Abstract
RUNX1/AML1 regulates lineage-specific genes during hematopoiesis and stimulates G1 cell-cycle progression. Within RUNX1, S48, S303, and S424 fit the cyclin-dependent kinase (cdk) phosphorylation consensus, (S/T)PX(R/K). Phosphorylation of RUNX1 by cdks on serine 303 was shown to mediate destabilization of RUNX1 in G2/M. We now use an in vitro kinase assay, phosphopeptide-specific antiserum, and the cdk inhibitor roscovitine to demonstrate that S48 and S424 are also phosphorylated by cdk1 or cdk6 in hematopoietic cells. S48 phosphorylation of RUNX1 paralleled total RUNX1 levels during cell-cycle progression, S303 was more effectively phosphorylated in G2/M, and S424 in G1. Single, double, and triple mutation of the cdk sites to the partially phosphomimetic aspartic acid mildly reduced DNA affinity while progressively increasing transactivation of a model reporter. Mutation to alanine increased DNA affinity, suggesting that in other gene or cellular contexts phosphorylation of RUNX1 by cdks may reduce transactivation. The tripleD RUNX1 mutant rescued Ba/F3 cells from inhibition of proliferation by CBFβ-SMMHC more effectively than the tripleA mutant. Together these findings indicate that cdk phosphorylation of RUNX1 potentially couples stem/progenitor proliferation and lineage progression.
Introduction
The core binding factor (CBF) transcription factor family consists of heterodimers forming between 1 of 3 CBFα subunits, RUNX1/AML1, RUNX2, or RUNX3 and CBFβ.1 RUNX1 contacts CBFβ and binds DNA via its Runt domain.2,3 CBF activities are commonly reduced in acute myeloid leukemia cases due to expression of AML1-ETO or CBFβ-SMMHC or to deletion or point mutation of the RUNX1 gene, and TEL-AML1 dominantly inhibits CBF in a subset of pediatric acute lymphoblastic leukemias.1 RUNX1 regulates genes specific to the lymphoid, myeloid, and megakaryocyte lineages,4–7 and mice lacking RUNX1 do not develop definitive hematopoiesis, indicating a role in adult hematopoietic stem cell (HSC) formation.8,9 In contrast, deletion of the RUNX1 gene in adult mice leads to increased HSCs, a reduction in common lymphoid progenitor numbers and in platelet formation, and increased myelopoiesis.10
In addition to regulating lineage-specific genes and cytokine receptors,1 RUNX1 directly stimulates G1 to S cell-cycle progression. CBFβ-SMMHC or AML1-ETO dominantly inhibit RUNX1 and slow G1 progression in hematopoietic cell lines or in murine or human marrow progenitors,11–14 cdk4, cyclin D2, or c-Myc overcome inhibition of proliferation by these CBF oncoproteins,13,15,16 exogenous RUNX1 stimulates G1 progression,14,15,17 and stimulation of G1 via deletion of p16INK4a or expression of E7 cooperates with CBFβ-SMMHC or TEL-AML1 to induce acute leukemia.18,19 Induction of cdk4 or cyclin D3 transcription may underlie stimulation of G1 progression by RUNX1.15,20
Not only does RUNX1 regulate cell-cycle progression, but in addition RUNX1 levels increase as hematopoietic cells progress from G1 to S and from S to G2/M.20 Phosphorylation of S303 by cdks leads to RUNX1 degradation during G2/M.21 We now provide additional evidence that cdks phosphorylate S303 and demonstrate that S48 and S424 are also substrates of cdk1/cyclin B and cdk6/cyclin D3. Moreover, phosphorylation of S48, S303, and S424 strengthens the ability of RUNX1 to activate transcription and to stimulate hematopoietic cell proliferation.
Methods
Plasmids
Human RUNX1 cDNA segments were subcloned downstream of glutathionine S-transferase (GST) in pGEX-4T-1 (Amersham, Piscataway, NJ). GST-RUNX1(28-86), containing RUNX1 amino acids 28-86, was generated using an XbaI/SmaI RUNX1 cDNA fragment. GST-RUNX1(28-290), GST-RUNX1(290-480), GST-RUNX1(86-217), GST-RUNX1(320-480), and GST-RUNX1(410-480) were generated by ligation of XbaI/BamHI, BamHI/EcoRI, SmaI, SalI/EcoRI, or NcoI/EcoRI fragments into the pGEX-4T-1 vector. Mutations S424A, T41A, S48A, and T41A/S48A were introduced into these plasmids by polymerase chain reaction (PCR)–based site-directed mutagenesis. RUNX1 in the context of CMV-Myc-RUNX1 was also subjected to site-directed mutagenesis to generate S48A, S303A, S424A, S48D, S303D, and S424D. All mutants were confirmed by DNA sequencing. DNA subcloning allowed the generation of mutant combinations in CMV-Myc-RUNX1, and the S48A/S303A/S424A (tripleA) or S48D/S303D/S424D (tripleD) mutants were transferred to the pBabePuro-RUNX1-ER retroviral vector or to pMTCB6 downstream of the zinc-responsive metallothionein (MT) promoter. CMV-Flag-p300 was kindly provided by P. Cole (Johns Hopkins University, Baltimore, MD).
