Abstract
Data sparsity and schema evolution issues affecting clinical informatics and bioinformatics communities have led to the adoption of vertical or object-attribute–value-based database schemas to overcome limitations posed when using conventional relational database technology. This paper explores these issues and discusses why biomedical data are difficult to model using conventional relational techniques. The authors propose a solution to these obstacles based on a relational database engine using a sparse, column-store architecture. The authors provide benchmarks comparing the performance of queries and schema-modification operations using three different strategies: (1) the standard conventional relational design; (2) past approaches used by biomedical informatics researchers; and (3) their sparse, column-store architecture. The performance results show that their architecture is a promising technique for storing and processing many types of data that are not handled well by the other two semantic data models.
Introduction
Researchers at the Yale Center for Medical Informatics (YCMI) have worked for many years developing database approaches to accommodate the fields of clinical medicine, neuroscience, and molecular biology. As part of these efforts, they have created databases that store and integrate diverse types of heterogeneous data, including models and properties of neurons and subcellular components, clinical patient data, genomic sequence, gene expression, and protein expression data, and the results of many different neuroscience experiments. These databases have properties that make them difficult to implement and maintain using conventional relational databases that rely on horizontal data storage, including the use of sparse and heterogeneous data, the frequency of schema changes, and the extensive use of metadata.
This paper focuses on two of these databases: TrialDB, 1 which stores clinical patient data, and SenseLab, 2 which stores information related to neurons and neuronal properties. TrialDB and SenseLab are implemented using the semantic data models known as EAV 3 and EAV/CR, 4 respectively, which make use of vertical storage to store data values, plus an extensive set of metadata to allow generalized tools to be developed to query and maintain the data. Unfortunately, these tools must also re-implement certain features commonly taken for granted in a database engine, including typical methods of querying the logical schema of the data. Furthermore, the use of vertical storage can cause performance problems, particularly with attribute-centered queries. 5
To alleviate some of the limitations imposed by vertical schemas, we propose a database system based on the use of sparse, column-based storage, which we call dynamic tables. The use of a column store, also known as decomposed storage, was first proposed by Copeland and Khoshafian 6,7 and is also used by the Sybase IQ 8 database engine. Our use of decomposed storage is to replace vertical schema at the level of the database engine itself, enabling the key advantages of vertical storage while maintaining the clarity and maintainability of querying a horizontal schema. Our storage implementation presents a standard horizontal view of the data to the database user, and was designed as an addition to—not a replacement of—the database engine’s storage architecture. As a result, dynamic tables can be flexibly and transparently mixed with standard horizontally stored tables in queries.
The remainder of the paper is structured as follows. We first describe the two databases, the data model they are represented with, the tools developed to query them, and motivation for our architecture. Next, we present our architecture, describe its implementation, and discuss the optimizations we employ to improve query performance. We then give performance results comparing the performance of queries and schema modifications using horizontal schema, vertical schema, and our decomposed schema. Finally, we discuss other implementation strategies we tried, related work, and give our concluding remarks.
Background
TrialDB is a clinical study data management system (CSDMS) created at Yale that contains information on clinical trials from several organizations. TrialDB relies on the Entity-Attribute-Value (EAV) semantic data model, a type of vertical schema in which attributes are divided into tables based on type—there is a vertical table for string-valued attributes, another for floating-point-valued attributes, and so on. Attributes are stored as integers for space efficiency, and some EAV tables have a fourth column with a time-stamp value to support versioning. Metadata tables are used to catalog which entities possess which attributes, as well as to provide a mapping between attribute numbers and names. Trial/DB is implemented using the EAV model.
The Entity-Attribute-Value (EAV) data model, also known as the Object-Attribute-Value model, dates back to the use of association lists in LISP, where it was used as a general means of information representation in the context of artificial intelligence research. It was subsequently adapted for use in clinical patient record management systems, including the HELP system, 9, 10 the Columbia-Presbyterian clinical data repository, 11, 12 and the ACT/DB Client—Server Database System. 13
In clinical trial databases, as in electronic medical records systems, a given patient-event requires only a very limited subset from a myriad of possible clinical attributes. These attributes also tend to change as new medical procedures are introduced and others are retired. Storing patient-event data using straight relational tables and columns for attributes would produce extremely sparse tables as well as increased schema change overhead.
