Abstract
Background
This work was carried out to assess the patterns and prevalence of resistance to chloroquine (CQ) and sulphadoxine-pyrimethamine (SP) in Iran.
Methods
The prevalence of pfcrt K76T, pfmdr1 N86Y, pfdhfr N51I, C59R, S108N/T and I164L and codons S436F/A, A437G, K540E, A581E, and A613S/T in pfdhps genes were genotyped by PCR/RFLP methods in 206 Plasmodium falciparum isolates from Chabahar and Sarbaz districts in Sistan and Baluchistan province, Iran, during 2003–2005.
Results
All P. falciparum isolates carried the 108N, while 98.5% parasite isolates carried the 59R mutation. 98.5% of patients carried both 108N and 59R. The prevalence of pfdhps 437G mutation was 17% (Chabahar) and 33% (Sarbaz) isolates. 20.4% of samples presented the pfdhfr 108N, 59R with pfdhps 437G mutations. The frequency of allele pfcrt 76T was 98%, while 41.4% (Chabahar) and 27.7% (Sarbaz) isolates carried pfmdr1 86Y allele. Eight distinct haplotypes were identified in all 206 samples, while the most prevalent haplotype was T76/N86/N51R59N108/A437 among both study areas.
Conclusion
Finding the fixed level of CQ resistance polymorphisms (pfcrt 76T) suggests that CQ must be withdrawn from the current treatment strategy in Iran, while SP may remain the treatment of choice for uncomplicated malaria.
Background
Drug resistance is the most serious problem in achieving control of malaria. The spread of Plasmodium falciparum resistance to available cheap drugs, the increased cost of insecticides, the vector's resistance to insecticides and the lack of an effective vaccine, together with a socio-economic instability in many malaria-endemic region, impact on malaria control. Therefore, surveillance and prevention of drug resistance and also effective curative chemotherapy have become more important to be considered as the primary approach to malaria control. For decades, chloroquine (CQ) was an efficacious antimalarial, however malaria parasite resistance to treatment with chloroquine has already complicated malaria management and has been associated with increased malaria morbidity and mortality [1,2]. The antimalarial compounds sulphadoxine (S) and pyrimethamine (P) are usually given together as a synergistic combination, known commercially as Fansidar® [3], and represent an effective agent against chloroquine-resistant P. falciparum. However, resistance to this drug has also been reported from parts of South East Asia, Latin America and Africa [4-6].
In Iran, malaria was a serious health problem in the past and according to the records of Center for Diseases Management and Control (CDMC), Ministry of Health and Medical Education (MHME), during the last decade (1990–1999) the annual malaria cases have decreased from 96,340 in 1991 to 24,000 in 2005. Malaria transmission mostly occurs in the south-eastern parts of the country in Sistan-Baluchistan, Hormozgan and Kerman (mainly in Kahnoudj) provinces. Resistance of P. falciparum to chloroquine has increased since it was first reported in 1983 in Sistan-Baluchiatan province and later in Hormozgan province in 1986 [7], and currently accounts for more than 78.5% of treatment failures in south-eastern provinces of Iran [8]. Resistance to SP was confirmed in malaria-endemic areas of Iran by in vitro and in vivo tests [9]. Despite the report of resistance to these drugs in Iran, CQ and SP are still used as antimalarial drugs in this region.
Plasmodium falciparum resistance to CQ and SP has been associated with combinations of the presence of single nucleotide polymorphisms (SNPs) in a number of P. falciparum genes. CQ resistance is associated with polymorphisms in the P. falciparum chloroquine resistance transporter (pfcrt) gene [10-12], while polymorphisms in the P. falciparum multidrug resistance 1 (pfmdr1) gene have been shown by transfection to modulate higher levels of CQ resistance [13]. In this regards, K76T mutation of the pfcrt gene is strongly associated with CQ resistance [10], while N86Y mutation of pfmdr1 gene may modulate its degree [14]. SP resistance is associated with polymorphisms in the dihydropteroate synthetase gene (dhps) and the didydropolate reductase gene (dhfr) [15,16]. Among SNPs currently identified, the N51I, C59R, I164L with S108N/T in dhfr confer increasing levels of pyrimethamine resistance [17]. Similarly, polymorphisms at positions S436A/F, A437G, L540E, A581G and S613A/T in dhps are the key mutations associated with sulphadoxine resistance. The polymorphism 437G in dhps gene appears to be selected first by drug pressure and accompany with polymorphisms at other positions confers increasing levels of resistance to this drug [18]. Several studies have revealed that the differing degrees of antimalarial drug resistance are dependent upon the number and combination of mutations present in aforementioned genes [19-21]. Initial clinical failures to SP usually become evident when an isolate carries a triple mutant, 51I, 59R and 108N in the pfdhfr gene with or without additional mutations in pfdhps [22-24], however, the quintuple mutant carrying all of dhfr triple mutant at codons 51I, 59R and 108N also in combination with double mutant at codons 437G and 540E in dhps are associated with SP treatment failure [20,24-27]. These SNPs could be used as rapid molecular marker for detecting these mutations, therefore represent molecular epidemiology surveillance tools of antimalarial resistance, which may replace the conventional in vitro or in vivo phenotyping assays [28,29].