Bacterial expression and in vitro kinase assay
pGEX-4T-1 plasmids were transformed into BL21(DE3) bacteria (Novagen, La Jolla, CA) and grown to log phase. Isopropyl-β-d-thiogalactopyranoside was then added to 0.25 mM for 4 hours. Bacteria were collected, frozen overnight, thawed, and resuspended at 5 mL per gram in 50 mM Tris, pH 7.5, 50 mM NaCl, 5% (wt/vol) glycerol, containing 10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 μg/mL pepstatin A, 0.1 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF). Lysonase (3 μL; Novagen), containing lysozyme and DNaseI, was added per milliliter, followed by incubation at 4°C for 3 hours. After centrifugation at 30 000g for 30 minutes, supernatant was collected and 1 mL was added per 0.2 mL washed GST-Bind Resin (Novagen). After incubation at 4°C for 3 hours, the resin was washed 3 times with phosphate-buffered saline (PBS), and GST proteins were eluted with 10 mM glutathione, 50 mM Tris, pH 8.0. GST-retinoblastoma protein (GST-Rb) was obtained commercially (Santa Cruz Biotechnology, Santa Cruz, CA). Kinase reactions were carried out at 30°C for 30 minutes using cdk1/cyclinB or cdk6/cyclin D3 complexes (Upstate Biotechnology, Lake Placid, NY) in 8 mM MOPS, pH 7.0, 0.2 mM EDTA, using either 10 μCi (0.37 MBq) γ-ATP (Amersham) or 10 mM ATP. Reactions with cdk1 also contained 14 mM MgCl2, 1 mM EGTA, and reactions with cdk6 also contained 10 mM magnesium acetate. Reactions were stopped by addition of Laemmli sample buffer. Radioactive samples were subject to acrylamide gel electrophoresis, followed by gel drying and autoradiography.
Cell culture and gel shift assay
293T cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% heat-inactivated fetal bovine serum (HI-FBS), and Ba/F3 cells were cultured in RPMI 1640 with 10% HI-FBS and 1 ng/mL murine IL-3 (Peprotech, Rocky Hill, NJ). 293T cells were transiently transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). For expression and coimmunoprecipitation studies, 3 μg plasmid DNA was used per 100-mm dish. Cycloheximide (50 μg/mL) was added to 293T cells in 60-mm dishes transfected 24 hours earlier with 1 μg plasmid DNA, and samples were collected 0, 4, 8, or 16 hours later. Roscovitine (100 μM; Calbiochem, San Diego, CA) was added to 293T cells 24 hours after transfection, and samples were collected 16 hours later. For retroviral packaging, 2 μg pBabePuro vectors were cotransfected with 8 μg pkat2ecopac.22 Supernatants collected 2 and 3 days later were adsorbed to retronectin-coated dishes (Takara, Shiga, Japan), after which Ba/F3 cells expressing CBFβ-SMMHC from the MT promoter11 were added with 4 μg/mL polybrene for 2 days. Pools of transductants were subsequently obtained by culture in 2 μg/mL puromycin. Viable cell numbers were enumerated using a hemocytometer and trypan blue dye, after culture in the presence of 200 nM 4-hydroxytamoxifen (4HT), with or without 100 μM zinc chloride (Zn). For reporter studies, 293T cells were transfected in 6-well dishes with 750 ng (CBF)4TKLUC, 50 ng CMV-Myc-RUNX1 plasmids, and 5 ng CMV-βGal. Luciferase and β-galactosidase activities were assessed in cell extracts obtained 2 days later.23 MT-RUNX1 plasmids were linearized with ScaI and introduced into Ba/F3 cells by electroporation, and subclones were isolated by limiting dilution.11 Ba/F3 cells expressing wild-type RUNX1 from the MT promoter were incubated overnight in zinc chloride prior to cell-cycle fractionation via elutriation.20 Cell-cycle distributions were obtained by staining with 11 μg/mL Hoechst 33258 dye in 0.7% NP-40, 4.7% formaldehyde followed by fluorescence-activated cell sorting (FACS) analysis. Nuclear extracts were prepared from transfected 293T cells and subjected to gel shift assay using a CBF site from the myeloperoxidase (MPO) promoter as described.5
Western blotting and coimmunoprecipitation
Total cell extracts were prepared by washing cell pellets with PBS followed by resuspension in Laemmli sample buffer. Western blotting was carried as described,11 using GST-Tag monoclonal antibody (Novagen), 9E10 c-Myc tag monoclonal antibody or rabbit ERα MC-20 antiserum (Santa Cruz Biotechnology), RUNX1 monoclonal antibody or CBFβ rabbit antiserum (kindly provided by N. Speck), or rabbit anti-RUNX1 phospho-S48, phospho-S303, or phospho-S424 antisera (Proteintech Group, Chicago, IL). The peptides used to raise phospho-specific antisera were PPSTALS(phos)PGKMSEA for S48P, HPATPIS(phos)PGRASGM for S303P, and FSMVGGRS(phos)PPRILP for S424P. Band intensities were quantified using NIH ImageJ (National Institutes of Health, Bethesda, MD). For coimmunoprecipitation, 293T cells were washed with 150 mM NaCl, Tris, pH 7.5, and cell lysates were then prepared by incubation at 4°C for 20 minutes at a ratio of 1 mL per 10 × 106 cells with 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.2% NP-40, 0.2% Triton X-100, 0.2% deoxycholate, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 μg/mL pepstatin A, 0.1 mM NaF, 0.1 mM sodium orthovanadate, and 1 mM PMSF, followed by freeze-thaw using dry ice/ethanol. Aliquots were saved as “input,” and the supernatant was precleared using 50 μL of 50% protein A/G-Sepharose. The supernatants were then split and incubated with 2 μg rabbit IgG or rabbit anti-Flag antiserum F7425 (Sigma, St Louis, MO) for 3 hours at 4°C, followed by addition of 50 μL protein A/G–Sepharose for 1 hour. The beads were then washed 3 times with lysis buffer, and the samples were eluted in Laemmli sample buffer at 95°C and subjected to Western blotting.