The SenseLab project has built several neuroinformatics databases to support both theoretical and experimental research on neuronal models, membrane properties, and nerve cells using the olfactory pathway as one model domain. SenseLab is part of the national Human Brain Project which seeks to develop enabling informatics technologies for the neurosciences.
Neuroscience is a discipline characterized by continual evolution. Underlying hypotheses may change in the course of ongoing research, and knowledge bases may be difficult to maintain due to evolving conceptualization of the domain. Due to these issues, experimental databases can be expensive to maintain and can easily become out of date. At the same time, neuroscience data are far more complexly interrelated than patient data, requiring more than one type of entity, and relationships between entities. For this reason, EAV/CR was created by extending the EAV model with classes and relationships. In this context, classes mean that the values stored in the EAV table are no longer required to be simple values but instead can be complex objects. The addition of relationships allows data values to reference other entities in the database, representing relationships amongst data items. SenseLab is implemented using the EAV/CR model.
In addition to the data models described above, YCMI researchers have created a set of tools to query and manage data stored in the EAV and EAV/CR models. At the same time, their use of these semantic data models has come with a cost. 1) The limited number of tables, coupled with the need to retrieve data stored vertically, results in longer query execution time compared to conventional relational approaches, particularly for multi-attribute queries. 2) Vertical systems require additional storage overhead when storing dense data due to the cost of storing an object identifier and an attribute tag for each stored attribute value. 3) The creation of ad hoc queries becomes more complex, since they have to be devised using a virtual schema but translated into the physical vertical schema.
The inability to execute standard ad-hoc relational queries against an EAV or EAV/CR database’s logical schema is particularly troublesome—queries that are relatively simple to express in a horizontal schema become significantly more complex in a vertical schema. Consider the example in ▶. Part of the complexity comes from the fact that when querying multiple attributes in a vertical schema, we need to join the vertical table with itself once for each desired attribute. Another loss of clarity comes from EAV’s use of integers to identify attributes (if strings were used instead, the string value of each attribute would have to be repeated for each value in the vertical table, adding an unacceptable amount of storage overhead).
Figure 1.
A typical query in the SenseLab database expressed using horizontal and vertical data representations.
Even if the database engine contains extensions such as PIVOT 14 and UNPIVOT operators to simplify the translation between vertical and horizontal schema, if the operators are implemented at a high enough level such that the translated queries are processed by the query planner, the resulting queries will involve a large number of joins. Most modern database engines perform poorly with queries that contain large numbers of project-join operations. 15
In order to overcome the problems imposed by the vertical storage approach, we have devised our dynamic table implementation as described below.
Dynamic Table Implementation and Optimizations
The design goals of our approach are to support the simplicity of querying a horizontal schema and the flexible and efficient schema manipulations of a vertical schema, while maintaining reasonable query performance. To achieve these goals, we have implemented a sparse, column-based storage architecture within the PostgreSQL 16 database engine. Our current implementation does not implement enriched metadata or value-timestamp features found on EAV systems.
We first present the system from the database user’s point of view—how to create and manipulate dynamic tables. We then show how dynamic tables are implemented within the database engine, followed by several optimizations we have employed to improve query performance.
Interface
A table’s storage format is specified at creation time by the presence of the DYNAMIC flag in the CREATE TABLE statement in SQL:
Table foo1.
| CREATE DYNAMIC TABLE table_name |
|---|
| (column_1 type_1, |
| (column_2 type_2, |
| … |
| (column_n type_n) |
The DYNAMIC keyword indicates that the table is to be created using the decomposition storage model. From the database user’s point of view, after the table has been created, operations on the table are semantically indistinguishable from those on a standard, horizontally represented table—the user queries and updates the table as if it were stored horizontally. Dynamic tables also fully support indexes, primary and foreign-key constraints, and column constraints.
The addition of dynamic tables to the database engine does not affect the availability of regular, horizontally stored tables. The user is free to choose the storage format of each individual table based on the sparsity of the data to be stored and the expected frequency of modifications to the table’s schema. Dynamic tables can also be freely mixed with standard tables within queries.