The aim of the present study was to complement existing data on molecular studies by determining the frequency of known mutations in dhfr and dhps genes of P. falciparum isolates from south-eastern region of Iran. In addition, we investigated the possibility of an association between CQ and SP resistance by analysing pfcrt and pfmdr1 mutations on the same P. falciparum isolates. The data presented here revealed the prevalence of mutations associated to CQ and SP resistance in P. falciparum isolates collected at health centers in Chabahar and Sarbaz districts in Sistan and Baluchistan province, Iran and therefore, will be contributed to the development of strategies for therapeutic intervention of malaria in Iran.
Materials and methods
Study sites and samples collection
Chabahar and Sarbaz districts in Sistan and Baluchistan province in south-eastern part of Iran have been selected as study areas. Both areas are malaria endemic and the patients have access to antimalarial drugs through local health centers. In Chabahar district samples were collected from Chabahar, Nowbandiyan and Dargas health centers, however, in Sarbaz district most of the samples were collected from Pishin health center (5 Km far from border line with Pakistan), where, there is a lot of population movement to Pakistan and vice versa (Figure 1). In these regions, malaria transmission is year-round with two peaks, the first in May to August with P. vivax as the predominant species and the second peak from October to November, when both P. falciparum and P. vivax infections are generally equally recorded. 206 blood samples were collected from P. falciparum-infected patients, aged from 1 to >60 years old with Iranian, Afghani and Pakistani nationals from Chabahar (n = 152) and Sarbaz (n = 54) districts in Sistan and Baluchistan during 2003 to October 2005.
Figure 1.
Map of Iran indicating the location of the study area in Chabahar and Sarbaz districts situated in the south-eastern corner of Baluchistan Province, from where the P. falciparum isolates were collected. S&B: Sistan and Baluchistan province, Ch: Chabahar, P: Pishin
Thin and thick blood films were stained with Giemsa and examined microscopically for detection of P. falciparum parasite. Approximately 1 ml venous blood was obtained in tube containing EDTA from patients who were confirmed to be positive for the presence of P. falciparum parasites. Then all patients were treated with CQ (25 mg/kg body weight over 3 days) plus primaquine (P) (0.75 mg/kg single dose in third day) as first line drug and if needed treated with single dose of SP as second line drug for treatment of uncomplicated malaria cases. Patients' or parents' informed consent was obtained before inclusion in the study. The study was reviewed by, and received Ethical Clearance from Pasteur Institute of Iran. Blood samples were collected in tube containing EDTA, stored at 4°C, and then transported to the main laboratory in Tehran.
Parasite DNA extraction and nested polymerase chain reaction amplification of pfdhfr and pfdhps genes
The Parasite genomic DNA was extracted from infected red blood cells using phenol/phenol-chloroform followed by ethanol precipitation as described previously [30]. Nested PCRs were performed for dhfr and dhps genes, and all reactions were carried out in 25 μl reaction mixtures containing 1.5–3 mM MgCl2, 200 μM dNTP mixture (invitrogene, USA), 1 unit of Taq polymerase (invitrogen, USA), and a pair of primers (10 pmol each). For both dhfr and dhps one to two microliters of DNA was used as template in the first reaction and for second reaction, 1 μl of first PCR product, if no band was seen from first round PCR product, whereas 1 μl of a 1/100 dilution of the samples with an intense band was used as template for secondary PCR. The negative (water) controls were used in all PCRs. The PCR primers and conditions for both genes were those published by Duraisingh et al. [31]. Secondary PCR products were resolved by electrophoresis on 1–2% agarose gels and visualized by staining with ethidium bromide.
Restriction Fragment Length Polymorphism-Polymerase Chain Reaction of pfdhfr and pfdhps genes
The mutation specific restriction endonuclease digestion was used to detect SNPs in dhfr at positions N51I, C59R, S108N/T, I164L, and in dhps at position S436F/A, A437G, K540E, A581E, A613S/T [31]. A number of restriction enzymes were used for RFLP of PCR products. For dhfr, the PCR products were digested with TasI and TaqI to determine the polymorphisms at codons 51 and 59, respectively. Three enzymes, AluI, BsrI, and MvaI were used to identify wild and mutant dhfr allele at codon 108 and DraI used to detect mutation at position I164L. For dhps, the PCR products were digested with HhaI, MnlI and HindIII to determine the polymorphisms at codon 436, AvaII and MwoI for codon 437, FokI, BstUI and MwoI to determine the polymorphisms at codons 540, 580 and 613, respectively. Digestions were done in 20 μl reactions containing 10 μl of PCR fragments according to the manufacturer's instructions (New England Biolab, Beverly, MA, USA; Roche, Germany; Invitrogen, Carlsbad, CA). Digested products were subjected to electrophoresis on 1.5–2% agarose or 2–3% Metaphor agarose gels, and visualized by ultraviolate (UV) transillumination.
