 μg/ml G418 (Sigma). Colonies surviving G418 selection were expanded and tested for the presence of Fra-1 or ERK5 as an indicator of plasmid activity.
siFra-1 and siMEK5 RNA interference (RNAi) duplexes were constructed from sequence information on mature mRNA extracted from EST database (www.ncbi.nlm.nih.gov). The siFRA-1 and siMEK5 duplexes are from exon 2 of the Fra-1 gene, and exon 3 (NM_002757) of the MEK5 gene. The siRNA pool sequences targeting Fra-1 corresponded to the 107–126, 124–143, 230–249, and 276–295 coding regions relative to the first nucleotide of the start codon, and the sequence targeting MEK5, corresponded to the 193–212, 201–220, and 236–255 coding regions relative to the first nucleotide of the start codon. The sequences were BLAST-searched (NCBI database) against EST libraries to ensure the specificity of the siRNA molecule. The siRNA duplexes or a scramble control were transfected into MM lines using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Cells were incubated with complexes overnight, and the medium was replaced the next day. Cells were allowed to recover for 48 hours before treatments.
Enzyme-Linked Immunosorbent Assay–Based PI3K Assay
The effectiveness of the LY290042 inhibitor on PI3K activity was assessed by enzyme-linked immunosorbent assay. The production of PI(3,4,5)P3 from PI(4,5)P2 by PI3K was used to measure PI3K activity. MM controls and MM treated with different concentrations of LY290042 (10, 20, and 50 μM) for 3 hours were stimulated with 10 ng/ml HGF for 30 minutes at 37°C to induce PI3K activity. Cells were then lysed in 1 ml of NP-40 containing lysis buffer with protease inhibitors and protein concentration determined using Bradford reagent (Bio-Rad Laboratories, Inc.). The amount of PI (3–5) P3 produced was quantified by a competition enzyme immunoassays according to the manufacturer's protocol (Echelon, Inc., Salt Lake City, UT). In brief, PI3K was immunoprecipitated from equal protein cell lysates using 4 mg rabbit anti-PI3K (p85) antibody (Upstate Biotechnology, Lake Placid, NY). The immunoprecipitated PI3K was mixed and incubated with a PI (3–5) P3 detector protein and then added to a PI (3–5) P3-coated microplate for competitive binding. A peroxidase-linked secondary detection reagent was used to detect PI (3–5) P3 detector protein binding to the plate, and the amount of PI (3–5) P3 produced measured by absorbance at 450 nm was calculated from a standard curve prepared from known concentrations of product. The enzyme activity was expressed as amounts of PIP3 (picomoles per milliliter) and then converted to % inhibition with respect to the control cells.
MTS Cell Viability Assay
Cell viability was measured with a CellTiter 96 (Promega) colorimetric assay, using an MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) assay, as per manufacturer's instructions. In brief, MM cells were plated in a 96-well microtiter plates at a concentration of 5 × 104 cells per well and incubated in complete medium overnight. Medium was aspirated and replaced with DMEM/F12 medium + 0.5% FBS containing different concentrations of HGF (1–100 ng/ml) or control vehicle (PBS). The absorbance of the wells was read at 492 nm as a measure of cell viability at different time points. Fold changes were calculated with respect to the control.
Annexin V/PI Staining
Apoptosis was determined in control scramble, positive control (Onconase 10 μg/ml), and cells transfected with siMEK5 or siFra-1 constructs for 66 hours (48 h required for the siRNA construct to knockdown Fra-1 and MEK5 plus 18 h used in the longest experiment) by staining with annexin V–fluorescein isothiocyanate (FITC) and PI labeling. In brief, cells were washed twice with cold PBS and then resuspended in 500 μl of binding buffer (10 mM HEPES/NaOH [pH 7.4], 140 mM NaCl, 2.5 mM CaCl2) at a concentration of 1 × 106 cells/ml. Five microliters of annexin V–FITC (PharMingen, San Diego, CA) and PI (1 μg/ml) were added to cells and analyzed by flow cytometry (Coulter EPICS XL / XL-MCL; Beckman Coulter, Fullerton, CA).
Statistical Analyses
In all experiments, three or six replications were conducted for each group per time point. Experiments were performed in triplicate. Results were evaluated by one-way ANOVA using the Student-Newman-Keuls procedure for adjustment of multiple pairwise comparisons between treatment groups. Differences with P values less than or equal to 0.05 were considered statistically significant.