Implementation
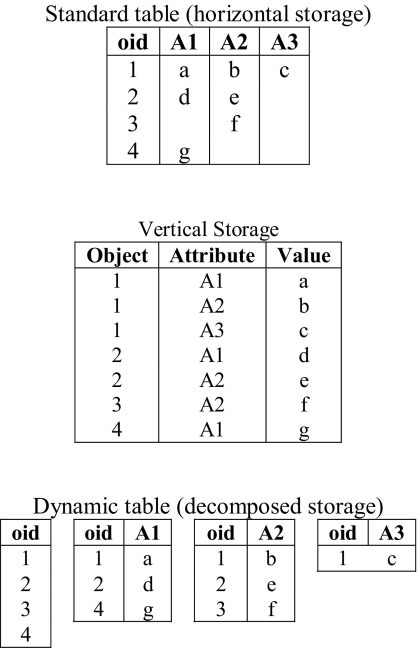
Let r be an n+1-ary relation consisting of n attributes plus an object identifier. To implement the decomposed storage of r, we create n two-column “attribute” tables, and a single one-column “object” table. Each two-column table stores pairs of object identifiers and attribute values. NULL values of an attribute are not explicitly stored; their presence is inferred by the absence of an object-attribute pair in the attribute table. The one-column table stores a list of all object identifiers present in the relation. Finally, the correspondence between r, r’s object table, and r’s attribute tables is stored in a system catalog. ▶ shows the layout of a standard table compared to a dynamic table.
Figure 2.
Horizontal, vertical, and decomposed storage models.
The object table was not present in Copeland and Khoshafian’s original decomposed storage model. The addition of the object table allows us to quickly determine if a row is present in a relation, avoiding a potential scan of each of the attribute tables. It also allows us to maintain a single virtual location of a tuple set to the physical location of it’s object identifier in the object table, and enables us to use a left outer join instead of a full outer join when computing query results. Additionally, the object table was also devised to allow future versions of the dynamic table infrastructure to provide row level security and versioning, which are present in the EAV/CR data model.
When handling sparse data, the dynamic table-based approach is more space efficient than EAV because the attribute columns are not needed.
We implemented dynamic tables at the heap-access layer of PostgreSQL, which implements operations on individual tuples in a relation, namely inserting, updating, deleting, and retrieving a tuple. When the database user creates a new dynamic table, we create the attribute and object tables in a hidden system namespace, exposing only the virtual, horizontal schema given as the table’s definition in the CREATE TABLE statement.
To retrieve a tuple for a query on r, we first scan the object table to find a qualifying object identifier, o. If found, we use o to query each of r’s attribute tables. If the pair (o, a) is present in an attribute table for some value a, we set the corresponding attribute in the returned tuple equal to a. Otherwise, the tuple’s attribute value is set to NULL. To facilitate the efficient lookup of object IDs in the attribute tables, we maintain a B-tree index on object IDs for each table.
Likewise, inserting a new tuple into r is implemented by inserting a new identifier into r’s object table, then for each of the new tuple’s attributes, if the new attribute value is non-NULL, we insert the new object ID and attribute value pair into r’s corresponding attribute table. For deletes, given an object ID, we simply delete each tuple from r’s object and attribute tables. Updates are slightly more complex in that the operation to perform is dependent on both the old and new values of each attribute. The semantics for updating an attribute a to a’ in r are listed in ▶.
Figure 3.
Action performed when updating attribute a to a’ on a dynamic table.
Given the evolving nature of the data we intend to store, it is important that schema modifications on dynamic tables are implemented efficiently and that we avoid the cost of copying an entire table whenever possible. We support the schema operations of adding and removing columns, and changing a column’s type, in addition to trivial schema operations such as renaming a column or renaming the entire table. We implement the non-trivial operations as follows:
• ADD COLUMN col type: To add a new column col to r, we create a new, empty attribute table for col and update the system catalog to associate it with r.
• DROP COLUMN col: To drop a column from r, we find and drop the attribute table associated with the column and remove r’s association with the dropped attribute table from the system catalog.
• ALTER COLUMN col TYPE type: Changing a column’s type requires us to copy and convert the data in the column’s associated attribute table to the new type. However, this is still significantly cheaper than copying and converting an entire horizontal schema, particularly when the schema has a large number of attributes.
