Abstract
DNA topoisomerase II—an essential nuclear enzyme in DNA replication and transcription, chromatin segregation, and cell cycle progression—is also a target of clinically useful anticancer drugs. Preliminary observations of a positive correlation between the expression of topoisomerase (topo) IIα and the retinoblastoma protein (Rb) in a series of rhabdomyosarcoma cells prompted us to ask whether these two proteins interact in vivo. Using human rhabdomyosarcoma and leukemic cell lines, we found a physical association between topo IIα and Rb protein by reciprocal immunoprecipitation and immunoblotting, in which topo IIα appeared to interact primarily with the underphosphorylated form of Rb. Experiments with truncated glutathione S-transferase-Rb fusion proteins and nuclear extracts of Rh1 rhabdomyosarcoma cells indicated that topo IIα binds avidly to the A/B pocket domain of Rb, which contains the intact spacer amino acid sequence. To determine whether this interaction has functional consequences in vivo, we expressed wild-type and mutant Rb in human cervical carcinoma cells lacking functional Rb. Wild-type, but not mutant, Rb inhibited topo II activity in nuclear extracts of these transfected cells. Moreover, purified wild-type Rb inhibited the activity of purified human topo IIα, indicating a direct interaction between these two proteins. We conclude that topo IIα associates physically with Rb in interactions that appear to have functional significance.
The essential nuclear enzyme DNA topoisomerase II (topo II) unknots and decatenates DNA through an ATP-dependent double-strand break followed by strand passing and religation (1), functions that are required in DNA replication and transcription, chromosome segregation, cell cycle progression, and, possibly, DNA repair (2, 3). Topo II is one of the major components of the nuclear matrix (4) and the target of clinically important anticancer drugs (5, 6). In mammalian cells, topo II exists as two isoforms, α (170 kDa) and β (180 kDa), which are encoded by different genes (7). The expression of topo IIα is cell cycle-dependent, reaching a maximum in G2/M phase (8). Because of its role in critical cell cycle events (9–12), topo II offers an attractive target to regulatory proteins.
The retinoblastoma protein (Rb), a 105- to 110-kDa nuclear phosphoprotein, regulates the cell cycle, cell proliferation, differentiation, and, possibly, apoptosis (13–15). When hypophosphorylated (in the G0 and G1 phases of the cell cycle), Rb blocks cell cycle progression by binding to and suppressing transcription factors of the E2F family (16, 17). Phosphorylation of Rb during late to mid-G1 and continuing through S and G2/M (18, 19), leads to inactivation of Rb and the release of bound E2F proteins, which, in turn, activate genes involved in cell cycle progression (15). Inactivating deletions or mutations in the Rb gene have been observed in a variety of cancers (14). More recently, additional functions have been suggested for Rb on the basis of its binding to other cellular proteins (20, 21).
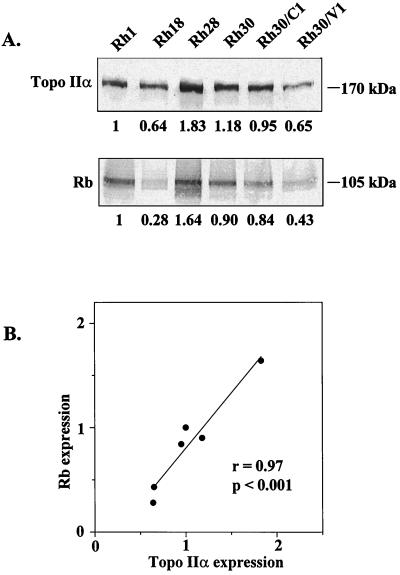
In preliminary experiments (Fig. 1), we observed a positive correlation between the constitutive expression of topo IIα and Rb in six human rhabdomyosarcoma (RMS) cell lines. Because of the known regulatory roles of Rb, we postulated that the two phosphoproteins interact with each other. The data reported here support this hypothesis and suggest that the association of topo IIα and Rb is physiologically relevant.
Figure 1.
Constitutive expression of topo IIα and Rb proteins in RMS cell lines. (A) Representative immunoblots based on analysis of total cell extracts with specific antibodies (Ab-284, topo IIα; G3–245, Rb). Equal loading of the proteins was confirmed by Ponceau-S staining (Materials and Methods). Values beneath the lanes are mean levels of topo IIα or Rb expression (relative to the levels in Rh1 cells) from five separate experiments. SDs ranged from 0.08 to 0.47. (B) Correlation of topo IIα expression with Rb expression in RMS cell lines. r = Pearson’s correlation coefficient; the P value was determined by Student’s t test.
MATERIALS AND METHODS
Cell Lines and Culture Conditions.