RESULTS
Activation of the PI3K Pathway by HGF in Some MM Lines Causes Increased Fra-1 Expression that Is Accompanied by Increased Levels of Phosphorylated AKT and ERK5
To address the hypothesis that phosphorylation of AKT and ERK5 is linked to increases in Fra-1, LP9 human mesothelial and seven MM cell lines were evaluated for Fra-1, p-ERK5, and p-AKT expression after addition of HGF, a factor known to activate the PI3K pathway in MMs (16). In nontransformed LP9 mesothelial cells, HGF (10 ng/ml) did not increase levels of Fra-1 protein significantly (Figure 1A), but p-AKT/AKT (Figure 1B) and p-ERK5/ERK5 (Figure 1C) ratios were elevated (P ⩽ 0.05). HGF increased expression of Fra-1 in 3 (MC1, MC2, MP5) of seven cell lines (Figure 1A). These patterns correlated with significantly (P ⩽ 0.05) increased p-AKT/AKT (Figure 1B) and p-ERK5/ERK5 (Figure 1C) levels in these lines. Subsequently, experiments were performed with MM lines expressing elevated levels of p-AKT, p-ERK5, and Fra-1 after addition of HGF.
Figure 1.
Levels of fos-related antigen 1 (Fra-1) and phosphorylated AKT, extracellular signal–regulated kinase (ERK)5 are increased after addition of hepatocyte growth factor/scatter factor (HGF) (10 ng/ml for 30 min) (+ lane) to three of seven malignant mesothelioma (MM) cell lines (MC1, MC2, and MP5). Western blot analysis of (A) Fra-1/α-Tubulin expression, (B) p-AKT/AKT expression, and (C) p-ERK5/ERK5 expression. Data in graphs are expressed as mean ± SEM (n = 3). *P ⩽ 0.05 compared with respective untreated control group (−).
Fra-1 Expression Is Decreased after Inhibition of PI3K Activity and by Knocking Down MEK5
We first determined whether inhibition of basal and HGF-induced Fra-1 expression was reduced after inhibition of the PI3K pathway by LY290042 (20 μM), or after knockdown of MEK5 expression using a siRNA construct (siMEK5). Figure 2 shows that approximately 85% knockdown of MEK5 expression was achieved using this construct at the mRNA level as verified by RT-QPCR (Figure 2A) and the protein level (Figure 2B). As shown in Figure 2, addition of LY290042 at a concentration of 20 μM, a concentration found to decrease significantly PI3K activity (Figure 2C) and phosphorylation of AKT (Figure 2D), reduced both basal (group 2 versus 1) and HGF-induced (groups 4 versus 3) levels of Fra-1 in MM scramble control cells (P ⩽ 0.05), and HGF-induced Fra-1 significantly. Knockdown of MEK5 with siMEK5 did not affect basal levels of Fra-1 (group 5 versus 1), but reduced HGF-induced Fra-1 expression (group 7 versus 3). MM cells transfected with siMEK5 after addition of LY290042 did not contribute further to Fra-1 down-regulation (group 6 versus 5 and group 8 versus 7). RT-QPCR revealed increases in Fra-1 mRNA levels after addition of HGF to scramble control cells, which were decreased by siMEK5. However, although basal Fra-1 protein levels were not significantly affected by siMEK5 (Figure 2E), Fra-1 mRNA levels were increased (P ⩽ 0.05) in siMEK5-transfected MM cells (Figure 2F), suggesting a compensatory mechanism at the mRNA level for MEK5 modulation of Fra-1 expression.
Figure 2.
HGF-dependent Fra-1 expression is modulated by inhibition of the PI3K pathway and MEK5. (A) RT-QPCR and (B) Western blot showing approximately 85% knockdown expression of MEK5 after siMEK5 transfection for 48 hours. (C) Enzyme-linked immunosorbent assay phosphatidylinositol 3-kinase activity assay and (D) Western blot showing efficacy of the LY29042 inhibitor in blocking PI3K activity and AKT phosphorylation, respectively. (E) Western blot shows that HGF-induced Fra-1 protein levels in the MP5 cell line are decreased after treatment with the PI3K inhibitor LY294002 (20 μM) or knockdown of MEK5 with an RNAi construct. (F) RT-QPCR showing HGF-increased mRNA levels of Fra-1 in siMEK5-transfected cells, and abrogation of HGF-induced Fra-1 mRNA expression in MEK5 knockdown cells. *P ⩽ 0.05 compared with respective control (scramble or siMEK5); +P ⩽ 0.05 siMEK5 compared with its equivalent in the scramble control group. All results are graphed as the mean ± SEM (n = 3).