Detection of pfcrt and pfmdr1 mutations by restriction digestion of PCR products
For mutation detection at codon K76T in pfcrt and N86Y in pfmdr1 genes one to two microliters of DNA was used as template in the first reaction (if no band was seen), whereas 1 μl of a 1/100 to 1/500 dilution of the samples with an intense band from first round PCR product was used as template for the secondary PCR. The PCR primers and conditions for both genes were those published previously [10,32]. Purified genomic DNA from P. falciparum clones HB3 (Chloroquine sensitive) and Dd2 (chloroquine resistant) were used as positive controls, and water, extracted uninfected blood used as negative controls. PCR products were resolved by electrophoresis on 1–2% agarose gels and visualized by staining with ethidium bromide.
Following amplification of the fragments concerned, polymorphisms in the pfcrt and pfmdr1 genes were assessed by the mutation specific restriction endonuclease digestion to detect Single Nucleotide Polymorphisms (SNPs) in pfcrt at positions K76T, and in pfmdr1 at position N86Y/H [10,32]. A number of restriction enzymes were used for RFLP of PCR products. For pfcrt, the PCR products were digested with ApoI, determine the polymorphisms at codons 76. For pfmdr1, three enzymes, ApoI, NsiI and AFlIII were used to identify polymorphisms at codon 86. Digestions were done in 20 μl reactions containing 10 μl of PCR products according to the manufacturer's instructions (New England Biolab, Beverly, MA, USA; and/or Fermentase, Vilnius, Lithuania; New England Biolabs, Beverly, MA). If there was doubt about a complete digestion, reactions were repeated overnight. Digested products were subjected to electrophoresis on 1.5–2% agarose or 2–3% Metaphor agarose gels, and visualized by ultraviolate (UV) transillumination.
Results
Pfdhfr and pfdhps genotypes
All 206 P. falciparum isolates were successfully genotyped for the detection of dhfr and dhps mutations associated to SP resistance. All P. falciparum isolates (100%) from both Chabahar and Sarbaz districts were found to carry the mutant type 108N, and 98.5% of them carried the 59R mutation, however the 51I mutation was present in 0.97% of examined samples (Table 1). The majority of the Patients (98.5%) were found to carry both 108N and 59R in pure form, while retaining a wild-type mutation at position 51 and 164 (Table 2).
Table 1.
Frequency distribution of mutations conferring resistance to chloroquine and pyrimethamine-sulphadoxine in Plasmodium falciparum isolates from southeastern Iran
| Gene | Codon | Chabahar (n = 152) | Sarbaz (n = 54) | |
| Mutation (%) | Mutation (%) | |||
| Choloroquine | pfcrt | 76T | 149 (98%) | 53 (98%) |
| pfmdr 1 | 86Y | 63 (41.4%) | 15 (27.7%) | |
| 86N/Y | 1 (0.77%) | 1 (1.96%) | ||
| Pyrimethamine | pfdhfr | 51I | 1 (0.77%) | 1 (1.96%) |
| 59R | 150 (98.6%) | 53 (98%) | ||
| 108N | 152 (100%) | 54 (100%) | ||
| 108T | - | - | ||
| 164L | - | - | ||
| Sulphadoxine | pfdhps | 436A | - | - |
| 436F | - | - | ||
| 437G | 26 (17%) | 18 (33%) | ||
| 437A/G | 3 (1.9%) | 3 (5.5%) | ||
| 540E | - | - | ||
| 581G | - | - | ||
| 613N | - | - | ||
Table 2.
The frequency distribution of SNPs combinations of pfcrt and pfmdr1 associated to chloroquine resistance, plus pfdhfr and pfdhps alleles associated with pyrimethamine-sulphadoxine resistance in clinical isolates of Plasmodium falciparum in southeastern Iran
| Gene | Codon/Mutation | Chabahar | Sarbaz | Total |
| n = 152 | n = 54 | n = 206 | ||
| pfcrt + pfmdr 1 | 76T + 86Y | 63 (41.4%) | 16 (29.6%) | 79 (38.3%) |
| Pfdhfr | 59R + 108N | 150 (98.7%) | 53 (98%) | 203 (98.5%) |
| Pfdhfr | 51I + 59R + 108N | 1 (0.77%) | 1 (1.96%) | 2 (1.1%) |
| Pfdhfr + pfdhps | 59R + 108N + 437G | 24 (15.8%) | 18 (35.3%) | 42 (20.4%) |
| pfcrt + pfmdr 1 + pfdhfr + pfdhps | 76T + 86Y + 59R + 108N +437G | 13 (8.55%) | 9 (17.6%) | 22 (10.7%) |
In case of the dhps gene, polymorphisms in different loci of dhps (S436F/A, A437G, K540E, A581E and A613S/T)) were investigated. All isolates were found to carry wild-type amino acids at positions 436, 540, 581, and 613, while 437G mutation in pur form was detected among 17% and 33% examined samples collected from Chabahar and Sarbaze districts, respectively (Table 1). Mutations at codons 108N, 59R of pfdhfr with pfdhps 437G were detected in 20.4% of examined samples (Table 2).