The xenograft-derived pediatric RMS cell lines Rh1, Rh18, Rh28, and Rh30 were established from patients before they received chemotherapy (kindly provided by P. J. Houghton, ref. 22). Rh30/C1 and Rh30/V1 were selected from the Rh30 parent cell line for resistance to camptothecin and VP-16 (etoposide), respectively, in our laboratory. C33A human cervical carcinoma cells were obtained from American Type Culture Collection (Manassas, VA). Cells were subcultured twice a week in either RPMI 1640 medium (RMS lines) or DMEM (C33A cells). Both media (BioWhittaker) were supplemented with 2 mM l-glutamine and 10% FBS.
Coimmunoprecipitation and Immunoblotting.
Cells were lysed by sonication in radioimmunoprecipitation assay (RIPA) buffer (PBS + 1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS) containing protease inhibitors (1 mM PMSF/1 mM benzamidine/10 μg of soybean trypsin inhibitor per ml/50 μg of leupeptin per ml/1μg of pepstatin per ml/20 μg of aprotinin per ml). Lysates were precleared for 2 h with either mouse IgG (Rb experiments) or rabbit IgG (topo IIα experiments) and protein G plus protein A-agarose (Oncogene Research Products, Cambridge, MA). Topo IIα- or Rb-specific antibodies (5 μg) or control IgG and protein G plus protein A-agarose (25 μl) were added to the precleared lysates and gently rotated overnight at 4°C. The following antibodies were used for immunoprecipitation: Rb, XZ-77 (amino acids 387–928, mouse monoclonal; Upstate Biotechnology, Lake Placid, NY) and G3–245 (amino acids 300–380, mouse monoclonal; PharMingen); topo IIα, Ab-284 (amino acids 1–15, rabbit polyclonal; a generous gift from F. Boege, University of Würzburg, Germany) and Ab-1 (amino acids 1513–1530, rabbit polyclonal; NeoMarkers/LabVision, Fremont, CA); and E2F-1, KH-95 (mouse monoclonal; PharMingen). The immunoprecipitates were washed four times with EBC1 buffer (23) (50 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.1% Nonidet P-40/1 mM EDTA) containing protease inhibitors and then were resuspended in 2× Laemmli sample buffer (24), boiled for 5 min, and separated by 7.5% SDS/PAGE (24). The proteins were transferred to nitrocellulose (Schleicher & Schuell) by use of the Milliblot SDE system (Millipore). Blots were stained with Ponceau-S to confirm equal loading of proteins. Blots were incubated overnight at 4°C with the primary antibody (Ab-284, 1:4,000 dilution for topo IIα and G3–245, and 1 μg/ml for Rb) in 5% nonfat dry milk in Tris-buffered saline. After incubation with horseradish peroxidase-labeled anti-mouse (for Rb) or anti-rabbit (for topo IIα) IgG (both from Amersham), the blots were developed by the enhanced chemiluminescence system (ECL; Amersham). When required, dephosphorylation was performed by treating the cell lysate in EBC1 buffer with 3 units of bacterial alkaline phosphatase, followed by incubation at 37°C for 20 min.
Preparation of Glutathione S-transferase (GST)-Rb Fusion Proteins and in Vitro Binding Assay.
pGEX-2T (Pharmacia) plasmids containing Rb cDNA inserts encoding truncated Rb proteins [e.g., GST-Rb (amino acids 379–928), GST-Rb (amino acids 379–792), GST-Rb (amino acids 792–928] were generous gifts of David Livingston (Dana–Farber Cancer Institute, Boston, MA). These and other truncated GST-Rb plasmids were transformed in competent Escherichia coli DH5α (GIBCO/BRL), and the fusion proteins were expressed by stimulation with 0.2 mM isopropyl-β-d-thio-galactopyranoside. Bacteria were lysed, and the fusion proteins were purified by binding to glutathione-Sepharose beads (Pharmacia) as described (25). The purity and relative amounts of the GST fusion proteins bound to the beads were checked by 7.5–14% SDS/PAGE followed by staining with Coomassie brilliant blue. Nuclear extracts of Rh1 cells were prepared by lysis in a buffer containing 0.5% Nonidet P-40 followed by extraction of the nuclei in a 0.4 M NaCl buffer (with the same protease inhibitors described earlier) (26). Extracts (≈1 mg total protein) were diluted 1:5 in EBC1 buffer (with protease inhibitors) and clarified by centrifugation. The diluted lysates were added to equivalent amounts of GST-Rb or control GST proteins bound to glutathione-Sepharose beads in 1.5-ml microfuge tubes, with overnight rotation of the tubes at 4°C. Beads then were washed four times with EBC1 buffer and resuspended in 50 μl of Laemmli sample buffer. Samples were boiled for 5 min and separated by 7.5% SDS/PAGE. Proteins then were transferred to nitrocellulose, immunoblotted with Ab-284 antibody, and developed by the ECL method.