To further address the effect of MEK5 expression on HGF-induced p-AKT and Fra-1 expression, a dnMEK5-expressing MM line was evaluated in comparison to its empty vector (EV) control. A time course study revealed that HGF-dependent Fra-1 expression in EV control cells was increased significantly 1 hour after addition of HGF, and at 24 hours (Figures 3A and 3B). Increases in Fra-1 expression were accompanied by higher phosphorylation levels as previously demonstrated by the upward mobility shift of Fra-1 in Western blots 30 minutes after addition of HGF (24). In the dnMEK5 MM line, there was a significant increase in Fra-1 expression at 1 hour but not later (Figures 3C and 3D). The patterns of phosphorylation of AKT were also different in both cell lines, the EV line showing a significant increase from 20 minutes to 2 hours, while in the dnMEK5 line, phosphorylation of AKT was less protracted and significant only at 1 and 2 hours.
Figure 3.
HGF-dependent phosphorylation and expression of Fra-1 are lower in MM transfected with a dominant-negative (dn) MEK5 construct compared with empty vector (EV) control cells. Western blots show the expression of p-AKT and Fra-1 in a time course study using MM transfected with an EV (A, B) or a dn MEK5 construct (C, D) after addition of HGF (10 ng/ml). Detection of α-Tubulin was used as a control for protein loading in lanes. Mean ± SEM of n = 3. *P ⩽ 0.05 compared with control (0) group.
HGF-Induced ERK5 and AKT Phosphorylation Are Decreased by Overexpression of Fra-1
To address whether HGF-dependent Fra-1 up-regulation modulated expression of p-ERK5 or p-AKT, a stable Fra-1–overexpressing cell line (MM wtFra-1) was used. As shown in Figure 4A, in comparison to EV control and vehicle controls (PBS+.1%BSA added to medium), HGF (10 ng/ml) added to EV control cells induced increased amounts of phosphorylated ERK5 and AKT after 30 minutes. These effects were blocked by overexpression of wtFra-1. To verify that the effect we were observing on wtFra-1 transformed cells were due to increases in Fra-1 binding to the AP-1 complex, chromatin binding assays were performed. As shown in Figure 4B, chromatin-bound and total Fra-1 were increased in the AP-1 complex after addition of HGF to EV cells. Furthermore, wtFra-1 cells exhibited more strikingly increased levels of chromatin-bound Fra-1.
Figure 4.
HGF-dependent phosphorylation of ERK5 and AKT are blocked by overexpression of Fra-1. (A) Western blots show increases in p-ERK5 and p-AKT after addition of HGF (10 ng/ml) for 30 minutes in MM cells transfected with an EV. Controls included untreated (EV) cells and EV cells after addition of PBS+0.1%BSA (final concentration in medium) (HGF vehicle). No increases in p-ERK5 or p-AKT are observed after addition of HGF (10 ng/ml) to a Fra-1–overexpressing permanent cell line, wtFra-1. (B) Chromatin binding assay showing increases in chromatin bound Fra-1 after HGF addition (10 ng/ml, 30 min) and in cells transfected with wtFra-1. The right three lanes show total Fra-1 in protein lysates. 1 = MC1, 2 = +HGF, and 3 = wtFra-1. (C) HGF increases expression of the proliferation marker, PCNA, at time points of maximum ERK5 and AKT phosphorylation. Pre-addition of the PI3K small molecule inhibitor, LY420029 (20 μM) for 1 hour reduced expression of HGF-induced p-ERK5, p-AKT, and PCNA at 30 minutes. Levels of total AKT were evaluated to control for protein loading of lanes. (D) Increased cell viability by HGF is blocked by knocking down Fra-1 or MEK5 with RNAi constructs. An MTS assay was performed using 96-well plates. Relative viabililty (absorbance at 492 nm) is graphed relative to untreated control groups. Mean ± SEM of n = 3 (6 repeats per experiment). *P ⩽ 0.05 compared with respective control (scramble or siMEK5); +P ⩽ 0.05 compared with its equivalent in the scramble control group.