Results
Cdk1 and cdk6 phosphorylate RUNX1 S48 and S424 in vitro
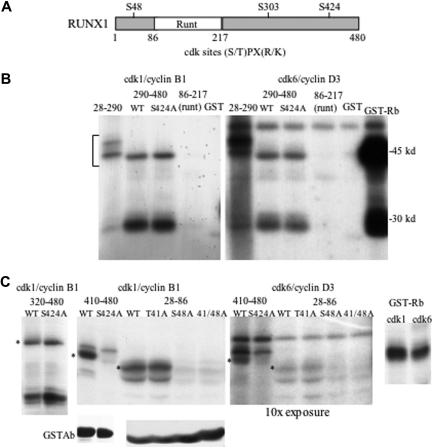
RUNX1 contains 3 cdk phosphorylation consensus sites, (S/T)PX(R/K) (Figure 1A).24 In addition, RUNX1 has multiple (S/T)P sites also potentially phosphorylated by cdks or by ERK. S303 was previously shown to be a cdk substrate and to regulate RUNX1 stability.21 To identify additional sites phosphorylated by cdks, we linked GST to RUNX1 segments 28-290, 290-480, or 86-217. Cdk1/cyclin B1 or cdk6/cyclin D3 modified GST-RUNX1(28-290) and GST-RUNX1(290-480), but not GST(86-217), or GST alone (Figure 1B). Reaction of GST-Rb, containing Rb residues 769-921, with cdk6 was assessed also as positive control. Mutation of S424 to alanine (S424A) within GST-RUNX1(290-480) did not prevent its phosphorylation. Cdk1 or cdk6 also modified RUNX1(320-480) or RUNX1(320-480, S424A) (Figure 1C left panel). On the other hand, whereas GST-RUNX1(410-480) was phosphorylated by cdk1 or cdk6, GST-RUNX1(410-480, S424A) was not (Figure 1C second and third panels, lanes 1-2). GST-RUNX1(410-480) and GST-RUNX1(410-480, S424A) were expressed similarly in the bacterial lysates, as detected by GST antibody (Figure 1C). These results, representative of 2 independent assessments, indicate that S424 is a direct substrate for cdk1 and cdk6 and also suggest that at least 1 of the 3 (S/T)P sites located between residues 320 and 410 is also modified by cdks in vitro. We focused our further efforts on S48, S303, and S424, as they match the more stringent cdk consensus.
Figure 1.
Cdk1 and cdk6 phosphorylate RUNX1 on S48 and S424 in vitro. (A) Diagram of RUNX1 showing its 3 cdk consensus phosphorylation sites. (B) In vitro kinase assay in which cdk1/cyclin B1 (left panel) or cdk6/cyclin D3 (right panel) were incubated with γ-ATP and GST-RUNX1 fusion proteins containing the RUNX1 amino acid segments 28-290, 290-480, and 290-480 with the S424A mutation, or the 86-217 Runt domain, GST alone, or GST-Rb protein (2 μg), followed by polyacrylamide gel electrophoresis, drying, and autoradiography. (C) In vitro kinase assays (top panels) using the indicated cdk/cyclin complexes and GST-RUNX1 fusion proteins 320-480, 320-480/S424A, 410-480, 410-480/S424A, 27-86, 27-86/T41A, 27-86/S48A, 27-86/T41A/S48A, or GST-Rb. Results are representative of 2 independent assessments for GST-RUNX1(410-480) and its S424A variant and 4 assessments for GST-RUNX1(28-86) and its 3 variants, with the data shown for the right 3 panels from the same experiment run on the same gel (Rb lanes are juxtaposed). Asterisks (*) indicate locations of relevant phosphorylated fragments. Equal volumes of bacterial lysates used for this experiment were also subjected to Western blotting using GST antibody (bottom panels).
Cdk1 or cdk6 phosphorylated either GST-RUNX1(28-86) or its T41A variant, whereas mutation of S48 to alanine (S48A) alone or with T41A prevented its modification (Figure 1C second and third panels, lanes 3-6). GST-RUNX1(28-86) and its 3 mutant variants were expressed similarly, as detected by GST antibody (Figure 1C). These results, representative of 4 independent assessments, indicate that S48 is also a direct in vitro substrate of cdk1 or cdk6.
Cdk1/cyclin B1 was approximately 2-fold more active against GST-Rb than cdk6/cyclin D3, under our reaction conditions (Figure 1C right panel). Cdk1 phosphorylated RUNX1 S48 or S424 with approximately 20-fold greater potency than cdk6, based on comparing the band intensities in the center panels, which representdifferent autoradiography exposures of the same gel. Thus, cdk1 has approximately a 10-fold selectivity compared with cdk6 toward S48 and S424, versus their relative ability to phosphorylate GST-Rb.
S48, S303, and S424 are modified in mammalian cells
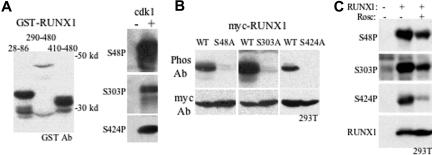
Phospho-specific antisera were raised against RUNX1 S48, S303, and S424 phosphopeptides. To verify the specificity of these reagents, GST-RUNX1(28-86), GST-RUNX1(290-480), and GST-RUNX1(410-480) were expressed in bacteria and isolated using glutathione resin (Figure 2A left panel). The lower band in each lane likely represents GST released by proteolysis. Each of these GST-RUNX1 proteins was incubated with cdk1, either in the absence or presence of unlabeled ATP. GST-RUNX1(28-86) was subjected to Western blotting with anti–phospho-S48 (Figure 2A right, top panel); GST-RUNX1(290-480), with anti–phospho-S303 (Figure 2A right, middle panel); and GST-RUNX1(410-480), with anti–phospho-S424 (Figure 2A right, bottom panel). The phosphopeptide antisera were highly specific, reacting only with the substrates modified by cdk1.
Figure 2.
RUNX1 S48, S303, and S424 phospho-specific antisera detect RUNX1 phosphorylation in mammalian cells. (A) Expression of GST-RUNX1(28-86), GST-RUNX1(290-480), and GST-RUNX1(410-480) in bacterial lysates was assessed by Western blotting with GST antibody (left panel). These proteins were incubated with cdk1 in the absence (−) or presence (+) or nonradioactive ATP and again subjected to Western blotting: GST-RUNX1(28-86), with anti–phospho-S48 antiserum (top, right panel); GST-RUNX1(290-480), with anti–phospho-S303 antiserum (center right panel), and GST-RUNX1(410-480), with anti–phospho-S424 antiserum (bottom right panel). (B) 293T cells were transiently transfected with CMV-Myc-RUNX1 (WT) or its S48A, S303A, or S424A variants. Total cell lysates prepared 2 days later were subjected to Western blotting with corresponding phospho-specific antisera (top panels) or c-Myc antibody (bottom panels). (C) Western blotting with phospho-specific or total RUNX1 antisera of extracts obtained from 293T cells transfected with CMV vector (lane 1) or CMV-RUNX1 (lanes 2-3) and treated 24 hours later with 100 μM roscovitine (lane 3), followed by culture for an additional 16 hours.