Transfection with Rb Expression Vectors and Decatenation Assay.
Cytomegalovirus expression vectors encoding wild-type Rb [pBC-Rb (379–928)] and mutant Rb [pBC-Rb (379–928), amino acid 706 C → F] (27) were transfected into Rb-mutant C33A human cervical carcinoma cells by the calcium phosphate method (28). After 16 h, the cells were washed twice, resuspended in fresh medium (DMEM +10% FBS), and incubated for 24 h. Nuclear protein extracts of the cells were prepared as described earlier (26) and analyzed for topo II activity by decatenation of kinetoplast DNA (k-DNA) by using a topo II assay kit (TopoGEN, Columbus, OH), according to the manufacturer’s instructions. Briefly, 2-fold dilutions of the nuclear extracts were incubated with 250 ng of k-DNA in the supplied reaction buffer (includes ATP) for 30 min at 37°C. “Stop” solution was added, and the reaction products were separated on a 1% agarose gel containing 0.5 μg/ml of ethidium bromide, destained, and photographed.
Treatment of Purified Topo IIα with Purified Rb Proteins and Decatenation Assay.
Wild-type [GST-Rb (379–928)] and mutant [GST-Rb (379–928), amino acid 706 C → F] Rb proteins were expressed in E. coli and purified by binding to glutathione-Sepharose beads followed by treatment with glutathione. The proteins (12 ng) then were added to 2-fold dilutions of purified human topo IIα (a generous gift from Neil Osheroff, Vanderbilt University School of Medicine, Nashville, TN) and incubated at 4° for 15 min. Topo II activity was determined by decatenation of k-DNA as described above.
RESULTS
Correlation Between Topo IIα and Rb Expression in RMS Cell Lines.
An association between topo IIα and Rb expression was suggested by studies to identify cell cycle-specific proteins that might regulate this topo II isoform. Fig. 1A shows the relative concentrations of these phosphoproteins in six lines of RMS cells with different levels of sensitivity to VP-16. As demonstrated in Fig. 1B, there was a strong, positive correlation (r = 0.97, P < 0.001) between topo IIα and Rb expression, suggesting a physical relationship between the two phosphoproteins.
Topo IIα Associates with Rb in Vivo.
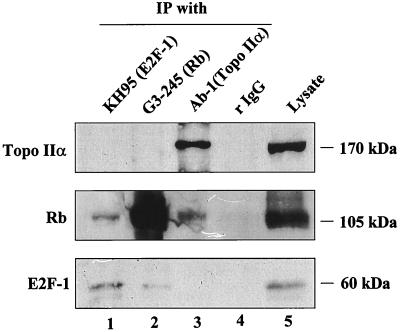
To test this hypothesis, we immunoprecipitated whole Rh1 cell lysates with antibodies specific for Rb and topo IIα. Fig. 2 (lane 3, Ab-1) shows clearly that Rb coimmunoprecipitates with topo IIα. An antibody (KH-95) against E2F-1, a known Rb-binding protein (15), coimmunoprecipitated Rb (lane 1), whereas rabbit IgG did not react with either Rb or topo IIα (lane 4).
Figure 2.
Coimmunoprecipitation of Rb by topo IIα-specific antibody in total extracts of Rh1 cells. Immunoprecipitations (IP) were performed with antibodies to the C-terminal region of topo IIα (Ab-1), Rb (G3–245), and E2F-1 (KH-95) or rabbit IgG. The immunoblots represent three separate experiments. (Top) Blotted with Ab-284. (Middle) Blotted with G3–245. (Bottom) Blotted with KH-95. Lanes 1–4, IP with the indicated antibodies; lane 5, direct immunoblots of the cell lysate used in IP.
