Abstract
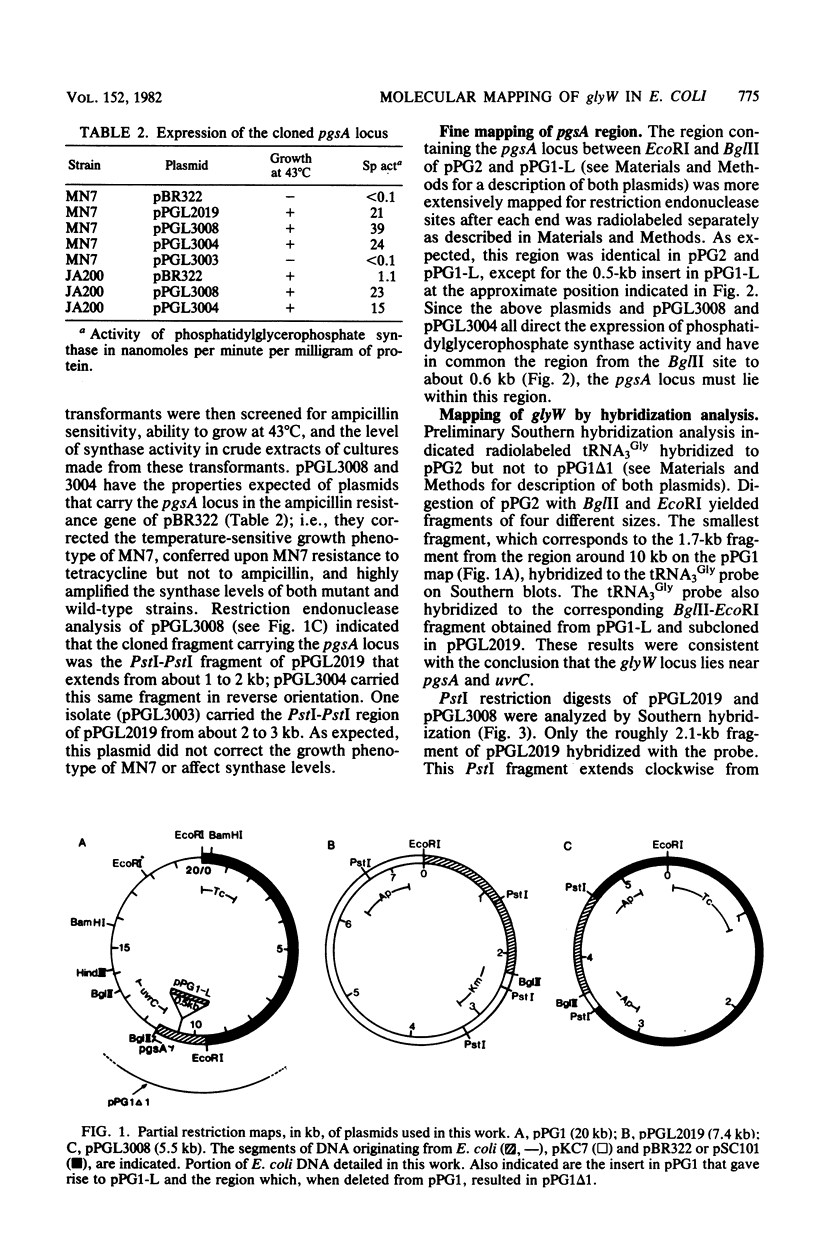
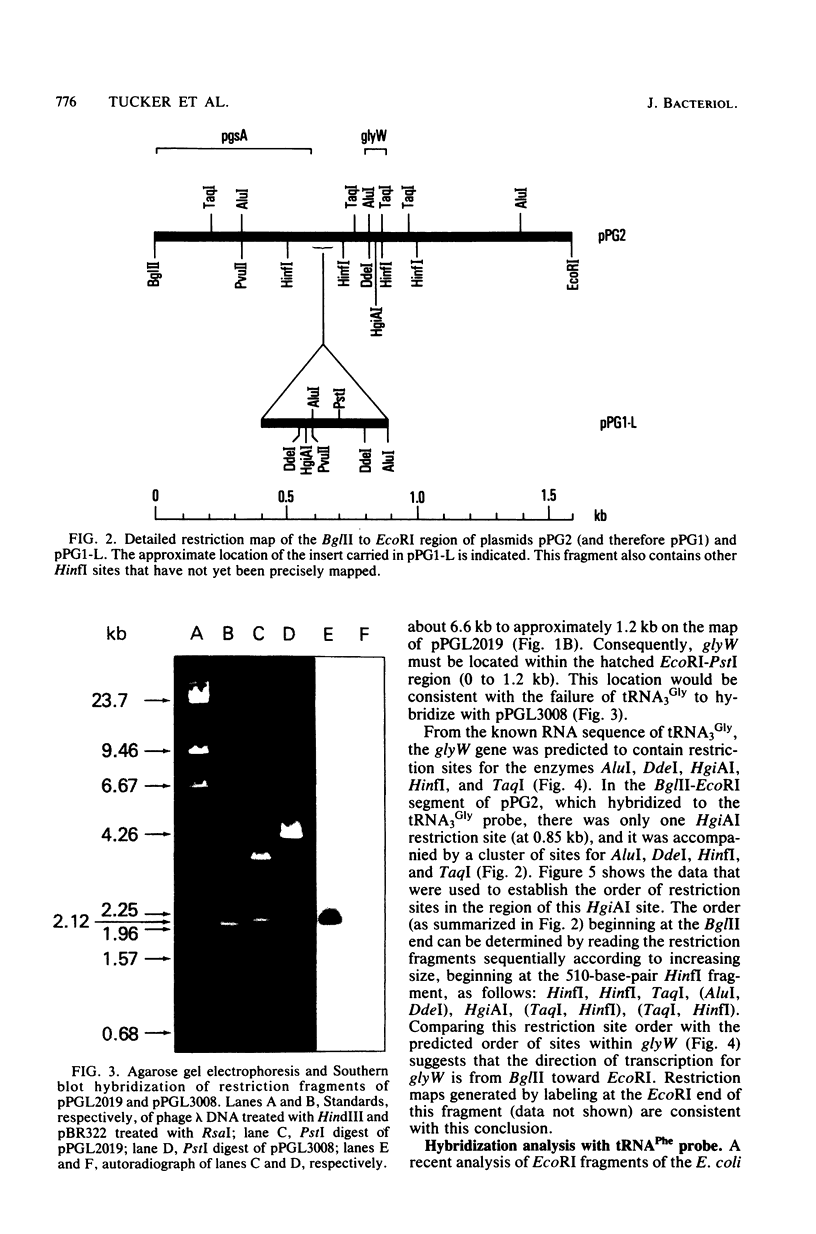
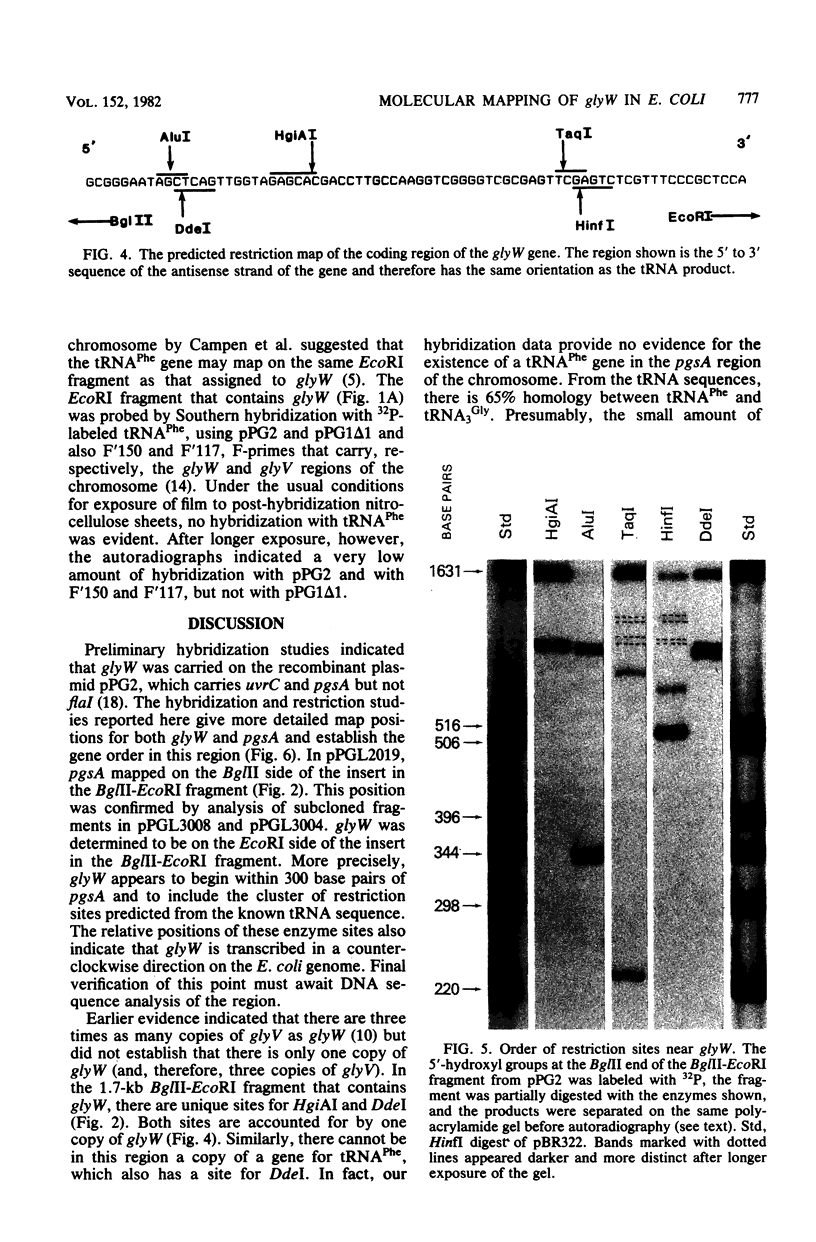
By the use of [5'-32P]tRNA3Gly from Escherichia coli as a hybridization probe, glyW was located on cloned fragments of the uvrC pgsA region of the bacterial chromosome. After determination of the sites of action of several restriction enzymes, glyW was found to be within approximately 300 base pairs of pgsA. The order of genes in this region is uvrC, pgsA, glyW, flaI. Comparison of the order of determined restriction sites with the sites predicted from the nucleotide sequence of tRNA3Gly indicates that the direction of transcription of glyW is counterclockwise on the circular E. coli map.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Gesteland R. F., Reid B. R., Anderson C. W. Normal tRNAs promote ribosomal frameshifting. Cell. 1979 Dec;18(4):1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi A., Bernardi F. Complete sequence of an IS element present in pSC101. Nucleic Acids Res. 1981 Jun 25;9(12):2905–2911. doi: 10.1093/nar/9.12.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2(2):75–93. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Campen R. K., Duester G. L., Holmes W. M., Young J. M. Organization of transfer ribonucleic acid genes in the Escherichia coli chromosome. J Bacteriol. 