Serines 48, 303, or 424 were mutated to alanine in the context of N-terminally Myc-tagged RUNX1. These proteins were expressed transiently from the CMV promoter in 293T cells and then subjected to Western blotting with the corresponding phosphopeptide antiserum and with c-Myc antibody (Figure 2B). The results indicate that S48, S303, and S424 are each phosphorylated in this cell line, as seen previously for S303.21
To demonstrate that RUNX1 is modified by cyclin-dependent kinases on these 3 serines, 293T cells were transfected with either CMV or CMV-RUNX1, and CMV-RUNX1–transfected cells were further cultured without or with roscovitine, a pan-cdk inhibitor (Figure 2C). Decreased phosphorylation of S48, S303, and S424 was observed, with total RUNX1 levels remaining constant. Similar findings had been observed using another S303P-specific antiserum.21
Cdk phosphorylation of RUNX1 increases transactivation
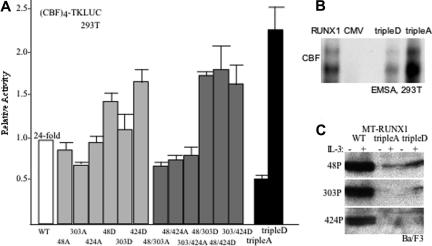
Serines 48, 303, or 424 in Myc-RUNX1 were mutated to alanine or to the phosphomimetic residue aspartic acid. In addition, the 3 combinations of double alanine or double aspartic acid mutations were introduced into RUNX1 as were the triple alanine (tripleA) and triple aspartic acid (tripleD) combinations. Each of these proteins was expressed in 293T cells along with the (CBF)4TKLUC reporter, containing 4 consensus CBF sites upstream of the thymidine kinase promoter and luciferase cDNA. Wild-type (WT) RUNX1 activated this reporter on average 24-fold. The S303A variant was approximately 30% less active and the S48D or S424D variants approximately 50% more active. More strikingly, each of the double alanine mutants was approximately 50% less active than WT and the double aspartic acid mutants 50% more active. The tripleA variant had approximately 40% of WT activity, whereas the tripleD variant was 2.2-fold more active (Figure 3A). These results indicate that phosphorylation of S48, S303, or S424 by cdks contributes to transactivation, with RUNX1(tripleD) having 5-fold greater potency than RUNX1(tripleA).
Figure 3.
Cdk phosphorylation of RUNX1 increases transactivation potency. (A) 293T cells were transiently transfected with (CBF)4TKLUC and either CMV, CMV-RUNX1 (WT), or the indicated RUNX1 mutant variants expressed from the CMV promoter. CMV-βGal was also included in each transfection as an internal control. TripleA and tripleD represent S48/303/424A or S48/303/424D, respectively. Induction by RUNX1 compared with empty CMV vector was set to 1 in each experiment. Relative activation by each mutant compared with RUNX1 is shown (mean and SE from 4 experiments). (B) Nuclear extracts from 293T cells transiently transfected on 100-mM dishes with 3 μg CMV-CBFβ together with 3 μg of either CMV-RUNX1, CMV, CMV-RUNX1(tripleD), or CMV-RUNX1(tripleA) were subjected to gel shift assay using a radiolabeled CBF-binding site from the MPO promoter. Input of RUNX1 or mutant RUNX1 proteins was normalized based on Western blot analysis. Data shown are representative of 2 assessments. (C) Ba/F3 cells stably transduced with MT-RUNX1, MT-RUNX1(tripleA), or MT-RUNX1(tripleD) were cultured for 24 hours with or without IL-3 and with zinc chloride. Total cell extracts corresponding to 106 cells were then subjected to Western blotting with anti-S48, anti-S303, or anti-S424 phospho-specific antiserum.
To assess the affect of cdk modification on RUNX1 DNA binding, nuclear extracts prepared from 293T cells transfected with CMV-CBFβ together with either CMV, CMV-RUNX1, CMV-RUNX1(tripleA), or CMV-RUNX1(tripleD) were subjected to gel shift assay using a consensus CBF site from the MPO promoter (Figure 3B). Western analysis of the extracts was used to normalize input of RUNX1 or its variants into this assay. The tripleD variant bound DNA with slightly reduced affinity compared with wild type, whereas the tripleA variant bound DNA with 2- to 3-fold greater affinity. The doublet gel shift species detected is similar to that obtained previously and may represent alternative phosphorylation at non-cdk substrate sites or use of alternative ATG codons to initiate translation.5 These gel shift findings indicate that changes in DNA affinity do not account for the increased transactivation potency seen with RUNX1(tripleD) and the reduced potency obtained with RUNX1(tripleA).
Aspartic acid, although negatively charged, does not perfectly mimic phosphorylation, leading to underestimation of the biologic effect of S48, S303, and S424 modification. To illustrate this limitation, extracts from Ba/F3 cells expressing RUNX1, RUNX1(tripleA), or RUNX1(tripleD) were compared for their reactivity with the 3 phospho-specific antisera (Figure 3C). RUNX1(tripleD) reacted more potently than RUNX1(tripleA), but far less strongly than wild-type RUNX1 with anti–phospho-S48 or anti–phospho-S303, and neither of the mutants reacted with anti–phospho-S424. These findings indicate that the phospho-specific antibodies bind phosphorylated S48, S303, or S424 far more effectively than S48D, S303D, or S424D. Culture in the absence of IL-3 overnight did not alter phosphorylation of RUNX1 on these serine residues. It is not clear why S303D was detected by the S303 phospho-antiserum in IL-3 but not after culture in its absence.