pfcrt K76T and pfmdr1 N86Y
The frequency of pure mutant of allele pfcrt 76T was 98% in both Chabahar and Sarbaz districts (Table 1). The frequency of the mutant pfmdr1 86Y allele was 41.4% and 27.7% among isolates from Chabahar and Sarbaz districts, respectively (Table 1).
Distribution of pfcrt, pfmdr1, pfdhfr and pfdhps haplotypes in Iran
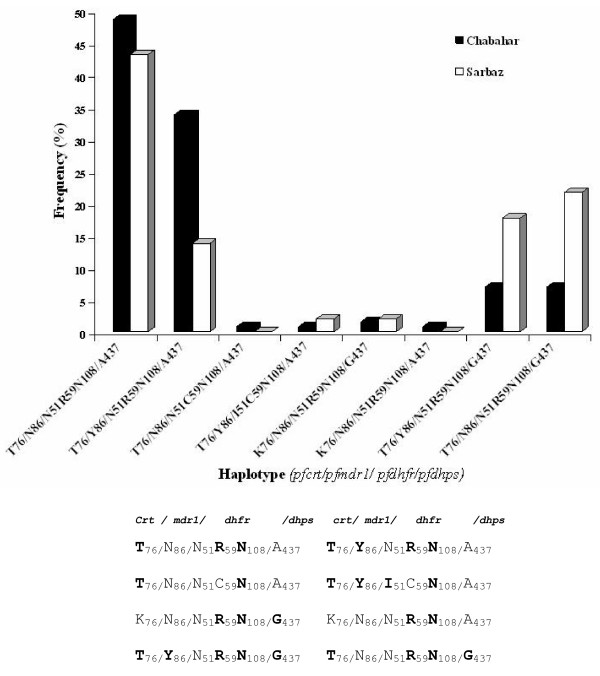
Combination of pfcrt, pfmdr1, pfdhfr and pfdhps haplotypes among all 206 samples in this study demonstrated 8 distinct haplotypes (Figure 2). The two most prevalent haplotypes among samples were T76/N86/N51R59N108/A437 (47%) and T76/Y86/N51R59N108/A437 (28.6%). In addition, the majority of isolates from Chabahar (48%) and Sarbaz (43%) were belonging to haplotype T76/N86/N51R59N108/A437 (Figure 2). pfcrt 76T and pfmdr1 86Y with the pfdhfr 59R, 108N and pfdhps 437G mutations was detected in 10.7% isolates from Chabahar (n = 13) and Sarbaz (n = 9) districts (Table 2).
Figure 2.
Frequency distribution of the combination pfcrt/pfmdr1/pfdhfr/pfdhps haplotypes obtained from 206 isolates collected in Sistan and Baluchistan of Iran. The haplotype T76/N86/N51R59N108/A437 was the most prevalent among Chabahar (48%) and Sarbaz (43%) P. falciparum isolates. Mutated amino acids are boldfaced.
Discussion
In Iran, chloroquine-resistant parasites were first observed in 1983 [33] and later reported for more than 78.5% of treatment failures in south-eastern provinces of Iran [8], but it is still under use as antimalarial drugs in these regions. In both Chabahar and Sarbaz districts pfcrt 76T polymorphism was fixed in parasites populations, as shown by the high rate of polymorphism and no mixed alleles, however, lower frequency of pfmdr1 86Y polymorphism has been detected among Sarbaz isolates. Based on the prevalence of pfcrt 76T (98%) and pfmdr1 86Y (37.8%) in all 206 isolates, it could be concluded that pfcrt76T but not pfmdr1 86Y mutation is strongly associated with CQ resistance and has the potential as a molecular predictor for CQ therapeutic treatment failure in Iran.
Furthermore, in vivo and in vitro study of clinical failures with SP reported since 1993 [9,34], but in vivo resistance of P. falciparum to SP is not yet common in malarious endemic area with a high level of CQ resistance in Iran. To investigate and complete molecular surveillance on SP efficacy in Iran, 206 collected P. falciparum isolates during 2003 to October 2005, have been analyzed. At the time that the present study initiated, SP was used for CQ treatment failures, as treatment failure of SP was not frequent in the region, at the end of this study, the CDMC and MHME revised the standard treatment recommendations for uncomplicated malaria to CQ plus SP as first line and Co-Artem, as second line anti malarial drugs, respectively.