Cell Proliferation in MM Cell Lines with HGF-Inducible ERK5 Activation Is PI3K Dependent
To determine if HGF-dependent ERK5 activation was PI3K dependent and impacted MM cell proliferation, a time course study was conducted using detection of PCNA by Western blot analysis (Figure 4C). Increased expression of PCNA, which peaked at 30 to 60 minutes after addition of HGF (10 ng/ml), followed the same pattern as increases in p-AKT and p-ERK5 expression. Although the pre-addition of LY294002 (20 μM) did not alter levels in untreated MM cells, p-AKT, p-ERK5, and PCNA expression at 30 minutes after exposure to HGF were decreased.
To determine if HGF-induced cell viability was altered by knockdown of MEK5 or Fra-1, an MTS assay was performed after addition of HGF at various concentrations (1, 5, and 10 ng/ml). As shown in Figure 4D, HGF (10 ng/ml) caused increases (P ⩽ 0.05) in cell viability of transiently transfected MM cells (scramble control) that were decreased (P ⩽ 0.05) in cells transfected with siFra-1 or siMEK5 constructs. After addition of HGF at 5 ng/ml, siFra-1 transfected cells also exhibited less viability than MM scramble cells, although levels were unchanged in control (0) groups.
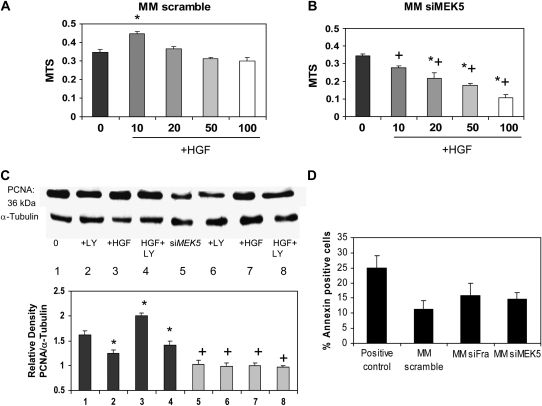
We next verified the effects of siMEK5 on blocking cell viability in response to HGF. Like effects shown in Figure 4, an increase in cell viability was observed after addition of HGF at 10 ng/ml, but not at higher concentrations (Figure 5A). HGF-induced proliferation was blocked by knocking down MEK5 with siMEK5, which also reduced cell viability at higher HGF concentrations (Figure 5B). Western blots showed that both addition of LY294002 (20 μM), and knockdown of MEK5 with siMEK5 reduced HGF-induced PCNA levels, but addition of LY294002 did not have additive effects with siMEK5 in diminishing HGF-induced PCNA expression (Figure 5C). To determine whether knocking down Fra-1 or MEK5 resulted in the induction of apoptosis, the early translocation of phosphatidyserine (PS) from the internal to external leaflet, a marker of early apoptosis, was used. As observed in Figure 5D, knocking down Fra-1 or MEK5 did not induce apoptosis in the time period in which these experiments were performed.
Figure 5.
Increased viability by HGF (10 ng/ml) is blocked by the use of siMEK5 as evaluated by a MTS assay after 18 hours. (A) Results using MM scramble control cells. Mean ± SEM of n = 6. (B) Results using MM siMEK5 cells. *P ⩽ 0.05 compared with untreated group (HGF 0), and +P ⩽ 0.05 compared with equivalent treatment in MM scramble group. Mean ± SEM of n = 3 (6 repeats per experiment). (C) Western blots showing decreases in PCNA protein after inhibition of PI3K by LY294002 (20 μM) added for 1 hour before addition of HGF (10 ng/ml) or after transient transfection with an siMEK5 RNAi construct. An antibody to α-tubulin was used as control to verify equal protein loading in lanes. Mean ± SEM of n = 3. *P ⩽ 0.05 compared with respective untreated group (MP5 or siMEK5), and +P ⩽ 0.05 compared with equivalent treatment in scramble control group. (D) Apoptotic cell distribution by annexin V–FITC staining quatitated by flow cytometry. Bars represent the mean ± SEM of n = 2. *P ⩽ 0.05.
DISCUSSION
The AP-1 family member, Fra-1, is up-regulated in several tumor types, including stomach cancer (25), esophageal tumors (26), squamous cell carcinomas (27), breast tumors (27), thyroid tumors (28), and mesotheliomas (1, 17). Although Fra-1 may play an important role in cell transformation (17, 28–33), little is known about how this important protein is regulated in human tumors.