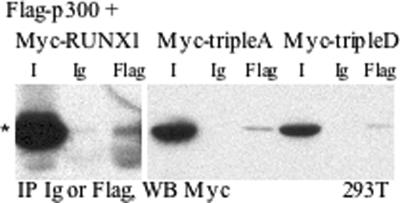
The p300 coactivator interacts with the C-terminal half of RUNX1, including segments containing S303 and S424.25 Myc-RUNX1 or its tripleA or tripleD variants were evaluated for interaction with Flag-p300 by coimmunoprecipitation, after transient expression in 293T cells (Figure 4). Wild-type RUNX1 interacted with p300, mutation of S48, S303, and S424 to alanine did not prevent this interaction, and mutation to aspartic acid residues did not strengthen the interaction.
Figure 4.
Cdk phosphorylation of RUNX1 does not increase its interaction with the p300 coactivator. Cell lysates prepared from 293T cells transiently transfected with Flag-p300 and Myc-RUNX1, Myc-RUNX1(tripleA), or Myc-RUNX1(tripleD) were subjected to immunoprecipitation (IP) with rabbit Ig or rabbit anti-Flag followed by Western blotting (WB) with c-Myc tag monoclonal antibody. I indicates input and represents 5% of lysate used for immunoprecipitation. * indicates location of specific bands.
S48, S303, and S424 are modified in hematopoietic cells and vary during the cell cycle
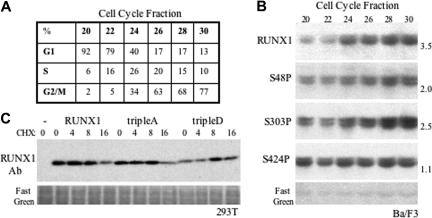
To assess phosphorylation of these 3 RUNX1 serine residues in a hematopoietic cell line and to assess their variation during the cell cycle, Myc-RUNX1 was expressed from the zinc-responsive MT promoter in Ba/F3 cells. These cells were cultured for 16 hours with zinc chloride and then subjected to elutriation to allow isolation of cell fractions enriched for G1 or G2/M phase cells (Figure 5A). Extracts corresponding to equal numbers of each fraction were subjected to Western blotting sequentially for RUNX1, phospho-S48, phospho-S303, or phospho-S424. S48, S303, and S424 were each phosphorylated in Ba/F3 cells. RUNX1 levels increased as the cells progressed through the cell cycle, as seen previously.20 Phosphorylation of S48 paralleled RUNX1 levels, modification of S303 was modestly more prominent in G2/M than S48P, and phosphorylation of S424 was fairly even in the various fractions and so markedly diminished as a percentage of total RUNX1 as the cell cycle progressed from G1 to G2/M (Figure 5B). As a further test of protein stability, 293T cells were transfected with either CMV, CMV-RUNX1, CMV-RUNX1(tripleA), or CMV-RUNX1(tripleD) followed by addition of cycloheximide to prevent new protein synthesis. Total cell extracts isolated 0, 4, 8, or 16 hours later were subjected to Western blotting for RUNX1 (Figure 5C). Endogenous RUNX1 was not detected in CMV-transfected cells, as was also apparent in Figure 2C lane 1. At the time of CHX addition, the steady-state level of RUNX1(tripleD) was lower than RUNX1 or RUNX1(tripleA), suggesting diminished stability in cycling cells, whereas addition of CHX allowed a similar rate of RUNX1 or RUNX1(tripleA) degradation with levels of RUNX1(tripleD) remaining unchanged.
Figure 5.
RUNX1 S48, S303, and S424 phosphorylation and cell-cycle variation in a hematopoietic cell line. (A) Ba/F3 cells expressing RUNX1 from the MT promoter were cultured in zinc chloride for 16 hours and then subjected to density fractionation by elutriation. An aliquot of each fraction was stained with Hoechst 33258 dye followed by FACS analysis to allow estimation of the percentage of cells in G1, S, or G2/M, as shown. (B) Total cell extracts corresponding to 2 × 106 cells were subjected to Western blotting. The blot was stained sequentially with anti–phospho-S48, anti–phospho-S303, anti–phospho-S424 antisera, and anti-RUNX1 antibody and was also stained with Fast Green to confirm equivalent loading. Relative expression in fraction 30 compared with the mean of fractions 20 and 22 is shown to the right of each panel. (C) Cycloheximide (CHX) was added to 293T cells 24 hours after transfection with CMV, CMV-RUNX1, CMV-RUNX1(tripleA), or CMV-RUNX1(tripleD), and total cell proteins prepared 0, 4, 8, or 16 hours after CHX addition were subjected to Western blotting using anti-RUNX1 antiserum. The blot was also stained with Fast Green.
Cdk phosphorylation of RUNX1 increases stimulation of proliferation
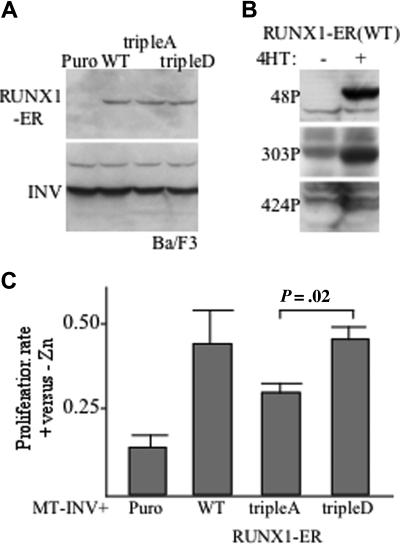
Exogenous RUNX1 only minimally stimulates proliferation of hematopoietic cell lines, whereas inhibition of RUNX1 markedly slows their growth. To assay cell-cycle stimulation by RUNX1 or its mutants, we previously used an assay in which these proteins were linked to the estrogen receptor (ER) ligand-binding domain and then coexpressed with CBFβ-SMMHC, a dominant inhibitor of RUNX1.26 The tripleA or tripleD RUNX1 mutations were transferred into the pBabePuro-RUNX1-ER retroviral vector. These variants, RUNX1-ER, or empty pBabePuro was transduced into Ba/F3-MT-INV cells expressing CBFβ-SMMHC (designated as INV for brevity). Western blotting using equal cell numbers of pooled transductants confirmed similar levels of each ER fusion protein and of INV (Figure 6A). The nonspecific band in the lower blot above the INV band confirms equal loading. The ER segment linked to RUNX1 in these proteins responds to 4-HT but not estradiol. Ba/F3 cells expressing wild-type RUNX-ER were cultured with or without 4-HT for 1 day, and cell extracts were subjected to Western blotting with the 3 phospho-specific antisera (Figure 6B). As with RUNX1, RUNX1-ER is phosphorylated on S48, S303, and S424. The 4 cell populations were cultured with 4-HT and either with or without zinc chloride for 4 days, at which time cell counts were enumerated. The ratio of viable cells in the presence of zinc divided by the number obtained in the absence of zinc is shown for 3 experiments (Figure 6C). Induction of INV in cells transduced with the empty pBabePuro vector reduced proliferation 8-fold during this interval. Activation of wild-type RUNX1-ER partially rescued this inhibition, as did activation of the tripleA or tripleD variants. However, RUNX1(tripleD)-ER was significantly more effective than RUNX1(tripleA)-ER at rescuing inhibition of proliferation of Ba/F3 cells by INV.