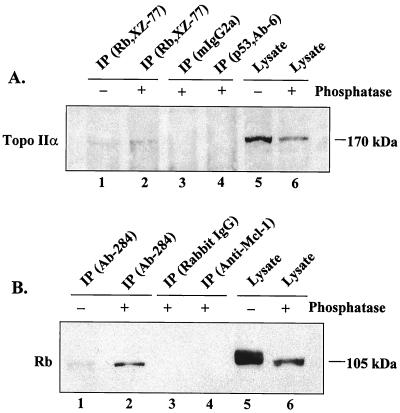
The Rb-specific antibody G3–245 coimmunoprecipitated E2F-1 but not topo IIα (lane 2), causing us to modify the immunoprecipitation procedure. Topo IIα, like other Rb-binding proteins (20, 29), appears to associate with the underphosphorylated form of Rb, which represents only a fraction of the total Rb in cells. Hence, we lysed Rh1 cells in a low-stringency EBC1 buffer, treated the lysates with bacterial alkaline phosphatase, and immunoprecipitated with XZ-77, an Rb-specific antibody that recognizes a longer epitope than does G3–245. In the assay shown in Fig. 3A, a weak band of topo IIα coimmunoprecipitated with Rb from untreated lysate probed with a topo IIα-specific antibody (lane 1); a denser band was seen when the experiment was performed with phosphatase-treated lysate (lane 2). Neither mouse IgG2a (lane 3) nor an irrelevant, isotype-matched mouse antibody (lane 4) reacted with topo IIα in phosphatase-treated lysate. In a reverse experiment with phosphatase-treated and untreated Rh1 lysates (Fig. 3B), using Ab-284 for immunoprecipitation and G3–245 for immunoblotting, we detected a more distinct band of coimmunoprecipitated Rb in phosphatase-treated (lane 2) than in untreated (lane 1) lysate. Again, Rb was not coimmunoprecipitated with either rabbit IgG (lane 3) or an irrelevant rabbit antibody (lane 4), nor did topo IIα-specific antibodies (either Ab-284 or Ab-1) recognize Rb protein directly in the extracts (data not shown). Moreover, Rb was not coimmunoprecipitated with topo IIα in extracts from Rb-null C33A or SAOS2 cells (data not shown).
Figure 3.
Reciprocal coimmunoprecipitations of topo IIα and Rb in Rh1 cell lysates treated with alkaline phosphatase (+) or untreated controls (−). Shown is a representative immunoblot of three separate experiments. (A) Coimmunoprecipitation of topo IIα by Rb-specific antibody (XZ-77) before (lane 1) and after (lanes 2–4) treatment with alkaline phosphatase. Total cell extracts were treated with alkaline phosphatase, immunoprecipitated with XZ-77, p53 (Ab-6, irrelevant isotype-matched antibody), or mouse IgG2a, and immunoblotted with Ab-284. Lanes 5 and 6, direct immunoblot of the lysates. (B) Coimmunoprecipitation of Rb by topo IIα-specific antibody Ab-284 before (lane 1) and after (lanes 2–4) treatment with alkaline phosphatase in Rh1 cells. Total cellular extracts were treated with alkaline phosphatase and immunoprecipitated with Ab-284, anti-Mcl-1 (an irrelevant rabbit antibody), or rabbit IgG, as indicated, and immunoblotted with G3–245 (Rb-specific). Lanes 5 and 6, direct immunoblot of lysates used in IP.
To exclude the possibility that topo IIα and Rb bind through a DNA intermediate, we performed coimmunoprecipitations in the presence of 100 μg of ethidium bromide per ml (29) or after treating the lysate with 200 units of DNase I per ml (30). Neither treatment interfered with the codetection of topo IIα and Rb (data not shown). Taken together, our findings demonstrate a physical association between topo IIα and Rb in vivo.
Topo IIα Binds to the A/B Pocket Domain of Rb Protein in Vitro.
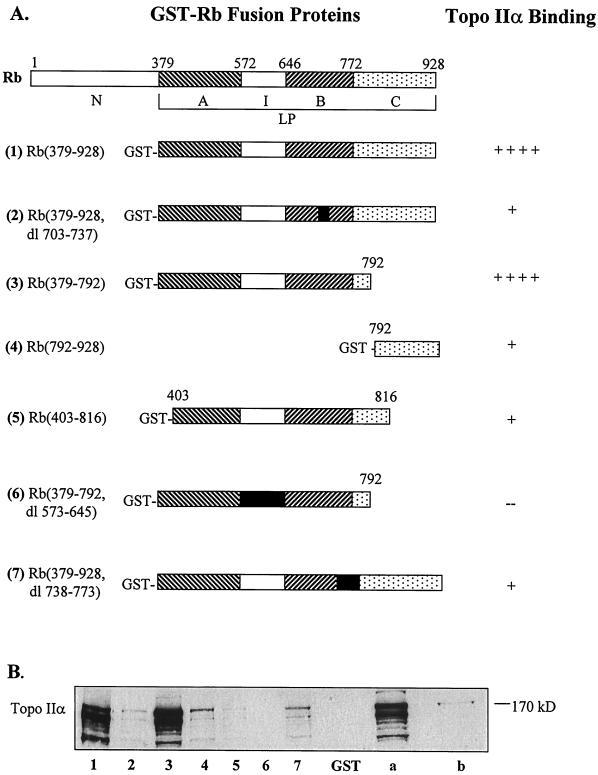
Rb is a 928-aa protein with functionally distinct protein-binding domains (31) (Fig. 4A Upper). The A/B pocket, which contains domains A (amino acids 379–572) and B (amino acids 646–772) separated by an insert (I) domain, is the binding site of several viral oncoproteins that inactivate Rb (32, 33). The C pocket (amino acids 772–870) binds c-Abl tyrosine kinase (34); MDM2 also recognizes the C-terminal region of Rb (23). The large pocket (LP), which includes the A, B, and C regions (amino acids 379–870), is the binding site of E2F transcription factors (31, 33).