1980 Dec;144(3):1083–1093. doi: 10.1128/jb.144.3.1083-1093.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon J., Squires C., Hill C. W. Glycine transfer RNA of Escherichia coli. II. Impaired GGA-recognition in strains containing a genetically altered transfer RNA; reversal by a secondary suppressor mutation. J Mol Biol. 1970 Sep 28;52(3):571–584. doi: 10.1016/0022-2836(70)90420-1. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck E. W., Carbon J. Multiple gene loci for a single species of glycine transfer ribonucleic acid. J Bacteriol. 1975 May;122(2):492–501. doi: 10.1128/jb.122.2.492-501.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson L., Rossi J., Landy A. Dual function transcripts specifying tRNA and mRNA. Nature. 1981 Dec 3;294(5840):422–427. doi: 10.1038/294422a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgen C., Kirk L. L., Carbon J. Isolation and characterization of large transfer ribonucleic acid precursors from Escherichia coli. J Biol Chem. 1976 Feb 25;251(4):922–929. [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J., Pagel F. T. Codon recognition by glycine transfer RNAs of Escherichia coli in vivo. J Mol Biol. 1980 Apr 25;138(4):833–844. doi: 10.1016/0022-2836(80)90067-4. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Prather N. E., Hadley K. H. Variations among glyV-derived glycine tRNA suppressors of glutamic acid codons. J Bacteriol. 