Summary
Many microtubule (MT) structures, such as the mitotic spindle, contain dynamic MTs that are bundled and stabilized in overlapping arrays. CLASPs are conserved MT-binding proteins that have been implicated in the regulation of MT plus ends. Here, we show that in the fission yeast Schizosaccharomyces pombe, the CLASP ortholog cls1p/peg1p mediates the stabilization of overlapping MT bundles. Cls1p localizes to regions of stable overlapping MTs within the mitotic spindle and interphase MT bundles, but was not detectable on growing interphase MT plus ends. Inactivation of cls1p leads to the rapid depolymerization of spindle midzone MTs, which normally persist through mitosis. Cls1p is also necessary for stabilization of a subset of MTs within overlap zones of interphase bundles and is critical for the maintenance of these bundles in the absence of MT nucleation. Loss-of-function and overexpression phenotypes indicate that cls1p functions by preventing MT disassembly while still allowing for MT plus end growth. Cls1p has no measurable effects on MT nucleation, polymerization, catastrophe, or bundling. A direct interaction between cls1p and the MT-bundling protein ase1p targets cls1p to regions of MT overlap. These findings show how a MT-stabilizing factor attached to specific sites on MTs can help to generate MT structures that have both dynamic and stable components.
Introduction
Spatial regulation of microtubule (MT) organization and dynamics is responsible for the complex assembly of MT-based structures such as the mitotic spindle (Karsenti et al., 2006; Nedelec et al., 2003). The stabilization of MTs underlies mechanisms of spindle assembly, chromosome segregation, cytokinesis, and polarization of interphase arrays in many cell types. Certain MT bundles contain MTs that are dynamic yet at the same time, resistant to complete depolymerization (Baas and Black, 1990; Eichenlaub-Ritter and Ruthmann, 1982; Salmon and Begg, 1980). Mechanisms for how MT dynamics may be regulated in this manner at only certain cellular locales are not understood.
The MT-binding protein CLASP is emerging as an important regulator of MT dynamics. In cultured mammalian cells, CLASP resides at plus ends of interphase MTs (Akhmanova and Hoogenraad, 2005; Akhmanova et al., 2001). Depletion studies suggest a role for CLASPs in enrichment of MT plus ends at the leading edge; time-lapse images suggest that they promote MT rescue and/or pause events at the cortex (Mimori-Kiyosue et al., 2005). “Pioneer” MTs at the leading edge of migrating cells, which grow through the dense actin-rich network without shrinking, are specifically decorated along the lattice with CLASP through a Rac-mediated pathway (Wittmann and Waterman-Storer, 2005). CLASP also functions during mitosis, where it is present on kinetochores and spindle MTs (Maiato et al., 2003). Loss of CLASP function causes many types of mitotic spindle failure, which vary according to cell type and methods of disrupting CLASP function (Hannak and Heald, 2006; Inoue et al., 2000; Inoue et al., 2004; Lemos et al., 2000; Maiato et al., 2003; Mimori-Kiyosue et al., 2006; Pasqualone and Huffaker, 1994; Pereira et al., 2006). In particular, CLASP is needed for promoting MT plus end growth at kinetochores in prometaphase (Maiato et al., 2005). The molecular mechanism of how CLASP regulates MT dynamics is still far from clear. Generally, domains of CLASP interact with MTs, EB1, and CLIP170 (Akhmanova et al., 2001; Mimori-Kiyosue et al., 2005; Wittmann and Waterman-Storer, 2005; Yin et al., 2002). CLASP also localizes in some cell types at sites of MT nucleation, near the minus end of MTs, at centrosomes, and also at sites on the Golgi apparatus (Akhmanova et al., 2001; Efimov et al., 2007; Lemos et al., 2000; Maiato et al., 2003).
The fission yeast Schizosaccharomyces pombe serves as a simple model cell for studying MT dynamics and organization. These cells exhibit two types of structures that contain bundles of stable overlapping anti-parallel MTs: the interphase MT bundles and the mitotic spindle. These structures are characterized by square-packed MTs linked by short fibrils of unknown composition (Ding et al., 1993; Hoog et al., 2007). During interphase, MTs are organized in 3–5 bundles, each containing on average 4–5 MTs, which are arranged in an anti-parallel configuration (Hoog et al., 2007; Sawin and Tran, 2006). The MT plus ends grow from the medial overlap zone to the cell tip, contact the cell tip and then exhibit catastrophe and shrink back to the overlap zone. These bundles are maintained by nucleation of new MTs from γ-tubulin complexes on the MTs, which move in a kinesin-dependent manner to the medial overlap zone (Carazo-Salas et al., 2005; Janson et al., 2007; Janson et al., 2005). In addition, these bundles have a short region of MT stabilization within the overlap zone, as shown by resistance to the MT inhibitor methyl-2-benzimidazole carbamate (MBC) (Tran et al., 2001). This stabilization zone does not appear to be a canonical MT-organizing center, as these bundles can “self-organize” and form this MBC-resistant stabilization zone even in anucleate cells (Carazo-Salas and Nurse, 2006; Daga et al., 2006).