Figure 4.
Topo IIα-binding domains in Rb protein determined by in vitro binding with GST-Rb fusion proteins. (A) Structures of the truncated GST-Rb fusion proteins used for in vitro binding with nuclear extracts from Rh1 cells. (Upper) Protein-binding domains illustrating the A/B and C pockets, insert (I) region, and the large pocket (LP) in human retinoblastoma protein. The numbers in parentheses refer to the amino acids from the N terminus; dl, deletion. The extent of binding of the proteins to topo IIα is indicated at the right; ++++, strong; ++, moderate; +, weak; −, none. (B) A representative immunoblot (of three separate experiments) illustrating the comparative binding of topo IIα from Rh1 nuclear extracts to the GST-Rb fusion proteins. The extracts were incubated with GST-Rb fusion proteins (equivalent amounts) bound to GST-Sepharose. The protein bound beads were separated by SDS/PAGE and immunoblotted with Ab-284. Lanes: 1–7, topo IIα binding to the respective GST-Rb fusion proteins (as numbered); GST, binding with GST alone; a, nuclear extract of Rh1 used for binding assay (no beads); b, total protein extract from HeLa cells (no beads; used as topo IIα marker).
To determine the topo IIα-binding domain in Rb (Fig. 4A Upper), we relied on several recombinant GST-Rb fusion proteins (Fig. 4A; ref. 25). These constructs, bound to glutathione-Sepharose beads containing equivalent amounts of proteins, then were used in an in vitro binding assay with nuclear extracts from Rh1 cells. GST-Rb (379–792), the fusion protein containing the A/B (small) pocket and an intact spacer region, bound most avidly with topo IIα (Fig. 4B, lane 3). GST-Rb (379–928), which contains the A, B, and C pockets (hence, the LP), also bound strongly to topo IIα (lane 1). By contrast, GST-Rb (792–928), which contains only the C pocket of Rb, bound weakly to topo IIα (lane 4), as did proteins with deletions in the A pocket [GST-Rb (403–816)] or B pocket [GST-Rb [379–928, deletion (dl) 703–737] and GST-Rb (379–928, dl 738–773)]. The spacer region separating the A/B pockets also appeared important for topo IIα binding, because the fusion protein with a deletion of this region [GST-Rb (379–792, dl 573–648)] precipitated very little topo IIα (lane 6). Neither of the negative controls [pure GST protein bound to glutathione-Sepharose beads (lane GST) and beads equilibrated with DH5α bacterial protein extracts (not shown)] bound to topo IIα. Similar results were obtained with nuclear extracts from CEM and HeLa cells (data not shown). The multiple bands of topo IIα observed in these experiments could reflect truncated isoforms of the enzyme (35) or its proteolytic products (36). Overall, the data indicate that the topo IIα-binding domain in Rb lies in the region of the A/B pocket that is also required for binding of viral oncoproteins (33, 37), although it is not yet clear whether the binding sites are the same.
Expression of Wild-Type Rb in C33A Cells Inhibits Topo II Activity.
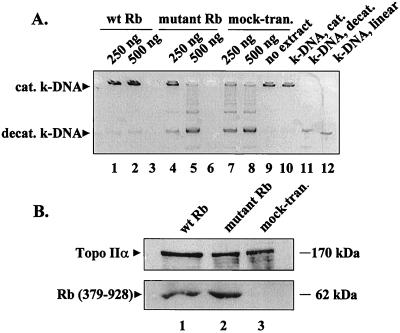
To investigate whether binding to Rb modulates topo II activity, we transiently expressed wild-type [Rb (379–928)] and mutant [Rb (379–928); amino acid 706, C → F] proteins (27) in C33A cells (which lack functional Rb) and compared the topo II activity in nuclear extracts by decatenation of k-DNA. As shown in Fig. 5A, less k-DNA was decatenated in nuclear extracts of cells expressing wild-type Rb (lanes 1 and 2) as compared with extracts from cells expressing mutant Rb (lanes 4 and 5) or in the extracts from mock-transfected cells (lanes 7 and 8). Immunoblots of the nuclear extracts with Ab-284 indicated that there was no decrease in topo IIα levels in C33A cells transfected with either wild-type Rb (Fig. 5B Upper, lane 1) or mutant Rb (Fig. 5B Upper, lane 2). Further, transient transfection of C33A cells with wild-type or mutant Rb did not alter their cell cycle distribution (data not shown). Finally, expression of wild-type or mutant Rb was confirmed by immunoblotting (Fig. 5B Lower). This experiment clearly shows that topo II activity is inhibited by wild-type but not mutant Rb.