Schema alteration operations in the dynamic approach have the advantage of intrinsic transactional support: schema modification statements are always executed atomically by the database engine and can be embedded seamlessly within other transactions. Vertical systems require careful SQL programming from the application programmers to avoid corrupting data and logical schema.
Optimizations
We employ two main optimizations to improve the performance of queries on dynamic tables: projection pushing and optimizing conditional selects. The use of decomposed storage gives us additional opportunities for projection pushing in comparison to a standard horizontal schema. Consider the query listed in ▶. If r is stored using decomposed storage, there is no reason to retrieve values from r’s attribute tables for a, c, or e. For the general case, we compute the set of attributes we are required to fetch by taking the union of the attributes present in the target list of the SELECT clause with the attributes present in the WHERE clause of a query.
Figure 4.
A good opportunity for projection pushing.
Next, consider the query listed in ▶. We cannot use projection pushing here because all r’s attributes are requested via “SELECT *”. However, when scanning r to find tuples that match the query condition, it would make sense to retrieve and test the values of attributes d and e against the condition, discarding the candidate tuple if the test fails before retrieving r’s remaining attributes. To implement this optimization in the general case, we divide the requested attributes from a query into two groups: those with conditions and those without conditions. When retrieving attributes, we first retrieve attributes with conditions, one at a time. If the attribute does not satisfy the condition, we immediately discard the tuple, skipping the remaining attributes. If all of the conditions are satisfied, we read the remaining attributes and return the tuple to the query executor.
Figure 5.
A candidate for conditional select optimization.
In both of these cases, these attribute-level I/O operations are not beneficial when using a standard horizontal schema because tuples are generally stored contiguously and packed into blocks, and sub-block I/O does not improve tuple throughput. However, with decomposed storage, attributes of each column are grouped together into blocks, giving us the opportunity to skip reading blocks for attributes we have determined that we don’t need. Of course, the other side of the story is that if we do end up needing all the attributes of a tuple, a horizontal schema that allows us to fetch the entire tuple at once will be more efficient, so the optimal choice of storage format is dependent on both the sparsity of the data being stored and the types of queries that will be run against the data.
Performance Results
To evaluate the performance of our implementation, we compared the performance of dynamic tables to both standard, horizontal storage and to storage using the EAV and EAV/CR vertical models. We test this using a typical series of queries executed by users of the SenseLab databases that retrieve a set of neurons and chemosensory receptors plus several of their attributes, filtering on between one and five attributes to narrow the number of results. These queries have a similar structure to the queries shown in ▶.
After each database was populated, the ANALYZE command was run to update the statistics used by the query planner. All tests were run on a 1.8Ghz Intel Pentium IV machine with 1GB of RAM running Fedora Core Linux version 4.01.
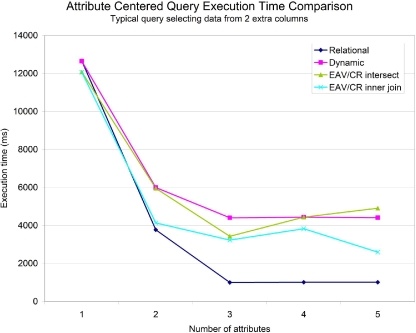
▶ shows the results of running each query using each of the three storage models. No indexes were used on the data items, representing the case where we do not know beforehand which attributes will be of interest to the database users. The number of attributes in this case refers to the number of values that are filtered on in the WHERE clause of the query.
Figure 6.
Comparison of query execution time for an attribute-centered query on SenseLab data without indexes.
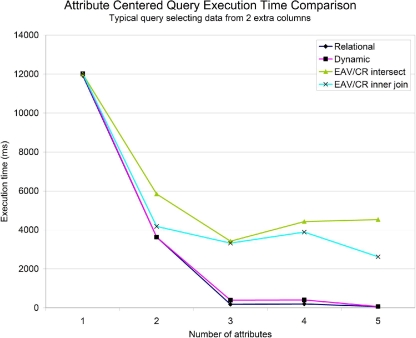
When we do know which attributes will commonly be queried on, we can create indexes on them to improve performance—this is the usual case for SenseLab data. ▶ shows the running time for each of the queries with indexes. Notice that the use of indexes virtually eliminates the performance difference between horizontal storage and dynamic tables, whereas the vertical approach does not benefit as much from using indexes.