During mitosis, MTs are organized from the spindle poles and bundled into an intra-nuclear spindle. In early mitosis, about half of the spindle MTs are overlapping interpolar MTs. At anaphase onset, approximately 18 kinetochore MTs segregate the 6 chromatids to the spindle poles within 1 minute of anaphase onset, and 10–20 interpolar MTs push the poles apart for anaphase B (Ding et al., 1993; Khodjakov et al., 2004; Nabeshima et al., 1998; Tolic-Norrelykke et al., 2004). These overlapping MTs represent a highly stable population: the same MTs that form during initial phases of spindle assembly in prometaphase persist through late anaphase (Khodjakov et al., 2004; Mallavarapu et al., 1999). Response to laser cuts suggests that stabilization zones in the spindle correspond to regions of overlapping MTs (Ding et al., 1993; Khodjakov et al., 2004). MT plus ends of these MTs still grow for spindle elongation and exhibit shrinkage events, but do not generally shrink past the midzone region until late anaphase (Sagolla et al., 2003), suggesting that MT rescue occurs in the midzone. In both the spindle and interphase arrays, the molecular bases for regulating these complex MT behaviors are not well understood.
One important factor for assembly of these overlapping MT arrays is a conserved MT-bundling protein, ase1p (PRC1/MAP65) (Loiodice et al., 2005; Yamashita et al., 2005). Ase1p decorates precisely regions of MT antiparallel overlap (Janson et al., 2007). ase1Δmutants are viable, but exhibit interphase MTs with reduced bundling and mitotic spindles that often fall apart in anaphase.
Fission yeast has a sole CLASP ortholog, cls1p/peg1p, which has significant homology to other CLASPs. In a recent study (Grallert et al., 2006), this fission yeast CLASP was found to localize to the spindle and to be necessary for spindle assembly. This study also claimed that CLASP localizes to growing MT plus ends and functions with dynein to induce MT catastrophe of plus ends at cell tips.
Here, in an independent study, we were surprised to find that this S. pombe CLASP homolog is generally not present at interphase MT plus ends, but resides in regions of stable MTs within the overlap zone of interphase bundles and the spindle midzone. We demonstrate that CLASP does not appear to affect general MT plus end dynamics, but is responsible for MT stabilization within these bundles. Further, a direct interaction with ase1p targets CLASP to these sites. This work describes a new mechanism for formation of a dynamic yet stable array of overlapping MTs.
Results
Cls1p Localizes to Regions of MT Overlap
To determine the localization of cls1p, we constructed a functional cls1-3GFP fusion expressed at endogenous levels from the cls1 chromosomal locus. In mitotic cells, cls1p was nuclear and accumulated along a subset of mitotic MTs (Figures 1A and S1A), similar to a previous report (Grallert et al., 2006). In pre-anaphase spindles, cls1p localized along the length of the spindle. In elongating anaphase B (phase III) spindles, cls1p was clearly concentrated to the spindle midzone region (average length of cls1p localization: 2.3±0.4μm; n=15). Short movements of cls1p were confined to this region (Figure S1B). Cls1p was not present in the region of the spindle poles or on cytoplasmic astral MTs during anaphase. This pattern of cls1p localization corresponded to predicted regions of MT stabilization and square-packed overlapping interpolar MTs (Ding et al., 1993; Khodjakov et al., 2004).
Figure 1. Cls1p localizes to overlapping MTs and kinetochores.