Previous studies by our laboratory and others show that Fra-1 expression is modulated by the ERK1/2 pathway (17, 34–36). We show here that Fra-1 expression is more complex in approximately half of the human MMs examined, also involving activation of PI3K and ERK5 pathways.
In human MM cell lines, the degree of basal and induced Fra-1 expression is tumor line dependent. In three of the MM lines we examined, HGF-induced activation of the PI3K pathway contributed to increases in p-AKT and p-ERK5, and increases in Fra-1 protein were induced after activation of the PI3K pathway by HGF. On the other hand, in MM cell lines where elevation of p-AKT or p-ERK5 levels was not achieved by HGF, basal Fra-1 expression was generally higher (Figure 1). The cell signaling differences among these two groups of MM cell lines requires more investigation, but simian virus 40 positivity was found in the three cell lines in which HGF induced the PI3K/MEK5/Fra-1 cascade, while all others were simian virus 40 negative.
HGF and its receptor Met, a proto-oncogene known to modulate cell growth and morphogenesis, are activated in many human MM cell lines (13, 37) and are present in many MMs (38–40). Our data show that HGF stimulates MM cell proliferation through a PI3K/ERK5/Fra-1 pathway in some MM lines. We have shown previously that HGF can phosphorylate AKT in MMs (16), and here we show for the first time that HGF phosphorylates ERK5.
Our previous work addressed the importance of the ERK1/ERK2 pathway in Fra-1 expression; here we focused on MM cell lines with PI3K-dependent induced Fra-1 expression. We demonstrated that HGF-dependant Fra-1 expression could be blocked by the PI3K inhibitor, LY294002 (20 μM), or by knocking down MEK5 with siMEK5. Furthermore, knocking down of MEK5 and inhibition of the PI3K pathway together (Figure 2E) affect not only HGF-induced Fra-1 levels, but also Fra-1 basal levels suggesting that PI3K could be acting above MEK5 in the regulation of Fra-1 expression. Surprisingly, the Fra-1 mRNA levels in cells transfected with siMEK5 were higher than respective scramble controls, but HGF-induced Fra-1 mRNA levels were lower, agreeing with Fra-1 protein levels (Figure 2). These data suggest that mRNA levels of basal Fra-1, but not HGF-induced Fra-1, might be up-regulated as a compensatory mechanism of Fra-1 post-transcriptional instability after MEK5 knockdown. This hypothesis is supported by studies showing that the MEK5/ERK5 pathway causes phosphorylation and stabilization of Fra-1 in COS-7 and HEK293 cells (41).
Use of dnMEK5 stable MM cell lines clearly showed that basal Fra-1 expression was significantly increased in dnMEK5 cells compared with EV controls. Studies also revealed that protracted (24 h) levels of Fra-1 protein and particularly phosphorylated Fra-1 protein were lower in dnMEK5 MM cells. These results suggest that basal levels of Fra-1 may be compensated upon down-regulation of the MEK5 pathway, but that the phosphorylation and stability of Fra-1 is reduced in down-regulated MEK5 cells. Phosphorylation of Fra-1 is known to be important for its activation and stability. For example, Fra-1 is phosphorylated by ERK1/2, and phosphorylation of rat Fra-1 at Thr231 increases its transactivation activity (36).