Figure 6.
Cdk phosphorylation of RUNX1 increases stimulation of proliferation. (A) Ba/F3 cells expressing CBFβ-SMMHC (INV) were transduced with empty pBabePuro (Puro) or pBabePuro encoding RUNX1-ER (WT) or its tripleA or tripleD mutants. Total cell extracts were subjected to Western blotting using ER antiserum (top panel) or CBFβ antiserum (bottom panel) to detect INV. (B) Ba/F3 cells expressing INV and RUNX1-ER were cultured in the absence (−) or presence (+) or 4HT for 24 hours. Total cell extracts were then subjected to Western blotting with anti-S48, anti-S303, or anti-S424 phospho-specific antisera. (C) The 4 indicated Ba/F3 cells lines were seeded at 5 × 103 cells/mL and cultured with 4HT and with or without zinc chloride for 4 days. Viable cell counts were then enumerated, and the ratios (+Zn)/(−Zn) are shown (mean and SE from 3 determinations). P value is from paired Student t test.
Discussion
This study demonstrates that cdks phosphorylate RUNX1 on S48 and S424, in addition to the previously defined S303 cdk substrate, and that phosphorylation of S48, S303, and S424 progressively increases RUNX1 transactivation potency. RUNX1 regulates HSC formation during development and participates in maturation along several lineages in adult marrow. Activation by cdks potentially coordinates RUNX1 control of stem/progenitor differentiation and cell-cycle control.
RUNX1 has 14 (S/T)P motifs representing potential sites for both cdk and ERK modification, of which only S48, S303, and S424 match the more restricted (S/T)PX(R/K) cdk phosphorylation motif.24 S276 and S293 are kinased in vitro by ERK, and mutation of these serines obviates induction of RUNX1 transcriptional activity by coexpressed ERK.27 Mutation of S276 and S293 to alanine does not alter RUNX1 variation during the cell cycle.20 Of the 14 (S/T)P motifs in RUNX1, S41 and S48 are N-terminal to the RUNX1 domain, S94 is located in the DNA-binding domain, and the remaining 11 are more C-terminal. Exposure of K562 cells to phorbol ester, which activates protein kinase C and thus indirectly ERK, JNK, and p38 kinases, induces phosphorylation of RUNX1 on serines 276, 293, 300, 303, and 462, but not on S48 or S424.28 Phosphorylation of RUNX1 by ERK increases its transactivation potency by preventing interaction of S276 and S293 with the mSin3A corepressor.28,29
Cdk inhibitors markedly reduced phosphorylation in vivo of both S276 and S303 within a RUNX1 segment containing residues 267-315, and the wild-type fragment but not the S276/303A double mutant was modified by cdk1/cyclin B in vitro.21 We have extended these findings by using our S303P phospho-specific antiserum to demonstrate that cdk1 phosphorylates S303 within RUNX1(290-480) in vitro, further implicating this serine as a direct cdk1 target. In addition, we demonstrate direct, in vitro phosphorylation of S48 and S424 by cdk1 and cdk6 and have also detected their in vivo modification in a hematopoietic cell line and in 293T cells, as well as inhibition of modification by a cdk inhibitor, roscovitine. Of note, neither of these serines has been implicated as an ERK target. Within RUNX2, S451 lies in a region homologous to RUNX1 and corresponds to RUNX1 S424. Phosphorylation of S451 in RUNX2 has been observed in an osteosarcoma cell line, and cdk1 phosphorylates RUNX2 on S451 in vitro.30,31 RUNX2 also demonstrates prominent phosphorylation of S14 and S104,30 corresponding to S41 and S94 in RUNX1, but these serines were not modified by cdk1 or cdk6 within RUNX1 in our in vitro kinase assays. On the other hand, we identified S48 as a novel RUNX1 phosphorylation site in vivo and as a substrate for cdks in vitro. In sum, we find that all 3 serines that fit the (S/T)PX(R/K) consensus within RUNX1 are cdk substrates.
The quadruple RUNX1 mutant S276/293/300/303A has increased stability in mammalian cells, mutation of these 4 residues to aspartic acid leads to decreased stability, and phosphorylation of S303 is associated with RUNX1 degradation by the mitotic anaphase promoting complex.21 Consistent with these findings, we detected a modest preference for phosphorylation of S303 compared with S48 in G2/M, whereas S424 was modified to a much greater extent during G1. When expressed transiently in 293T cells, the tripleD mutant demonstrated a diminished steady-state level but a prolonged half-life in CHX. Perhaps this behavior reflects reduced stability in cycling cells due to increased G2/M degradation, as a consequence of the S303D mutation, but increased stability in cells arrested in G1 and S by CHX,32 as a result of the S48D/S424D alterations. The protein interactions mediating increased phosphorylation of RUNX1 S424P during the G1 cell-cycle phase and the increased stability of RUNX1(tripleD) in arrested cells will be sought in future studies.