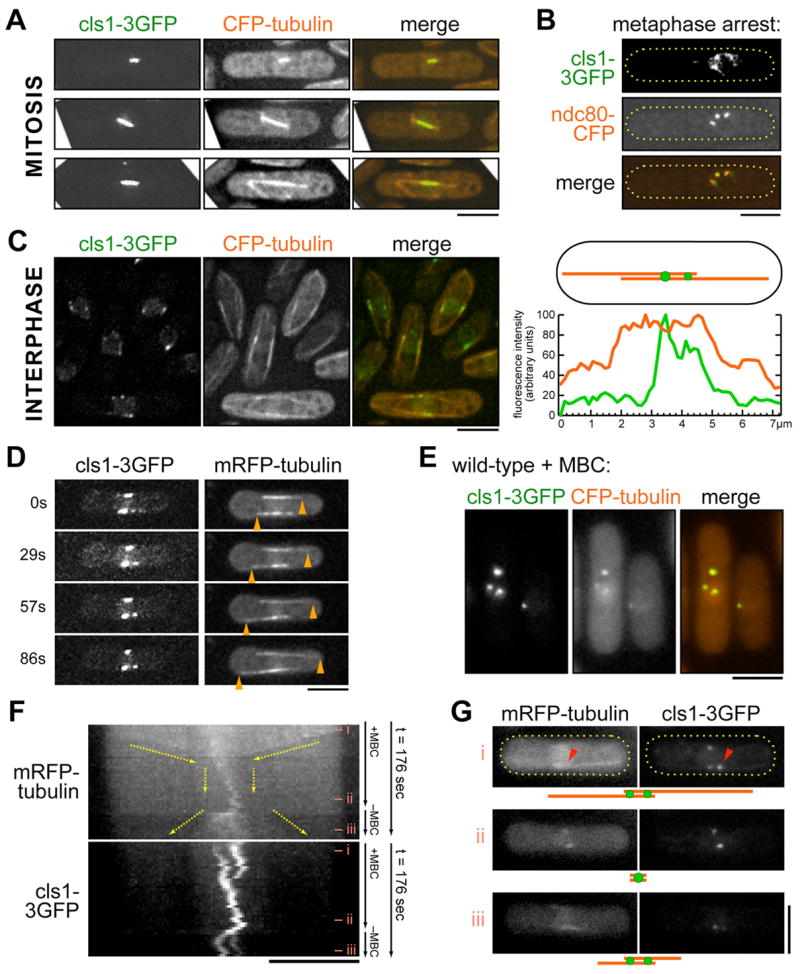
(A) Mitotic cells expressing cls1-3GFP and CFP-tubulin in early mitosis (top), metaphase (middle), and anaphase (bottom). Images in (A-D) are maximum projections of deconvolved stacks. Scale bar: 5μm.
(B) Cells expressing cls1-3GFP and the kinetochore marker ndc80-CFP in a tubulin mutant nda3-311, which arrests in metaphase without a spindle.
(C) Interphase cells expressing cls1-3GFP and CFP-tubulin. The graph shows fluorescence intensity profile of cls1-3GFP and CFP-tubulin along one representative bundle. Note localization of cls1p to dot or dots within MT overlaps, as shown by medial regions of increased tubulin fluorescence.
(D) Time-lapse images of interphase cell expressing cls1-3GFP and mRFP-tubulin. Arrowheads mark growing MT plus ends.
(E) Interphase cells expressing cls1-3GFP and CFP-tubulin were treated with MBC for 10 min. Maximum projection images are shown.
(F,G) Time-lapse of cell expressing cls1-3GFP and mRFP-tubulin and treated with MBC in a flow chamber. Kymographs of the indicated bundle (arrowheads)—constructed from single-plane time-lapse images (0.25 fps)—show MT plus ends (yellow arrows) shrink toward cls1p dots within the overlap zone. A region within the overlap zone is stable and can support MT regrowth once MBC is removed. For selected time points, raw images and schematic interpretations are shown (G).
Cls1p also localized to pre-anaphase kinetochores. As it was not possible to distinguish kinetochore staining from MT staining in pre-anaphase spindles, we used the nda3-311 (β-tubulin) strain, which arrests in metaphase without any spindle MTs (Hiraoka et al., 1984). These arrested cells exhibited three nuclear dots of cls1p, which colocalized with a kinetochore marker ndc80-CFP (Figure 1B). During anaphase, cls1p was not detectable on kinetochores, which are situated in wild-type cells near the spindle poles (Figures 1A and S1A; Nabeshima et al., 1998). These data are consistent with findings of cls1p association with CEN DNA by CHIP analyses (Grallert et al., 2006).