Figure 5.
(A) Topo II activity in C33A cells transfected with wild-type or mutant Rb. The cytomegalovirus expression vectors were transfected in C33A cells, nuclear proteins were extracted, and topo II activity was measured by decatenation of k-DNA. Displayed here is a representative (negative) image of a 1% agarose gel (ethidium bromide-stained) showing the decatenation of k-DNA by the indicated amounts of nuclear extracts. Lanes: 1 and 2, extracts from wild-type Rb transfected cells; 4 and 5, extracts from mutant Rb-transfected cells; 7 and 8, extracts from mock-transfected cells; 9, no extract control; 10, catenated k-DNA marker; 11, decatenated k-DNA marker; 12, linear k-DNA marker. Lanes 3 and 6 were left blank. (B) Immunoblots of nuclear extracts from C33A cells transfected with wild-type Rb (379–928) (lane 1) or mutant Rb (379–928) (lane 2) or from mock-transfected (lane 3) cells showing topo IIα (Upper) and Rb (Lower) expression. wt, wild type; mock-tran, mock-transfected; cat, catenated; decat, decatenated.
Direct Inhibition of Purified Topo IIα by Purified Wild-Type Rb.
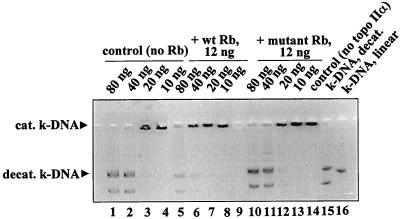
To address the possibility that inhibition of topo II activity in wild-type Rb-transfected C33A cells may be an indirect effect, we investigated whether Rb directly inhibits the activity of topo IIα in vitro. Fig. 6 compares the effects of purified wild-type [GST-Rb (379–928)] and mutant [GST-Rb (379–928), amino acid 706 C → F] Rb proteins on the decatenation activity of purified human topo IIα. Less k-DNA was decatenated when equal amounts of topo IIα were treated with wild-type Rb (lanes 5–8), compared with mutant Rb (lanes 12–14) or untreated controls (lanes 1–4). This result demonstrates direct inhibition of topo II activity by active Rb.
Figure 6.
Effect of purified wild-type and mutant Rb on activity of purified human topo IIα. Indicated amounts of purified human topo IIα were incubated with 12 ng of purified wild-type [GST-Rb (379–928)] or mutant [GST-Rb (379–928), amino acids 706 C → F] Rb. Topo II activity was measured by decatenation of k-DNA. A representative (of three separate experiments) negative image of 1% agarose gel (ethidium bromide-stained) is shown. Lanes: 1–4, untreated control (no Rb); 5–8, treated with wild-type Rb; 10–13, treated with mutant Rb; 14, control (no topo IIα); 15, decatenated k-DNA marker; and 16, linear k-DNA marker. Lane 9 was left blank.
DISCUSSION
In this report, we demonstrate a physical and functional relationship between topo IIα and Rb in vivo. Most likely, the underphosphorylated form of Rb has the major role in this interaction (20), although we have not yet determined the phosphorylation states of topo IIα and Rb involved in binding. Our limited ability to detect interactions between these phosphoproteins probably reflects the instability of topo IIα in vivo, as well as differences in the abundance of functional Rb and topo IIα during specific phases of the cell cycle (8, 19). The likelihood that such interactions involve a DNA intermediate is remote, because neither ethidium bromide (29) nor DNase I (30) abolished or diminished this binding. Moreover, purified wild-type Rb directly inhibited the catalytic activity of purified topo II. However, the participation of a protein intermediate cannot be ruled out.