In addition to the role of S303 phosphorylation in the regulation of RUNX1 stability, we now find that RUNX1 modification by cdks on S48, S303, and S424 increases transactivation potency. Moreover, the 5-fold increase observed in the tripleD variant likely underestimates the effect of phosphorylation, as aspartic acid is not a perfect phosphomimetic. Interaction with p300 does not account for the effect of cdk phosphorylation on transactivation, as the tripleA variant retained interaction with this coactivator. In addition, the tripleD mutations actually reduced DNA affinity mildly, and the tripleA mutations increased DNA affinity, further supporting the conclusion that in the context of 293T cells cdk modification of RUNX1 increases its intrinsic ability to activate transcription. RUNX1 residues 183-291 have the capacity to mask the Runt DNA-binding domain, but this autoinhibition is obviated in the presence of CBFβ.33 As we assessed DNA binding in the presence of coexpressed CBFβ, this mechanism does not account for the increased affinity of the tripleA mutation, which may reflect subtle alterations in the RUNX1 structure. In some cellular or gene contexts expressing a specific repertoire of coactivator, corepressors, or cooperating transcription factors, RUNX1(tripleD) may actively repress transcription more effectively than RUNX1(tripleA), and in that setting the effect of cdk modification on DNA affinity may amplify the negative effect on gene expression.
Cyclin D3 inhibits RUNX1 transactivation by competing for CBFβ interaction with the Runt domain.34 As cyclin D proteins are induced by cytokine signals, this inhibition may serve as a feedback to prevent excessive growth stimulation, as those authors suggested. On the other hand, our findings indicate that at least for a subset of stem/progenitors that continue to proliferate at a steady-state level, cdk6/cyclin D3 and to a greater extent cdk1/cyclin B modify RUNX1 to increase its ability to both retain the proliferative state and to induce lineage-specific genes and transcription factors.
Given the role of RUNX1 in HSC formation during development and in cell-cycle control, it is tempting to speculate that emergence of HSCs from quiescence under normal or stress conditions requires activation of RUNX1, and that this activation is either initiated or maintained by cdk modification. Once HSCs mature into transient amplifying progenitors, ongoing proliferation and cdk modification may induce RUNX1 to activate lineage-specific genes such as those encoding myeloperoxidase, M-CSF receptor, or PU.1.5,6,35,36 In contrast, RUNX1 might be expected to be less active in terminally differentiating cells undergoing cell- cycle arrest. Recent studies regarding the C elegans RUNX1 ortholog, RNT-1, indicate that RNT-1 stimulates G1 cell cycle progression and coordinates stem-cell proliferation and differentiation in this lower metazoan.37 Although RUNX1(tripleD) did not induce an increased number of myeloid colony-forming units (CFUs) compared with RUNX1(tripleA) when transduced into lineage-negative murine marrow cells, approximately 10% of the CFUs obtained with RUNX1(tripleD) were macroscopically visible, compared with only an occasional large colony with the RUNX1(tripleA) variant; furthermore, upon morphologic evaluation, full granulocytic and monocytic maturation was evident, although an increase in immature cells was seen within some CFUs (H.G. and A.D.F., unpublished data, October 2007). These data suggest that cdk modification of RUNX1 increases its ability to stimulate proliferation not only of Ba/F3 cells but also of myeloid progenitors. In future experiments, we intend to compare these and additional RUNX1 variants also in the context of RUNX1−/− hematopoietic stem and progenitor cells. Further elucidating the regulation of RUNX1 phosphorylation by cdks and the consequences for RUNX1 interactions and control of specific target genes should provide important insights into hematopoiesis and into transformation by CBF oncoproteins.
Acknowledgments
This work was supported by the National Institutes of Health (grant CA098805), by the Children's Cancer Foundation (A.D.F.), and by the Lauri Strauss Leukemia Foundation (F.B.F.).
We thank N. Speck for providing RUNX1 antibody.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.Z., F.B.F., H.G., and A.D.F. performed the research and analyzed the results; A.D.F. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan D. Friedman, Johns Hopkins University, CRB I, Rm 253, 1650 Orleans St, Baltimore, MD 21231; e-mail: afriedm2@jhmi.edu.