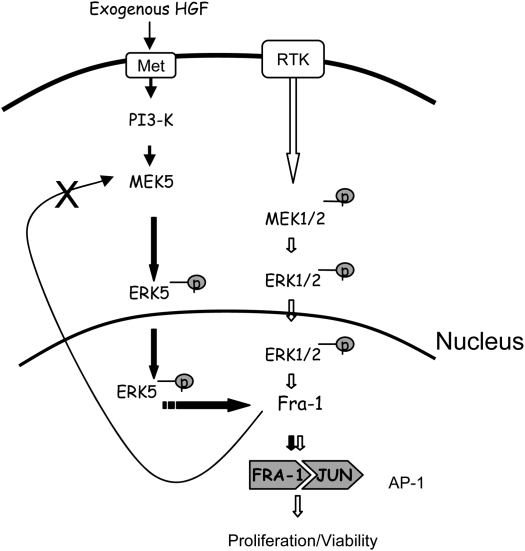
Figure 6 shows a hypothetical model of Fra-1 regulation in MMs. This model depicts on the right the importance of ERK1/2 phosphorylation in regulation of the Fra-1 pathway as previously reported in our studies (17) and in a number of cell types (34–36). In novel observations here, a second pathway is proposed (Figure 6, left) whereby HGF stimulates the PI3K pathway and the phosphorylation of MEK5 in some of the human MMs examined. Even though the phosphorylation of AKT was used as a measure of the PI3K pathway activation in these experiments, the involvement of AKT in Fra-1 regulation needs to be further addressed and therefore AKT was not incorporated in the hypothetical model. In support of our findings, activation of the PI3K pathway by HGF also have been reported in some human MM lines that respond differentially to inhibition of cell growth by the small molecule c-Met inhibitor, SU11274 (40). As shown in our studies here, overexpression of Fra-1 regulates HGF-induced phosphorylated ERK5 and phosphorylated AKT in a negative feedback control loop in MM cell lines with PI3K-dependent Fra-1 expression. This feedback control may govern responsiveness of MMs to HGF. For example, we reveal that knocking down MEK5 or Fra-1 with siRNA constructs or use of a dnMEK5 construct reduces HGF-dependent proliferation in MM. Even though proliferation is affected by knocking down Fra-1 or MEK5, apoptosis was not observed in the time period in which the experiments were performed. These data suggest that the ERK5/MEK5 pathway and Fra-1 may be important targets in HGF-dependent MM proliferation. Furthermore, the HGF-dependent Fra-1 expression, together with our previous observation that Fra-1 regulates the HGF receptor c-Met (1), suggests a self-regulatory mechanism of Fra-1 expression.
Figure 6.
Pathways of proposed Fra-1 regulation in human MMs. In cell lines with HGF-induced PI3K/MEK5-dependent Fra-1 expression, Fra-1 modulates its own pathway in a negative feedback loop that modulates p-ERK5 expression (left). In previous studies we have shown a ERK1/2-dependent Fra-1 expression (right) (17).
Acknowledgments
The authors acknowledge Timothy Hunter, Scott Tighe, Mary Lou Shane, and Meghan Brown from the Vermont Cancer Center (VCC) DNA Analysis Facility at the University of Vermont, where Real Time-Quantitative PCR and Flow cytometry analysis were performed.
This research was supported by NCI grants K01 CA104159–01 (M.E.R.-N.) and PO1CA114047 (M.C., H.P., J.T., and B.T.M.).
Originally Published in Press as DOI: 10.1165/rcmb.2007-0206OC on September 13, 2007
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Ramos-Nino ME, Scapoli L, Martinelli M, Land S, Mossman BT. Microarray analysis and rna silencing link fra-1 to cd44 and c-met expression in mesothelioma. Cancer Res 2003;63:3539–3545. [PubMed] [Google Scholar]
- 2.Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med 2005;353:1591–1603. [DOI] [PubMed] [Google Scholar]
- 3.Mossman B, Gee J. Asbestos related disease. N Engl J Med 1989;320:1721–1730. [DOI] [PubMed] [Google Scholar]
- 4.Mossman B, Gruenert D. SV40, growth factors, and mesothelioma–another piece of the puzzle. Am J Respir Cell Mol Biol 2002;26:167–170. [DOI] [PubMed] [Google Scholar]
- 5.Pass HI, Mew DJ, Carbone M, Matthews WA, Donington JS, Baserga R, Walker CL, Resnicoff M, Steinberg SM. Inhibition of hamster mesothelioma tumorigenesis by an antisense expression plasmid to the insulin-like growth factor-1 receptor. Cancer Res 1996;56:4044–4048. [PubMed] [Google Scholar]
- 6.Lee T, Zhang Y, Aston C, Hintz R, Jagirdar J, Perle M, Burt M, Rom W. Normal human mesothelial cells and mesothelioma cell lines express insulin-like growth factor i and associated molecules. Cancer Res 1993;53:2858–2864. [PubMed] [Google Scholar]
- 7.Versnel M, Claesson-Welsh L, Hammacher A, Bouts M, van der Kwast T, Eriksson A, Willemsen R, Weima S, Hoogsteden H, Hagemeijer A, et al. Human malignant mesothelioma cell lines express pdgf β-receptors whereas cultured normal mesothelial cells express predominantly pdgf α-receptors. Oncogene 1991;6:2005–2011. [PubMed] [Google Scholar]