All 206 P. falciparum isolates examined in this study carry pfdhfr 108N mutation (100%) with no evidence of clinical failure of SP in patients. Although it has been postulated that pfdhfr 108N mutation might be a good marker of clinical SP resistance [35], but high prevalence of this mutation in areas with low clinical failure to SP may be associated with prior primaquine exposure [36] and this could explain the fixed prevalence of this mutation among our isolates. Further study is needed to clarify the association of primaquine exposure and pfdhfr 108N mutation rates.
It was also found that polymorphism in the pfdhps gene were found less frequent in P. falciparum population in Iran. The only mutation detected in P. falciparum isolates from both study areas was pfdhps 437G. This mutation was rather more prevalent in Sarbaz (33%), in border area with Pakistan, while its prevalence in Chabahar isolates is almost half (17%), indicating that this mutant parasites might have been spread through the Indian subcontinent to Pakistan and further to Iran. However, this observation remains to be clarified by further study on Pakistani P. falciparum isolates. Also in comparison with our previous work [37], the results of this study showed that the rate of pfdhps 437G mutation has been increased from 17% mixed form to 21.3% pure form in Iran. Similarly, Heidari and co-workers [34] reported the increase of this mutation in Sistan and Baluchistan province.
Chabahar and Sarbaz isolates showed similar mutations pattern and combination at pfdhfr 51I, 59R, 108N and pfdhps 437G positions, but with varied mutation prevalence. The haplotype 59R, 108N (pfdhfr), 437G (pfdhps) was more prevalent in Sarbaz (35.3%) than Chabahar (15.8%) districts. It is worth to note that, 27.4% of Sarbaz isolates with these three mutations isolated from patients who either had trip to or came from Pakistan two-three weeks prior to blood sampling. Hence to control of the disease, it should be kept in our mind the possibility of the spread of these alleles from neighbouring countries to malaria settings in Iran. Therefore, the predominant pfdhfr haplotype in Iran seems to be N51,59R,108N rather than 51I,59R,108N. Low prevalence of triple mutation in examined isolates in this study was similar with finding from India [38,39], Sri Lanka [40] and Papua New Guinea [41], however was different with Vietnam [16,42]; Malaysian [43], Gaboni [44] and Brazilian [45] isolates, where the predominant haplotype was 51I,59R,108N. This may suggested that the pfdhfr allele has evolved independently due to drug pressure in geographically isolates regions of the world. Regarding CQ resistant P. falciparum, it was believed that resistant parasites emerged in South East Asia and then spread to Africa via the Indian subcontinent; however the results of recent works [46,47] was contrary to such hypothesis. In case of SP resistant parasites, if the same hypothesis was true, the data from present study is not supporting a presumed spread of resistant parasites from South East Asia, and multiple geographic foci for the origin of both CQ and SP resistance mutations might be postulated. The high prevalence of double mutations at codon 59R/108N rather than 51I/108N in our parasite isolates suggested that 51I mutation would be a good molecular marker for the triple mutant and indicating failure to pyrimethamine in Iran and also Indian subcontinent. In addition, these two mutations with the mutation at 437G position in dhps indicate that the P. falciparum parasite populations have the potential to evolve into dhfr/dhps quintuple mutant polymorphism in near future, therefore, monitoring of the status of dhps alleles could be a high priority as a predictor of developing clinical resistance to sulphadoxine in this region.
Conclusion
In conclusion, the present results demonstrated the low progressing rate in SNPs frequencies in both pfdhfr and pfdhps genes since 2003 to 2005, more likely due to not having easily access to SP as antimalarial drug for self treatment by patients in these malaria-endemic areas of Iran. In addition, finding the fixed level of CQ resistance polymorphisms (pfcrt 76T) in our studied isolates suggested that the CQ must be withdrawn from the current treatment strategy in Iran. However, since October 2005, based on evidence of high rates of chloroquine treatment failure, Iran revised its national policy for treatment of malaria and SP combination therapy with CQ have replaced the CQ/P as first line antimalarial drug. Thus now with more availability of SP there is possibility of increasing resistance to SP from moderate to high and its rapid spread in this particular malaria setting. Therefore, although the results of this study suggested that SP may remain the treatment of choice for uncomplicated malaria in Iran, but due to high rates of treatment failures to CQ, serious consideration must now be given to replace SP/CQ combination therapy with SP/artemisinin as first line antimalarial drugs in near future in Iran, which has already been shown to be effective against CQ resistant isolates of P. falciparum.
Authours' contributions
SZ designed the study, and was responsible for supervision of laboratory work, development of the protocols, carrying out most parts of the laboratory experiments, analysis of data and writing up the paper with contribution of NDD. Also, MA was involved in laboratory work. AR coordinated for blood sampling and data processing. All authors read and approved the manuscript.