During interphase, cls1p localized to a small number of dots (2–8 per cell) on cytoplasmic MT bundles (Figure 1C), in addition to a faint, diffuse nuclear localization. Although Grallert et al. (2006) interpreted this cytoplasmic staining pattern to include growing MT plus ends, we did not detect cls1p on growing plus ends of interphase MTs (see below). Rather, the cytoplasmic dots localized within medial regions of antiparallel MT overlap, in the vicinity of MT minus ends. These dots decorated a region smaller than the overlap zone. Often multiple (2 or 3) cls1-3GFP dots of various intensities were present on a single MT bundle within the overlap zone. These dots moved in short excursions back and forth in the cell, and were constrained to the overlap zone (Figure S2A). Movements were MT-dependent, as they were abolished by treatment with the MT inhibitor MBC (data not shown). Within individual MT bundles, cls1p dots frequently coincided with edges of an overlap zone, suggestive of an association near MT minus ends. One dot always colocalized with the spindle pole body (SPB), as shown with a SPB marker, cut12-CFP (Figure S1A; Grallert et al., 2006).
S. pombe CLASP did not behave like bona fide +TIP proteins such as tip1p (CLIP170). Unlike +TIPs, cls1p dots did not exhibit directed movement all the way to the cell tips. Dual imaging of cls1-3GFP and MTs did not reveal any detectable cls1p on MT plus ends growing to cell tips (Figures 1D and S2A). Furthermore, cls1p did not colocalize with tip1p at MT plus ends (Figure S2C). To test whether cls1p levels were simply too low to visualize, or if the C-terminal fusion disrupted plus end localization, we mildly overexpressed (20–30 fold) GFP-cls1p (Figure S2D). This fusion, which was also functional, labeled more intensely dots near the nucleus and faintly along the rest of the MT lattice, but still no accumulation was observed at plus ends. Thus, fission yeast CLASP does not appear to track growing cytoplasmic MT plus ends.
Interphase MT bundles have a zone that is stabilized against MT depolymerization. In wild-type cells, treatment with MBC depolymerizes the MTs and leaves 2–5 stable MT remnants (or stubs) around the nucleus (Tran et al., 2001). These zones of stabilization were marked by cls1p (Figure 1E). By imaging cells before and during MBC treatment, we observed that within a minute of MBC treatment, MTs shrank toward regions marked by dots of cls1p within the overlap zone (Figures 1F and 1G). It is notable that this stable region marked by cls1p was often smaller than the whole overlap zone. Upon washout of the drug, the MT bundles regrew from this stub, and cls1p maintained its association with MTs at the edges of the overlap zone. Cls1p is the first protein demonstrated to be located specifically at this stabilization region. Thus, cls1p defines a stable region within the MT overlap.
Measurements of cls1-3GFP fluorescence intensities indicated that cls1p is a non-abundant protein with approximately 500 molecules per interphase cell. Each cls1p dot on MT bundles was estimated to represent a small number of molecules (mean=13±10, median=10, n=66; Figure S3). Total cls1p levels were 50% higher in mitotic cells. Within the spindle, the estimated number of cls1-3GFP molecules ranged from 83 to 389 (mean=227±94, median=216, n=13), representing on average ~30% of the total amount of cls1p in the cell. Assuming an average of 20 MTs per spindle (Ding et al., 1993; see Supplemental Methods), this suggested that there are around 11 molecules of cls1p per MT in spindles.
Cls1p is Required for the Stabilization of Spindle MTs
Cls1 is an essential gene, as cls1Δspores arrest in mitosis with spindle defects (Figure S4; Grallert et al., 2006). We therefore generated a set of conditional temperature-sensitive cls1 alleles (Figure S5). We used primarily the cls1-36 allele, which was inactivated within minutes at 30°C and was the most severe allele obtained. cls1-36 is a recessive loss-of-function allele, as a cls1+/cls1-36 heterozygous diploid was viable at 30°C and 36°C. This allele allowed us to examine the consequence of cls1p loss-of-function at different points in mitosis.
When cls1p was inactivated prior to mitosis, cells entered mitosis but then encountered severe defects in spindle assembly (50/50 cells; Figure 2A). Short spindles were unable to progress to a stable structure and rapidly collapsed. In these “monopolar” spindles, short dynamic MTs emanated from the clustered SPBs. The spindle poles repeatedly separated for short distances before collapsing together again. cls1-36 cells arrested in mitosis for an extended period (length of mitosis: 27±3 minutes for cls1+, 64±33 minutes for cls1-36; n=20, P<0.001) in a mad2-dependent manner (data not shown), suggesting that cls1p loss-of-function did not disrupt the spindle checkpoint.