- 8.Asplund T, Versnel MA, Laurent TC, Heldin P. Human mesothelioma cells produce factors that stimulate the production of hyaluronan by mesothelial cells and fibroblasts. Cancer Res 1993;53:388–392. [PubMed] [Google Scholar]
- 9.Harvey P, Warn A, Dobbin S, Arakaki N, Daikuhara Y, Jaurand MC, Warn RM. Expression of hgf/sf in mesothelioma cell lines and its effects on cell motility, proliferation and morphology. Br J Cancer 1998;77:1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heintz N, Janssen Y, Mossman B. Persistent induction of c-fos and c-jun expression by asbestos. Proc Natl Acad Sci USA 1993;90:3299–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janssen Y, Heintz N, Marsh J, Borm P, Mossman B. Induction of c-fos and c-jun proto-oncogenes in target cells of the lung and pleura by carcinogenic fibers. Am J Respir Cell Mol Biol 1994;11:522–530. [DOI] [PubMed] [Google Scholar]
- 12.Timblin C, Guthrie G, Janssen Y, Walsh E, Vacek P, Mossman B. Patterns of c-fos and c-jun protooncogene expression, apoptosis and proliferation in rat pleural mesothelial cells exposed to erionite or asbestos fibers. Toxicol Appl Pharmacol 1998;151:88–97. [DOI] [PubMed] [Google Scholar]
- 13.Mukohara T, Civiello G, Davis IJ, Taffaro ML, Christensen J, Fisher DE, Johnson BE, Janne PA. Inhibition of the met receptor in mesothelioma. Clin Cancer Res 2005;11:8122–8130. [DOI] [PubMed] [Google Scholar]
- 14.Klominek J, Baskin B, Liu Z, Hauzenberger D. Hepatocyte growth factor/scatter factor stimulates chemotaxis and growth of malignant mesothelioma cells through c-met receptor. Int J Cancer 1998;76:240–249. [DOI] [PubMed] [Google Scholar]
- 15.Tolnay E, Kuhnen C, Wiethege T, Konig J, Voss B, Muller K. Hepatocyte growth factor/scatter factor and its receptor c-met are overexpressed and associated with an increased microvessel density in malignant pleural mesothelioma. J Cancer Res Clin Oncol 1998;124:291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altomare DA, You H, Xiao GH, Ramos-Nino ME, Skele KL, De Rienzo A, Jhanwar SC, Mossman BT, Kane AB, Testa JR. Human and mouse mesotheliomas exhibit elevated akt/pkb activity, which can be targeted pharmacologically to inhibit tumor cell growth. Oncogene 2005;24:6080–6089. [DOI] [PubMed] [Google Scholar]
- 17.Ramos-Nino ME, Timblin CR, Mossman BT. Mesothelial cell transformation requires increased ap-1 binding activity and erk-dependent fra-1 expression. Cancer Res 2002;62:6065–6069. [PubMed] [Google Scholar]
- 18.Scapoli L, Ramos-Nino M, Martinelli M, Mossman B. Src-dependent erk5 and src/egfr-dependent erk1/2 activation is required for cell proliferation by asbestos. Oncogene 2004;23:805–813. [DOI] [PubMed] [Google Scholar]
- 19.Milde-Langosch K. The fos family of transcription factors and their role in tumourigenesis. Eur J Cancer 2005;41:2449–2461. [DOI] [PubMed] [Google Scholar]
- 20.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Human keratinocytes that express htert and also bypass a p16(ink4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 2000;20:1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao GH, Jeffers M, Bellacosa A, Mitsuuchi Y, Vande Woude GF, Testa JR. Anti-apoptotic signaling by hepatocyte growth factor/met via the phosphatidylinositol 3-kinase/akt and mitogen-activated protein kinase pathways. Proc Natl Acad Sci USA 2001;98:247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not rpa, is a component of the mammalian early g1-phase prereplication complex. J Cell Biol 1999;146:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato Y, Kravchenko VV, Tapping RI, Han J, Ulevitch RJ, Lee JD. Bmk1/erk5 regulates serum-induced early gene expression through transcription factor mef2c. EMBO J 1997;16:7054–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burch PM, Yuan Z, Loonen A, Heintz NH. An extracellular signal-regulated kinase 1- and 2-dependent program of chromatin trafficking of c-fos and fra-1 is required for cyclin d1 expression during cell cycle reentry. Mol Cell Biol 2004;24:4696–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsui M, Tokuhara M, Konuma Y, Nomura N, Ishizaki R. Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene 1990;5:249–255. [PubMed] [Google Scholar]