Acknowledgments
Acknowledgements
We are indebted to the patients and their families in Sistan and Baluchistan province for their willingness to parcipitate in this study. We also acknowledge with deep respect the co-operation of Center for Diseases Management and Control (CDMC), particularly Dr. M.M. Gouya, and are also grateful for the hospitality and generous collaboration of Zahedan University of Medical Sciences (Dr. M. Salehi), and the staff of the Public Health Department, Sistan and Baluchistan province, Chabahar and Sarbaz district (Dr. Mehdizadeh, Dr. Ebrahimpour, Dr. Kordi, Mr. Gorgij, Mr. Bra, Mr. Bram), for their assistance in collecting blood samples from the field. This work was supported by Iranian Ministry of Health and Medical Education and Pasteur Institute of Iran.
Contributor Information
Sedigheh Zakeri, Email: zakeris@yahoo.com.
Mandana Afsharpad, Email: afsharpad@yahoo.com.
Ahmad Raeisi, Email: ahmadraeisi@yahoo.com.
Navid Dinparast Djadid, Email: navid@pasteur.ac.ir.
References
- Greenberg AE, Ntumbanzondo M, Ntula N, Mawa L, Howell J, Davachi F. Hospital based surveillance of malaria related paediatric morbidity and mortality in Kinshasa, Zaire. Bull World Health Organ. 1989;67:189–196. [PMC free article] [PubMed] [Google Scholar]
- Trape JF, Pison G, Preziosi MP, Enel C, Desgreés du Loû A, Delaunay V, Samb B, Lagarde E, Molez JF, Simondon F. Impact of chloroquine resistance on malaria mortality. C R Acad Sci III. 1998;321:689–697. doi: 10.1016/s0764-4469(98)80009-7. [DOI] [PubMed] [Google Scholar]
- Chulay JD, Watkins WM, Sixsmith DG. Synergistic antimalarial activity of pyrimethamine and sulfadoxine against Plasmodiumfalciparum in vitro. Am J Trop Med Hyg. 1984;33:325–330. doi: 10.4269/ajtmh.1984.33.325. [DOI] [PubMed] [Google Scholar]
- Alecrim WD, Dourado H, Alecrim MG, Passos LF, Wanssa E, Albuquerque B. In vivo resistance of Plasmodium falciparum to the combination of sulfadoxine and pyrimethamine, at RIII level in Amazons, Brazil. Rev Inst Med Trop Sao Paulo. 1982;24:52–53. [PubMed] [Google Scholar]
- Hurwitz ES, Johnson D, Campbell CC. Resistance of Plasmodium falciparum malaria to sulfadoxine-pyrimethamine (Fansidar) in a refugee camp in Thailand. Lancet. 1981;16:1068–1070. doi: 10.1016/S0140-6736(81)92239-X. [DOI] [PubMed] [Google Scholar]
- Kilimali VAEB, Mkfuya AR. In vivo assessment ofthe sensitivity of Plasmodium falciparum tosulphadoxine/pyrimethamine combinations (Fansidar) in six locationsin Tanzania where chloroquine-resistant P. falciparum hasbeen detected. Trans R Soc Trop Med Hyg. 1985;79:482–483. doi: 10.1016/0035-9203(85)90071-9. [DOI] [PubMed] [Google Scholar]
- Edrissian GH, Shahabi S, Pishva E, Hajseyed-Javadi J, Khaleghian B, Ghorbani M, Emadi AM, Afshar A, Saghari H. Imported cases of chloroquine-resistant falciparum malaria in Iran. Bull Soc Path Exot. 1986;79:217–221. [PubMed] [Google Scholar]
- Raeisi A, Ringwald P, Safa O, Shahbazi A, Ranjbar M, Keshavarze H, Nateghpour M, Faraji L. Monitoring of the therapeutic efficacy of chloroquine for the treatment of uncomplicated, Plasmodium falciparum malaria in Iran. Ann Trop Med Parasitol. 2006;100:11–16. doi: 10.1179/136485906X86220. [DOI] [PubMed] [Google Scholar]
- Edrissian GH, Afshar A, Sayedzadeh A, Mohsseni GH, Satvat MT. Assessment of the response in vivo and invitro of Plasmodium falciparum to sulphadoxine-pyrimethamine in the malarious areas of Iran. J Trop Med Hyg. 1993;96:237–240. [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. A molecular marker for chloroquine- resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquineresistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Steketee RW, Plowe CV. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet. 2001;358:890–891. doi: 10.1016/S0140-6736(01)06040-8. [DOI] [PubMed] [Google Scholar]
- Sanchez C, Lanzer M. Changing ideas on chloroquine in Plasmodium falciparum. Curr Opin Infect Dis. 2000;13:653–658. doi: 10.1097/00001432-200012000-00013. [DOI] [PubMed] [Google Scholar]
- Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confersresistance to pyrimethamine in falciparum malaria. Proc Nat Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lee CS, Bayoumi R, Djimde A, Doumbo O, Swedberg G, Das LD, Mshinda H, Tanner M, Watkins WM, Sims PFG, Hyde JE. Resistance to antifolate in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origin. Mol Biochem Parasitol. 1997;89:161–177. doi: 10.1016/S0166-6851(97)00114-X. [DOI] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Peterson DS, Milhous WK, Wellems TE. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil I, Ronn AM, Alifrangis M, Gabar HA, Satti GM, Bygbjerg IC. Dihydrofolate reductase and dihydropteroate synthase genotypes associated with in vitro resistance of Plasmodium falciparum to pyrimethamine, trimethoprim, sulfadoxine and sulfamethoxazole. Am J Trop Med Hyg. 2003;68:586–589. doi: 10.4269/ajtmh.2003.68.586. [DOI] [PubMed] [Google Scholar]
- Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- Triglia TJ, Menting JG, Wilson C, Cowman AF. Mutations in dihydropteroate synthetase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talisuna AO, Nalunkuma-Kazibwe A, Langi P, Mutabingwa TK, Watkins WW, Marck EV, Egwang TG, D'Alessandro U. Two mutations in dihydrofolate reductase combined with one in the dihydropteroate synthase gene predict sulphadoxine-pyrimethamine parasitological failure in Ugandan children with uncomplicated falciparum malaria. Infect Genet Evol. 2004;4:321–327. doi: 10.1016/j.meegid.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Talisuna AO, Langi P, Mutabingwa TK, Watkins W, Van Marck E, Egwang TG, D'Alessandro U. Population-based validation of dihydrofolate reductase gene mutations for the prediction of sulfadoxine-pyrimethamine resistance in Uganda. Trans R Soc Trop Med Hyg. 2003;97:338–342. doi: 10.1016/S0035-9203(03)90163-5. [DOI] [PubMed] [Google Scholar]
- Nzila AM, Mberu EK, Sulo J. Towards an understanding of the mechanism of pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: genotyping of dihydrofolate reductase and dihydropteroate synthase of Kenyan parasites. Antimicrob Agents Chemother. 2000;44:991–996. doi: 10.1128/AAC.44.4.991-996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzila AM, Nduati E, Mberu EK, Hopkins SC, Monks SA, Winstanley PA, Watkins WM. Molecular evidence of greater selective pressure for drug resistance exerted by the long acting antifolatepyrimethamine/sulfadoxine compared with the shorter-actingchlorproguanil/dapsone on Kenyan Plasmodium falciparum. J Infect Dis. 2000;181:2023–2028. doi: 10.1086/315520. [DOI] [PubMed] [Google Scholar]
- Mutabingwa T, Nzila A, Mberu E. Chlorproguanil-dapsone for treatment of drug-resistant falciparum malaria in Tanzania. Lancet. 2001;358:1218–1223. doi: 10.1016/S0140-6736(01)06344-9. [DOI] [PubMed] [Google Scholar]
- Basco LK, Tahar R, Ringwald P. Molecular basis of in vivo resistance to sulfadoxine-pyrimethamine in Africa adult patients infected with Plasmodium falciparum malaria parasites. Antimicrob Agents Chemother. 1998;42:1811–1814. doi: 10.1128/aac.42.7.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe CV. Monitoring antimalarial drug resistance: making the most of the tools at hand. J Exp Biol. 2003;206:3745–3752. doi: 10.1242/jeb.00658. [DOI] [PubMed] [Google Scholar]
- Djimde A, Dolo A, Ouattara A, Diakite S, Plowe CV, Doumbo OK. Molecular diagnosis of resistance to antimalarial drugs during epidemics and in war zones. J Infect Dis. 2004;190:853–855. doi: 10.1086/422758. [DOI] [PubMed] [Google Scholar]
- Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-B. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Curtis J, Warhust DC. Plasmodium falciparum detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp Parasitol. 1998;89:1–8. doi: 10.1006/expr.1998.4274. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the antimalarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/S0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- Edrissian GH, Shahabi S. Preliminary study of the response of Plasmodium falciparum to chloroquine in Sistan and Baluchistan province of Iran. Trans R Soc Trop Med Hyg. 1985;79:563–564. doi: 10.1016/0035-9203(85)90101-4. [DOI] [PubMed] [Google Scholar]