Figure 7.
Comparison of query execution time for an attribute-centered query on SenseLab data with indexes on queried attributes.
Unlike many other relational database engines, PostgreSQL already has support for the efficient implementation of some schema modification commands. In particular, most cases of adding or removing a column from a table are executed lazily—the storage format of the table is not actually modified until the table is later compacted using the VACUUM command. Thus, in our implementation, adding a column to a table is a trivial operation using any of the three storage formats, as they only involve updating the system catalog (the metadata catalog in the case of EAV/CR), and are independent of the data currently stored in the table.
However, some schema modification operations do require the data in a table to be modified, including altering a column’s type and adding a new column with a default value. To implement altering a column’s type using horizontal schema, the entire table is copied, converting the data values for the modified column to the new type as they are copied. For dynamic tables, we only have to convert the single attribute table corresponding to the modified column. For tables stored using EAV or EAV/CR, we need to move the data values from the object-attribute-value table for the old type to the object-attribute-value table for the new type. ▶ shows the SQL statements used to change the data type of a column representing the year of publication of a paper from a string value to an integer. Dynamic tables are significantly faster than the other approaches for this operation: altering the data type for 20,988 values in the SenseLab database took 0.5 seconds using conventional tables, 0.29 seconds using dynamic tables, and 4.6 seconds using EAV-formatted tables.
Figure 8.
SQL queries to alter the type of a column representing the year of publication of a paper from a string value to an integer value.
We stress that adding and removing columns tend to be by far the most common types of schema changes in a database. When using a database engine that executes schema modifications eagerly, unlike PostgreSQL, the performance of these operations in a horizontal system is typically far worse than in a vertical system. In such a system the entire table needs to be translated to the new format, the execution time of which scales linearly with the size of the table, giving poor performance for large databases. Dynamic tables are particularly suited to efficiently implementing these operations in such systems as only the system catalog needs to be modified to complete the operation, hence the execution time is independent of the table’s size.
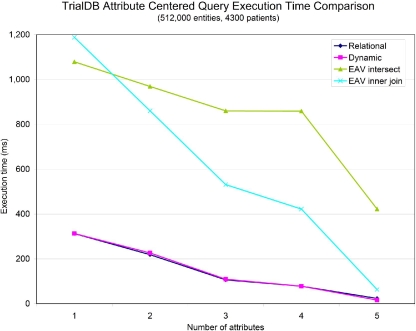
We also ran a set of queries on the TrialDB data to compare the performance of each storage model on a large, sparse dataset. Since the TrialDB data contain millions of records, querying the dataset without the use of indexes is too slow to be useful using any of the storage methods. ▶ shows the results of running a set of typical attribute-centered queries with indexes on each of the attributes in question. The performance difference between EAV and the other two storage methods is even more pronounced in this data set.
Figure 9.
Comparison of query execution time for an attribute-centered query on TrialDB data with indexes on queried attributes.
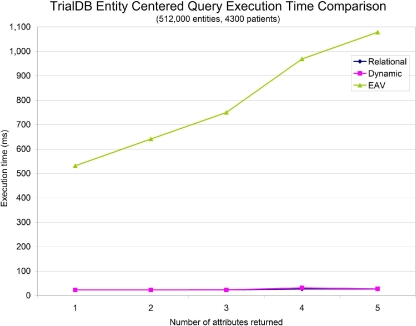
In addition to attribute-centered queries, we also compared the performance of the three storage models when evaluating entity-centered queries on TrialDB data. These queries have the form “retrieve a set of attributes for a every event relevant to a particular patient.” Instead of focusing on the execution time needed to fetch the entities alone, which is well known to be quite similar between vertical and relational systems, 5 we instead measured the execution time needed to retrieve the entity plus one to five additional attributes, which represent a set of typical real-world entity-based queries. ▶ shows that the execution time required for the vertical and dynamic storage mechanisms does not change significantly as additional attributes are requested, whereas the execution time for EAV scales linearly with the number of attributes.
Figure 10.
Comparison of query execution time for an entity-centered query on TrialDB data.
Sample clinical data from TrialDB used in this project were deidentified by removing all patient demographics data, scrambling integer and string data values, and replacing all internal system id’s (e.g., patient-event ids) with local sequential ones.