- 26.Hu Y, Lam K, Law S, Wong J, Srivastava G. Profiling of differentially expressed cancer-related genes in esophageal squamous cell carcinoma (escc) using human cancer cdna arrays: Overexpression of oncogene met correlates with tumor differentiation in escc. Clin Cancer Res 2001;7:2213–2221. [PubMed] [Google Scholar]
- 27.Zajchowski D, Bartholdi M, Gong Y, Webster L, Liu H, Munishkin A, Beauheim C, Harvey S, Ethier S, Johnson P. Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res 2001;61:5168–5178. [PubMed] [Google Scholar]
- 28.Chiappetta G, Tallini G, De Biasio MC, Pentimalli F, de Nigris F, Losito S, Fedele M, Battista S, Verde P, Santoro M, et al. Fra-1 expression in hyperplastic and neoplastic thyroid diseases. Clin Cancer Res 2000;6:4300–4306. [PubMed] [Google Scholar]
- 29.Kim YH, Oh JH, Kim NH, Choi KM, Kim SJ, Baik SH, Choi DS, Lee ES. Fra-1 expression in malignant and benign thyroid tumor. Korean J Intern Med 2001;16:93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kustikova O, Kramerov D, Grigorian M, Berezin V, Bock E, Lukanidin E, Tulchinsky E. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol Cell Biol 1998;18:7095–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belguise K, Kersual N, Galtier F, Chalbos D. Fra-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 2005;24:1434–1444. [DOI] [PubMed] [Google Scholar]
- 32.Debinski W, Gibo DM. Fos-related antigen 1 modulates malignant features of glioma cells. Mol Cancer Res 2005;3:237–249. [DOI] [PubMed] [Google Scholar]
- 33.Reddy S, Vuong H, Adiseshaiah P. Regulation of fra-1 gene expression by cigarette smoke in bronchial epithelium [abstract]. Am J Respir Crit Care Med 2002;165:A828. [Google Scholar]
- 34.Hoffmann E, Thiefes A, Buhrow D, Dittrich-Breiholz O, Schneider H, Resch K, Kracht M. Mek1-dependent delayed expression of fos-related antigen-1 counteracts c-fos and p65 nf-kappab-mediated interleukin-8 transcription in response to cytokines or growth factors. J Biol Chem 2005;280:9706–9718. [DOI] [PubMed] [Google Scholar]
- 35.Vial E, Sahai E, Marshall CJ. Erk-mapk signaling coordinately regulates activity of rac1 and rhoa for tumor cell motility. Cancer Cell 2003;4:67–79. [DOI] [PubMed] [Google Scholar]
- 36.Young M, Nair R, Bucheimer N, Tulsian P, Brown N, Chapp C, Hsu T-C, Colburn N. Transactivation of fra-1 and consequent activation of ap-1 occur extracellular signal-regulated kinase dependently. Mol Cell Biol 2002;22:587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cacciotti P, Libener R, Betta P, Martini F, Porta C, Procopio A, Strizzi L, Penengo L, Tognon M, Mutti L, et al. SV40 replication in human mesothelial cells induces hgf/met receptor activation: A model for viral-related carcinogenesis of human malignant mesothelioma. Proc Natl Acad Sci USA 2001;98:12032–12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thirkettle I, Harvey P, Hasleton PS, Ball RY, Warn RM. Immunoreactivity for cadherins, hgf/sf, met, and erbb-2 in pleural malignant mesotheliomas. Histopathology 2000;36:522–528. [DOI] [PubMed] [Google Scholar]
- 39.Harvey P, Warn A, Newman P, Perry LJ, Ball RY, Warn RM. Immunoreactivity for hepatocyte growth factor/scatter factor and its receptor, met, in human lung carcinomas and malignant mesotheliomas. J Pathol 1996;180:389–394. [DOI] [PubMed] [Google Scholar]
- 40.Jagadeeswaran R, Ma PC, Seiwert TY, Jagadeeswaran S, Zumba O, Nallasura V, Ahmed S, Filiberti R, Paganuzzi M, Puntoni R, et al. Functional analysis of c-met/hepatocyte growth factor pathway in malignant pleural mesothelioma. Cancer Res 2006;66:352–361. [DOI] [PubMed] [Google Scholar]
- 41.Terasawa K, Okazaki K, Nishida E. Regulation of c-fos and fra-1 by the mek5-erk5 pathway. Genes Cells 2003;8:263–273. [DOI] [PubMed] [Google Scholar]