- Heidari A, Dittrich S, Jelink T, Kheirandish A, Banihashemi K, Keshavarz H. Genotypes and in vivo resistance of Plasmodium falciparum isolates in an endemic region of Iran. Parasitol Res. 2007;100:589–592. doi: 10.1007/s00436-006-0291-z. [DOI] [PubMed] [Google Scholar]
- Peterson DS, Di SS, Povoa M, Calvosa VS, Do RV, Wellems TE. Prevalence of the dihydrofolate reductase Asn-108 mutation as the basis for pyrimethamine-resistant falciparum malaria in the Brazilian Amazon. Am J Trop Med Hyg. 1991;45:492–497. doi: 10.4269/ajtmh.1991.45.492. [DOI] [PubMed] [Google Scholar]
- Gasey GJ, Ginny M, Uranoli M, Mueller I, Reeder JC, Genton B, Cowman AF. Molecular analysis of Plasmodium falciparum from drug treatment failure patients in Papua New Guinea. Am J Trop Med Hyg. 2004;70:251–255. [PubMed] [Google Scholar]
- Zakeri S, Gil JP, Bereckzy S, Djadid ND, Bjorkman A. High prevalence of double Plasmodium falciparum dhfr mutations at codons 108 and 59 in the Sistan-Baluchistan province, Iran. J Infect Dis. 2003;187:1828–1829. doi: 10.1086/375250. [DOI] [PubMed] [Google Scholar]
- Ahmed A, Bararia D, Vinayak S, Yameen M, Biswas S, Dev V, Kumar A, Ansari MA, Sharma YD. Plasmodium falciparum isolates in India exhibit a progressive increase in mutations associated with Sulfadoxine-Pyrimethamine resistance. Antimicrob Agents Chemother. 2004;48:879–889. doi: 10.1128/AAC.48.3.879-889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Escalante A, Chaiyaroj S, Angkasekwinai P, Lal AA. Prevalence of point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum isolates from India and Thailand: a molecular epidemiologic study. Trop Med Int Health. 2000;5:737–743. doi: 10.1046/j.1365-3156.2000.00632.x. [DOI] [PubMed] [Google Scholar]
- Hapuarachchi HC, Dayanath MYD, Bandara KBAT, Abeysundara S, Abeyewickreme W, De Silva NR, Hunt SY, Sibley CH. Point mutations in the dihydrofolate reductase and dihydropteroate synthase genes of Plasmodium falciparum and resistance to sulfadoxine-pyrimethamine in Sri Lanka. Am J Trop Med Hyg. 2006;74:198–204. [PubMed] [Google Scholar]
- Mita T, Kaneko A, Hwaihwanje I, Tsukahara T, Takahashi N, Osawa H, Tanabe K, Kobayakawa T, Bjorkman A. Rapid selection of dhfr mutant allele in Plasmodium falciparum isolates after the introduction of sulfadoxine/pyrimethamine in combination with 4-aminoquinolines in Papua New Guinea. Infect Genet Evol. 2006;6:447–452. doi: 10.1016/j.meegid.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Wang P, Read M, Sims PFG, Hyde JE. Sulfadoxine resistancein the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979–86. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- Cox-Singh J, Zakaria R, Abdullah MS, Rahman HA, Nagappan S, Singh B. short report: differences in dihydrofolate reductase but not dihydropteroate synthetase alleles in Plasmodium falciparum isolates from geographically distinct areas in Malaysia. Am J Trop Med Hyg. 2001;64:28–31. doi: 10.4269/ajtmh.2001.64.1.11425158. [DOI] [PubMed] [Google Scholar]
- Aubouy A, Jafari S, Huart V, Migot-Nabias F, Mayombo J, Durand R, Bakary M, Bras JL, Deloron P. DHFR and DHPSgenotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine-pyrimethamine treatment efficacy. J Antimicrob Chemother. 2003;52:43–49. doi: 10.1093/jac/dkg294. [DOI] [PubMed] [Google Scholar]
- Vasconcelos KF, Plowe CV, Fontes CJ, Kyle D, Wirth DF, Pereira daSilva LH, Zalis MG. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthetase of isolates from the Amazon region of Brazil. Mem Inst Oswaldo Cruz. 2000;95:721–728. doi: 10.1590/S0074-02762000000500020. [DOI] [PubMed] [Google Scholar]
- Keen J, Farcas GA, Zhong K, Yohanna S, Dunne MW, Kain KC. Real-Time PCR assay for rapid detection and analysis of PfCRT haplotypes of chloroquine-resistant Plasmodium falciparum isolates from India. J Clin Microbiol. 2007;45:2889–2893. doi: 10.1128/JCM.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vathsala PG, Pramanik A, Dhanasekaran S, Devi CU, Pillai CR, Subbarao SK, Ghosh SK, Tiwari SN, Sathyanarayan TS, Deshpande PR, Mishra GC, Ranjit MR, Dash AP, Rangarajan PN, Padmanaban G. Widespread occurrence of the Plasmodium falciparum chloroquine resistance transporter(PFCRT) gene haplotype SVMNT in P. falciparum malaria in India. Am J Trop Med Hyg. 2004;70:256–259. [PubMed] [Google Scholar]