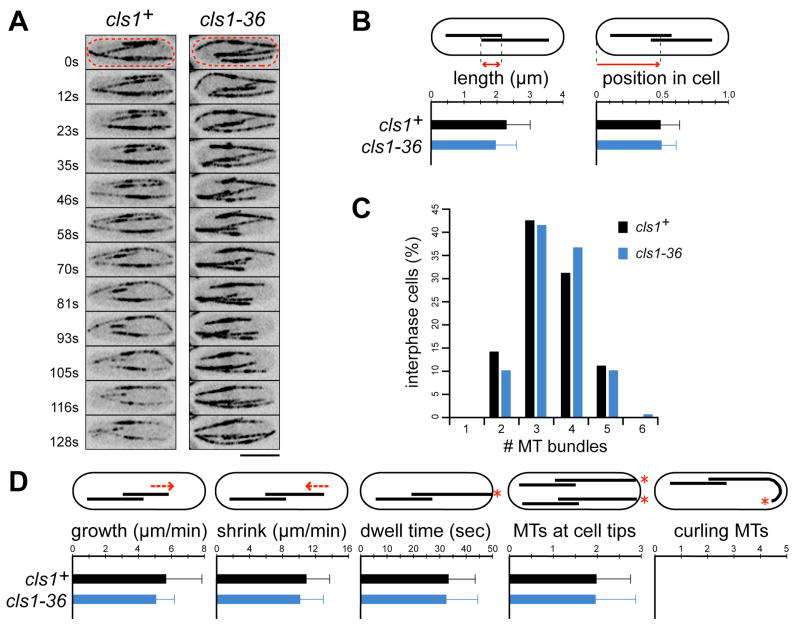
Figure 2. Cls1p is required for assembly and maintenance of a bipolar spindle.
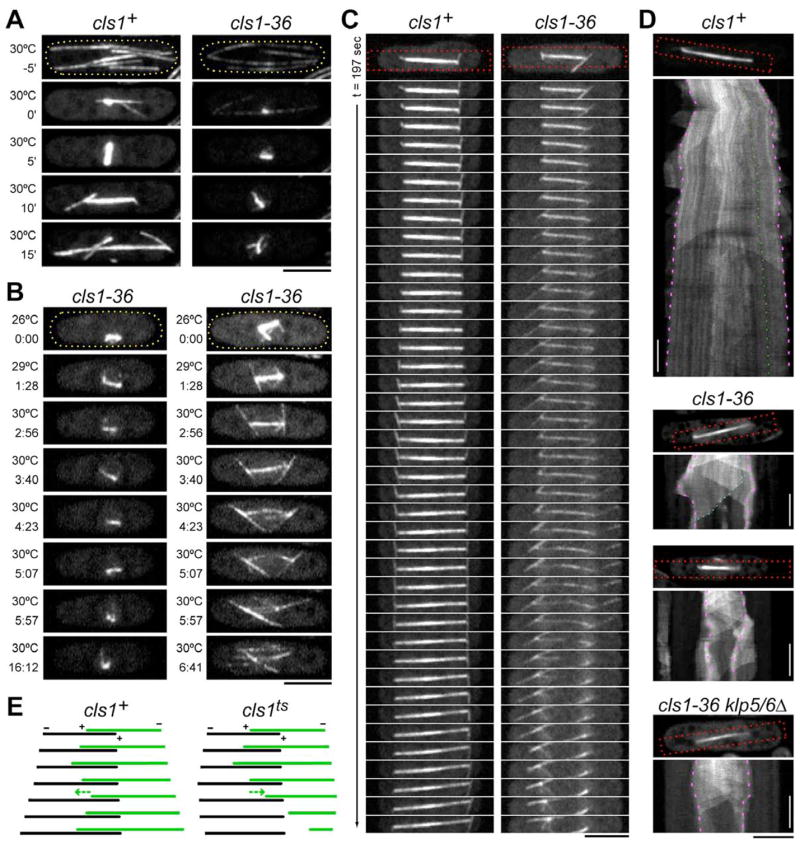
(A) Wild-type and cls1-36 cells expressing GFP-tubulin shifted to the restrictive temperature (30ºC) during interphase. Time-lapse images monitor cells as they go from interphase (top panel) into mitosis (subsequent panels). Time listed in minutes is relative to mitotic entry. Maximum projection confocal images are shown. Scale bar: 5μm.
(B) Defects in maintenance of mitotic spindle in cls1ts. cls1-36 cells were shifted to 30ºC in early mitosis (left panels) or late mitosis (right panels). Temperature and time (min:sec) are listed. (C) Anaphase spindle disassembly in cls1ts. Wild-type and cls1-36 cells were shifted to 30ºC and imaged immediately.