We used various truncated GST-Rb fusion proteins (25) to confirm the Rb–topo IIα interaction in vitro and to identify topo IIα-binding domains in Rb. Although the growth- suppressing activity of Rb exerted through E2F transcription factors is well known (20), the additional functions of Rb mediated through binding to other cellular proteins is only beginning to emerge (20). Rb interacts physically and functionally with a large number of cellular proteins (20, 38), a majority of which bind to either the A/B pocket domain (amino acids 379–792) or the large pocket (amino acids 379–928). In most instances, this binding causes either transcriptional activation or repression. Several viral oncoproteins, such as adenovirus E1A, simian virus 40 large T antigen, and papilloma virus E7, inhibit Rb function through binding to the A/B pocket domain (33, 37). In our study, topo IIα bound strongly to the GST-Rb fusion protein containing the A/B pocket and the spacer (I) domain (amino acids 573–648). However, further study is needed to determine whether this site is the same as that recognized by viral proteins or whether it is merely required for proper folding of Rb. Finally, the binding of topo IIα could displace other proteins whose interactions with Rb are essential for its normal function.
There are several precedents for the modulation of topo IIα activity by protein–protein interactions (39). For example, casein kinase binds strongly to topo IIα and phosphorylates it (40), whereas transcription factors such as CREB, ATF-2, and c-Jun appear to interact with topo IIα and stimulate its DNA decatenation activity (41). Topo IIα also forms heterodimers with topo IIβ in vivo as a means of maintaining its catalytic activity (42). Other proteins shown to interact with topo IIα include p53 (30), barren product (a chromosome-associated protein) (43), and sgs1, a eukaryotic homolog of E. coli Rec Q (44). Although several proteins isolated from HeLa cell nuclear extracts also interact with topo IIα, all but one (epsilon isoform of 14-3-3) remain to be characterized (39).
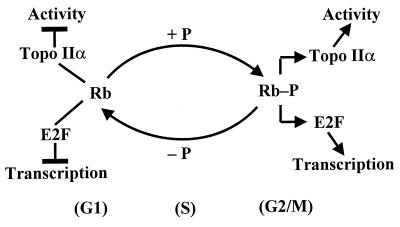
Our results with C33A cells showed that topo II activity is inhibited by ectopically expressed wild-type Rb but not by mutant Rb. Treatment of purified topo IIα with purified wild-type Rb clearly showed that this inhibition is direct and not mediated through growth control by Rb. Conceivably, underphosphorylated Rb could interact with both E2F transcription factors and topo IIα during late M and G1 phases of the cell cycle, inhibiting E2F-dependent transcription as well as topo II activity. During late G1 and early S phases, Rb may release the E2F factors and topo IIα, allowing the transcription of growth promoters and subsequent DNA replication and mitosis. Topo IIα also could be released from Rb as a consequence of competitive binding with E2F. We have proposed a model of these interactions in Fig. 7. The above hypothesis and model would accommodate the functional role of Rb during S phase progression (45).
Figure 7.
Suggested model for the regulatory role of topo IIα-Rb interactions in vivo.
In summary, we have shown that topo IIα physically associates with Rb in several cell types in a manner that suggests physiological significance. Most likely, underphosphorylated Rb inhibits topo II activity during late M and G1 phases of the cell cycle, curtailing the enzyme’s functional roles until S phase progression begins. Studies to pinpoint the consequences of topo IIα-Rb interaction are underway.
Acknowledgments
We thank Dr. David M. Livingston (Dana–Farber Cancer Institute) for providing several recombinant GST-Rb fusion plasmids and Dr. Neil Osheroff (Vanderbilt University Medical Center) for his generous gift of purified human topoisomerase II. We are grateful to Mr. John R. Gilbert (Cookeville, TN) for excellent editorial assistance. These studies were supported, in part, by Research Grants CA-40570 and CA-30103 from the National Cancer Institute, Bethesda, MD (to W.T.B.), by the American Cancer Society (BE-219A) (P. R), and by the Cancer Center of the University of Illinois at Chicago.