Other Implementation Strategies
The implementation strategy described above is actually our third refinement of alternate relational storage architectures. For database-specific reasons, our first two approaches encountered “mismatch impedance” when implemented in our PostgreSQL testbed, but we believe the ideas may well have merit if implemented in other database systems.
Our first approach was to use a dense column-store. In this architecture, a relation’s attribute values are densely packed into one-column tables. Tuples from the conceptual schema are returned by joining values from the attribute tables based on their index in the table: to retrieve the ith tuple from relation r, we simply take the ith entry from the first attribute table, the ith entry from the second attribute table, and so on. This approach still allows for the efficient schema modifications described above, and provides greater storage efficiency for dense data. However, we realized that this approach is incompatible with PostgreSQL’s use of multi-version concurrency control (MVCC). Using MVCC, PostgreSQL eliminates the need to hold locks on tables by storing multiple versions of tuples representing consistent snapshots of the data at different points in time, along with tuples that have been made obsolete by newer versions. When implementing the single-column store approach, even if we are careful to ensure that values from a tuple are inserted into the same logical position in each of the attribute tables, it is very difficult to ensure that values will remain in the correct logical order after a table compaction (VACUUM) operation.
Our second approach was to use sets of sparse columns to store attributes as with our final implementation, except we implemented the translation from decomposed storage to horizontal storage at the level of rewrite rules in the database engine instead of at the heap-access level. Our hypothesis was that rewrite rules would give the query planner maximal opportunity to optimize query plans involving dynamic tables. We found, however, that PostgreSQL’s query optimizer tried too hard to find the optimal join order when combining attribute tables, taking exponential time relative to the number of attributes to plan the query. The generated query plans were efficient, but for tables with large numbers of columns, query planning took several orders of magnitude longer than actually executing the query. By explicitly restricting the join order, we were able to marginally decrease planning time, with the tradeoff of generating inferior query plans. In the end, we found it was more efficient to implement decomposed storage at the heap-access level of the database, bypassing the query planner entirely.
Related Work
Beckmann et al. 17 propose an alternate storage format for tables on disk in which the tuple layout of each row is described by a variable-length, “interpreted” record. The interpreted records are implemented at a lower level in the database engine compared to dynamic tables (storage level vs. heap-access level), and provide many of the same benefits for storing sparse or heterogeneous data with potentially greater space efficiency. It is not clear, however, that interpreted records provide any benefit to the optimization of schema-modifying operations.
The C-Store 18 database engine is also based on a column-store architecture, but is designed for the application of read optimized databases, and thus may not be appropriate for typical neuroscience databases where writes are frequent.
The work of Agrawal et al. 19 on e-commerce data shows that the domain of e-commerce data shares many similarities with neuroscience and clinical informatics data, particularly with respect to schema evolution and heterogeneous data. They, however, advocate the use of vertical storage and show a performance advantage for vertical schema compared to horizontal schema for this domain.
Software
The PostgreSQL database engine and our modifications to support dynamic tables are open-source software distributed under the BSD 20 license, which allows the software to be freely distributed, modified, and included in commercial products, as long as the original copyright notice is preserved. Our software is available for download at the following address: http://crypto.stanford.edu/portia/software/postgredynamic.html.
We welcome feedback from anyone who makes use of our software as part of any research project or commercial solution.
Conclusion and Future Work
This paper presents the architecture of dynamic tables, which are based on implementing relational database storage using the decomposed storage model while maintaining a logical horizontal view of the data. Our implementation has significant advantages over past approaches used by the bioinformatics and medical informatics communities that were based on the use of vertical storage, including improved query performance on both attribute-centered and entity-centered queries, improved schema modification performance, and the greater manageability of working with a horizontal view of the data. Dynamic tables also maintain the capability of vertical schemas to efficiently store sparse data.
Our future work will continue our effort increasing the capabilities of database engines by adding features commonly needed by bioscience and clinical databases, including extensive support for metadata and a flexible system for row-level security.
Footnotes
This work was supported in part by the NSF Information Technology Research program under grant number 0331548, by NIH grants P01 DC04732 and R01 DA021253, and by NIH grant P20 LM07253 from the National Library of Medicine.