References
- 1.Friedman AD. Leukemogenesis by CBF oncoproteins. Oncogene. 1999;13:1932–1942. doi: 10.1038/sj.leu.2401590. [DOI] [PubMed] [Google Scholar]
- 2.Bae SC, Yamaguchi-Iwai Y, Ogawa E, et al. Isolation of PEBP2αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–814. [PubMed] [Google Scholar]
- 3.Meyers S, Downing JR, Hiebert SW. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redondo JM, Pfohl JL, Hernandez-Munain C, Wang S, Speck NA, Krangel MS. Indistinguishable nuclear factor binding to functional core sites of the T-cell receptor δ and murine leukemia virus enhancers. Mol Cell Biol. 1992;12:4817–4823. doi: 10.1128/mcb.12.11.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nuchprayoon I, Meyers S, Scott LM, Suzow J, Hiebert S, Friedman AD. PEBP2/CBF, the murine homolog of the human myeloid proto-oncogenes AML1 and PEBP2β/CBFβ, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol Cell Biol. 1994;14:5558–5568. doi: 10.1128/mcb.14.8.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang DE, Fujioka KI, Hetherington CJ, et al. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol Cell Biol. 1994;14:8085–8095. doi: 10.1128/mcb.14.12.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 8.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao W, Britos-Bray M, Claxton DF, et al. CBFβ-SMMHC, expressed in M4eo AML, reduced CBF DNA-binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene. 1997;15:1315–1327. doi: 10.1038/sj.onc.1201305. [DOI] [PubMed] [Google Scholar]
- 12.Cao W, Adya N, Britos-Bray M, Liu PP, Friedman AD. The core binding factor α interaction domain and the smooth muscle myosin heavy chain segment of CBFβ-SMMHC are both required to slow cell proliferation. J Biol Chem. 1998;273:31534–31540. doi: 10.1074/jbc.273.47.31534. [DOI] [PubMed] [Google Scholar]
- 13.Burel SA, Harakawa N, Zhou L, Pabst T, Tenen DG, Zhang DE. Dichotomy of AML1-ETO functions: growth arrest versus block of differentiation. Mol Cell Biol. 2001;21:5577–5590. doi: 10.1128/MCB.21.16.5577-5590.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Costa J, Chaudhuri S, Civin CI, Friedman AD. CBFβ-SMMHC slows proliferation of primary murine and human myeloid progenitors. Leukemia. 2005;19:921–929. doi: 10.1038/sj.leu.2403755. [DOI] [PubMed] [Google Scholar]
- 15.Lou J, Cao W, Bernardin F, Ayyanathan K, Rauscher FJ, III, Friedman AD. Exogenous cdk4 overcomes reduced cdk4 RNA and inhibition of G1 progression in hematopoietic cells expressing a dominant-negative CBF: a model for overcoming inhibition of proliferation by CBF oncoproteins. Oncogene. 2000;19:2695–2703. doi: 10.1038/sj.onc.1203588. [DOI] [PubMed] [Google Scholar]
- 16.Bernardin F, Yang Y, Civin CI, Friedman AD. c-Myc overcomes cell cycle inhibition by CBFβ-SMMHC, a myeloid leukemia oncoprotein. Cancer Biol Ther. 2002;1:494–498. doi: 10.4161/cbt.1.5.163. [DOI] [PubMed] [Google Scholar]
- 17.Strom DK, Nip J, Westendorf JJ, et al. Expression of the AML-1 oncogene shortens the G(1) phase of the cell cycle. J Biol Chem. 2000;275:3438–3445. doi: 10.1074/jbc.275.5.3438. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, Wang W, Cleaves R, et al. Acceleration of G1 cooperates with CBFβ-SMMHC to induce acute leukemia in mice. Cancer Res. 2002;62:2232–2235. [PubMed] [Google Scholar]
- 19.Bernardin F, Yang Y, Cleaves R, et al. TEL-AML1, Expressed from t(12;21) in human acute lymphocytic leukemia, induces acute leukemia in mice. Cancer Res. 2002;62:3904–3908. [PubMed] [Google Scholar]
- 20.Bernardin-Fried F, Kummalue T, Leijen S, Collector MI, Ravid K, Friedman AD. AML1/RUNX1 increases during G1 to S cell cycle progression independent of cytokine-dependent phosphorylation and induces cyclin D3 gene expression. J Biol Chem. 2004;279:15678–15687. doi: 10.1074/jbc.M310023200. [DOI] [PubMed] [Google Scholar]
- 21.Biggs JR, Peterson LF, Zhang Y, Kraft AS, Zhang DE. AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol Cell Biol. 2006;26:7420–7429. doi: 10.1128/MCB.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finer MH, Dull TJ, Quin L, Farson D, Roberts MR. Kat: a high-efficiency retroviral transduction system for primary human T lymphocytes. Blood. 1994;83:43–50. [PubMed] [Google Scholar]
- 23.Paz-Priel I, Cai DH, Wang D, et al. C/EBPα and C/EBPα myeloid oncoproteins induce bcl-2 via interaction of their basic regions with NF-κB p50. Mol Cancer Res. 2005;3:585–596. doi: 10.1158/1541-7786.MCR-05-0111. [DOI] [PubMed] [Google Scholar]
- 24.Moreno S, Nurse P. Substrates for p34cdc2: in vivo veritas? Cell. 1990;18(61):549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- 25.Kitbayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2294–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernardin F, Friedman AD. AML1 stimulates G1 to S progression via its transactivation domain. Oncogene. 2002;21:3247–3252. doi: 10.1038/sj.onc.1205447. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka T, Kurokawa M, Ueki K, et al. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol Cell Biol. 1996;16:3967–3979. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Biggs JR, Kraft AS. Phorbol ester treatment of K562 cells regulates the transcriptional activity of AML1c through phosphorylation. J Biol Chem. 2004;279:53116–53115. doi: 10.1074/jbc.M405502200. [DOI] [PubMed] [Google Scholar]
- 29.Imai Y, Kurokawa M, Yamaguchi Y, et al. The corepressor mSin3A regulates phosphorylation-induced activation, intranuclear location, and stability of AML1. Mol Cell Biol. 2004;24:1033–1043. doi: 10.1128/MCB.24.3.1033-1043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wee HJ, Huang G, Shigesada K, Ito Y. Serine phosphorylation of RUNX2 with novel potential functions as negative regulatory mechanisms. EMBO Reports. 2002;3:967–974. doi: 10.1093/embo-reports/kvf193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao M, Shapiro P, Fosbrink M, Rus H, Kumar R, Passaniti A. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J Biol Chem. 2006;281:7118–7128. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- 32.Burke DJ, Church D. Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3691–3698. doi: 10.1128/mcb.11.7.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanno T, Kanno Y, Chen LF, Ogawa E, Kim WY, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson LF, Boyapati A, Ranganathan V, et al. The hematopoietic transcription factor AML1 (RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol Cell Biol. 2005;25:10205–10219. doi: 10.1128/MCB.25.23.10205-10219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang G, Zhang P, Hirai H. PU. 1 is a major downstream target of AML1 (RUNX1) in adult hematopoiesis. Nature Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 36.Hoogenkamp M, Krysinska H, Ingram R, et al. The pu.1 locus is differentially regulated at the level of chromatin structure and noncoding transcription by alternate mechanisms at distinct developmental stages of hematopoiesis. Mol Cell Biol. 2007;27:7425–7438. doi: 10.1128/MCB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kagoshima H, Shigesada K, Kohara Y. RUNX regulates stem cell proliferation and differentiation: insights from studies of C. elegans. J Cell Biochem. 2007;100:1119–1130. doi: 10.1002/jcb.21174. [DOI] [PubMed] [Google Scholar]