1978 Jun;134(3):801–807. doi: 10.1128/jb.134.3.801-807.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijima M., Bulawa C. E., Raetz C. R. Two interacting mutations causing temperature-sensitive phosphatidylglycerol synthesis in Escherichia coli membranes. J Bacteriol. 1981 Jan;145(1):113–121. doi: 10.1128/jb.145.1.113-121.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta A., Waggoner K., Radominska-Pyrek A., Dowhan W. Cloning of genes involved in membrane lipid synthesis: effects of amplification of phosphatidylglycerophosphate synthase in Escherichia coli. J Bacteriol. 1981 Aug;147(2):552–562. doi: 10.1128/jb.147.2.552-562.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Ohta A., Dowhan W., Moses R. E. Cloning of the uvrC gene of Escherichia coli: expression of a DNA repair gene. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6033–6037. doi: 10.1073/pnas.78.10.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Squires C., Carbon J., Hill C. W. Glycine transfer RNA of Escherichia coli. I. Structural genes for two glycine tRNA species. J Mol Biol. 1970 Sep 28;52(3):557–569. doi: 10.1016/0022-2836(70)90419-5. [DOI] [PubMed] [Google Scholar]
- Squires C., Carbon J. Normal and mutant glycine transfer RNAs. Nat New Biol. 1971 Oct 27;233(43):274–277. doi: 10.1038/newbio233274a0. [DOI] [PubMed] [Google Scholar]