References
- 1.Nadkarni PM, Brandt C, Frawley S, et al. Managing attribute-value clinical trials data using the ACT/DB client-server database system J Am Med Inform Assoc 1998;5:139-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller PL, Nadkarni P, Singer M, Marenco L, Hines M, Shepherd G. Integration of multidisciplinary sensory data: a pilot model of the human brain project approach J Am Med Inform Assoc 2001;8:34-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nadkarni PM, Brandt C. Data extraction and ad hoc query of an entity-attribute-value database J Am Med Inform Assoc 1998;5:511-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marenco L, Nadkarni P, Skoufos E, Shepherd G, Miller P. Neuronal database integration: the Senselab EAV data model Proc AMIA Symp 1999:102-106. [PMC free article] [PubMed]
- 5.Chen RS, Nadkarni P, Marenco L, Levin F, Erdos J, Miller PL. Exploring performance issues for a clinical database organized using an entity-attribute-value representation J Am Med Inform Assoc 2000;7:475-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copeland GP, Khoshafian SN. A decomposition storage model. In: In Proceedings of the 1985 ACM SIGMOD International Conference on Management of Data May 28-31, 1985; Austin, Texas. p. 268–79.
- 7.Khoshafian S, Copeland G, Jagodis T, Boral H, Valduriez P. A query processing strategy for the decomposed storage model ICDE 1987:636-643.
- 8.Sybase Inc Sybase IQ: Query Search Test Software And Operational Data Store Warehouse Application. 2006. Available at: http://www.sybase.com/products/informationmanagement/sybaseiq. Accessed June 26, 2006.
- 9.Huff SM, Berthelsen CL, Pryor TA, Dudley AS. Evaluation of an SQL model of the help patient database Proc. 15th Symposium on Computer Applications in Medical Care; 1991. Washington, DC: IEEE Computer Press; 1991. pp. 386-390Los Alamitos, CA. [PMC free article] [PubMed]
- 10.Huff SM, Haug DJ, Stevens LE, Dupont CC, Pryor TA. HELP the next generation: a new client-server architecture Proc. 18th Symposium on Computer Applications in Medical Care; 1994. Washington, D.C: IEEE Computer Press; 1994. pp. 271-275Los Alamitos, CA. [PMC free article] [PubMed]
- 11.Friedman C, Hripcsak G, Johnson S, Cimino J, Clayton P. A generalized relational schema for an integrated clinical patient database Proc. 14th Symposium on Computer Applications in Medical Care; 1990. Washington, DC: IEEE Computer Press; 1990. pp. 335-339Los Alamitos, CA.
- 12.Johnson S, Cimino J, Friedman C, Hripcsak G, Clayton P. Using metadata to integrate medical knowledge in a clinical information system Proc. 14th Symposium on Computer Applications in Medical Care; 1990. Washington, D. C: IEEE Computer Press; 1990. pp. 340-344Los Alamitos, CA.
- 13.Nadkarni PM, Brandt C, Frawley S, et al. Managing attribute-value clinical trials data using the ACT/DB client-server database system J Am Med Inform Assoc 1998;5:139-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentin D, Nadkarni P, Brandt C. Pivoting approaches for bulk extraction of Entity-Attribute-Value data Comp Meth Programs Biomed 2006;82:38-43. [DOI] [PubMed] [Google Scholar]
- 15.McMahan BJ, Pan G, Porter P, Vardi MY. Projection Pushing Revisited EDBT 2004.
- 16.Group PGD. PostgreSQL. Available at: http://www.postgresql.org/. Accessed June 26, 2006.
- 17.Beckmann JL, Halverson A, Krishnamurthy R, Naughton JF. Extending RDBMSs To Support Sparse Datasets Using An Interpreted Attribute Storage Format ICDE 2006.
- 18.Stonebraker M, Abadi D, Batkin A, et al. C-Store: A Column Oriented DBMS. 2005. Proceedings of VLDB; August 2005; Trondheim, Norway.
- 19.Agrawal R, Somani A, Xu Y. Storage and querying of e-commerce data VLDB 2001:149-158.
- 20.Open Source Initiative. The BSD License. Available at: http://www.opensource.org/licenses/bsd-license.php. Accessed June 26, 2006.