ABBREVIATIONS
- RMS
rhabdomyosarcoma
- topo IIα
DNA topoisomerase IIα
- Rb
retinoblastoma protein
- GST
glutathione S-transferase
- k-DNA
kinetoplast DNA
References
- 1.Watt P M, Hickson I D. Biochem J. 1994;303:681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burden D A, Osheroff N. Biochim Biophys Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 3.Nitiss J L. In: DNA Topoisomerases in DNA and DNA Damage Tolerance. Nickloff J A, Hokstra M F, editors. Clifton, NJ: Humana; 1996. [Google Scholar]
- 4.Fernandes D J, Qiu J, Catapano C V. Adv Enzyme Regul. 1995;35:265–281. doi: 10.1016/0065-2571(94)00009-r. [DOI] [PubMed] [Google Scholar]
- 5.Nitiss J L, Beck W T. Eur J Cancer. 1996;32A:958–956. doi: 10.1016/0959-8049(96)00056-1. [DOI] [PubMed] [Google Scholar]
- 6.Beck W T. In: Principles of Antineoplastic Drug Development and Pharmacology. Schilsky R L, Milano G A, Ratain M J, editors. New York: Dekker; 1997. [Google Scholar]
- 7.Tan K B, Dorman T E, Falls K M, Chung T D Y, Mirabelli C K, Crooke S T, Mao J. Cancer Res. 1992;52:231–234. [PubMed] [Google Scholar]
- 8.Kimura K, Saijo M, Ui M, Enomoto T. J Biol Chem. 1994;269:1173–1176. [PubMed] [Google Scholar]
- 9.Downes C S, Mulliger A M, Johnson R T. Proc Natl Acad Sci USA. 1991;88:8895–8899. doi: 10.1073/pnas.88.20.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uemura T, Ohkura H, Adachi Y, Marino K, Shiozaki K, Yanagida M. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 11.Adachi Y, Luke M, Laemmli U K. Cell. 1991;64:137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- 12.Brill S J, DiNardo S, Voelket-Meiman K, Sternglanz R. Nature (London) 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 13.Riley D J Y, Lee H-P, Lee W-H. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 14.Sellers W R, Kaelin W G., Jr J Clin Oncol. 1997;15:3301–3312. doi: 10.1200/JCO.1997.15.11.3301. [DOI] [PubMed] [Google Scholar]
- 15.Weinberg R A. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 16.Bagchi S, Weinmann R, Raychaudhuri P. Cell. 1991;65:1063–1072. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- 17.Chellappan S P, Heibert S, Mudryj M, Horowitz J M, Nevins J R. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 18.Mittnacht S. Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 19.Ludlow J W, Shen J, Pipas J M, Livingston D M, DeCaprio J A. Cell. 1990;60:387–396. doi: 10.1016/0092-8674(90)90590-b. [DOI] [PubMed] [Google Scholar]
- 20.Taya Y. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 21.Wang J Y, Knudsen E S, Welch P J. Adv Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- 22.Hazleton B A, Houghton J A, Parham D M, Douglas E C, Torrance P M, Holt H, Houghton P J. Cancer Res. 1987;47:4501–4507. [PubMed] [Google Scholar]
- 23.Xiao Z-X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Nature (London) 1995;375:694–697. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli E K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Kaelin W G, Jr, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 26.Hamanaka R, Kohno K, Seguchi T, Okamura K, Morimoto A, Ohno M, Ogata J, Kuwano M. J Biol Chem. 1992;267:13160–13165. [PubMed] [Google Scholar]
- 27.Arroyo M, Raychaudhuri P. Nucleic Acids Res. 1992;20:5947–5954. doi: 10.1093/nar/20.22.5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wigler M, Siverstein S, Lee L S, Pellicer A, Cheng Y C, Axel R. Cell. 1977;11:223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]
- 29.Nead M A, Bagila L A, Antinore M J, Ludlow J W, McCance D J. EMBO J. 1998;17:2342–2352. doi: 10.1093/emboj/17.8.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuwen H, Hsia C C, Nakashima Y, Evangelista A, Tabor E. Biochem Biophys Res Commun. 1997;234:194–197. doi: 10.1006/bbrc.1997.6539. [DOI] [PubMed] [Google Scholar]
- 31.Knudsen E, Wang J Y. Mol Cell Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dyson N, Guida P, Munger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 33.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 34.Welch P J, Wang Y. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 35.Mo Y, Beck W T. Oncol Res. 1997;9:193–204. [PubMed] [Google Scholar]
- 36.Boege F. Eur J Clin Chem Clin Biochem. 1996;34:873–878. [PubMed] [Google Scholar]
- 37.Kaelin W G, Jr, Ewen M E, Livingston D M. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulligan G, Jacks T. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 39.Kroll D J. Arch Biochem Biophys. 1997;345:175–184. doi: 10.1006/abbi.1997.0267. [DOI] [PubMed] [Google Scholar]
- 40.Bojanowski K, Filhol O, Cochet C, Chambaz E M, Larsen A K. J Biol Chem. 1993;268:22920–22926. [PubMed] [Google Scholar]
- 41.Kroll D J, Sullivan D M, Gutierrez-Hartmann A, Hoeffler J P. Mol Endocrinol. 1993;7:305–318. doi: 10.1210/mend.7.3.8387155. [DOI] [PubMed] [Google Scholar]
- 42.Diersack H, Jensen S, Gromova I, Nielson I S, Westergaard O, Anderson A H. Proc Natl Acad Sci USA. 1996;93:8288–8293. doi: 10.1073/pnas.93.16.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhat M A, Philip A V, Glover D M, Bellen H J. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- 44.Watt P M, Louis E J, Borts R H, Hickson I D. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 45.Knudsen E S, Buckmaster C, Chen T T, Feramisco J R, Wang J Y. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]