(D) Kymographs of anaphase spindles. Cells were shifted to 30ºC and imaged by single-plane confocal time-lapse microscopy (0.5 fps). Kymographs were constructed from the red-boxed region. In wild-type GFP-tubulin speckles (one example indicated in green) track parallel to the spindle poles (purple). MT plus ends are dynamic within the midzone but rarely disassemble completely. In cls1-36 cells and cls1-36 klp5 klp6 cells, all interpolar MTs (one plus end indicated in blue) rapidly shrink to spindle poles. Vertical scale bar: 1 min.
(E) Schematic representation of interpolar MT behavior in anaphase spindle kymographs from cls1+ and cls1ts cells. For clarity, dynamics are highlighted in a single MT (green) within a simple antiparallel bundle. In cls1+, the MT plus end is dynamic, but depolymerization halts in the medial overlap zone (midzone) and the MT regrows (arrow); spindle poles (minus ends) continuously separate due to pushing forces in the midzone. In cls1ts, a shrinking interpolar MT will not stop depolymerizing (arrow); without a midzone, spindle pole separation ceases.
In order to determine whether cls1p is important for the maintenance of a bipolar spindle, cls1-36 cells were shifted to the restrictive temperature (30°C) after initial spindle assembly (Figure 2B). When cls1p was inactivated prior to anaphase, the spindle collapsed, lost bipolarity within 1–2 minutes, and persisted in a monopolar configuration. When the temperature was shifted during anaphase, MTs disappeared starting from the middle of the spindle in 2 minutes upon reaching 30ºC. This was followed by collapse of the spindle, as shown by movement of the spindle poles back together. Spindle MTs retained their linear organization and did not splay apart (Figure 2C and Movie 1).
We next analyzed more carefully the behavior of MTs within the spindle. In wild-type cells, kymographs revealed the presence of interpolar MTs that persisted throughout anaphase without minus end flux, as shown by the behavior of GFP-tubulin speckles (Figure 2D; Khodjakov et al., 2004; Mallavarapu et al., 1999). At this stage of mitosis, approximately 11–15 interpolar MTs overlap in the midzone (Ding et al., 1993). The plus ends of these MTs exhibited growth and shrinkage, but only rarely shrank past the midzone (Figure 2D; Sagolla et al., 2003). In cls1 mutants shifted in anaphase, we observed that individual interpolar MTs depolymerized one by one in a processive manner to the poles, without pause or rescue events in the midzone (Figure 2D). GFP-tubulin speckles did not move toward the poles, suggesting that MTs were depolymerizing from their plus ends rather than their minus ends. Astral MTs still exhibited dynamic growth during this period, suggesting that loss of cls1p did not affect general MT assembly in the cell. These results, together with its localization at the spindle midzone, show that cls1p maintains spindle stability by stopping interpolar MTs from disassembling past the midzone.
We tested whether cls1p halts MT disassembly by inhibiting a MT plus end depolymerase, kinesin-8 (Gupta et al., 2006; Varga et al., 2006). S. pombe kinesin-8 proteins klp5p and klp6p localize to the spindle midzone in anaphase and affect spindle length (Garcia et al., 2002; West et al., 2002). We found that deleting klp5 and klp6 did not alter the complete depolymerization of MT plus ends in the cls1-36 mutant (Figure 2D). Thus, depolymerization of interpolar MT plus ends in cls1 mutants is not dependent on kinesin-8 function.
CLASP does not Affect Interphase MT Plus End Dynamics or Bundling
We next examined the effects of cls1p loss-of-function on interphase MTs. In cls1 mutants, interphase MTs were arranged normally, with overlap zones near the nucleus at the cell center, and dynamic plus ends extending toward cell tips (Figure 3A). MT overlap zones in cls1-36 were of normal lengths and positioned near the nucleus (Figure 3B). The number of interphase bundles, and the rates of growth and shrinkage of MT plus ends were comparable to wild-type (Figures 3C and 3D).
Figure 3. Interphase MT bundles are properly organized in cls1ts.
(A) Wild-type and cls1-36 cells expressing GFP-tubulin were imaged at 30ºC. Inverted maximum projection confocal images are shown. Scale bar: 5μm.
(B) Length of interphase MT overlap zones and their relative positions in the cell (n=50 for each strain), as determined by regions of increased GFP-tubulin fluorescence intensity in bundles.
(C) Number of MT bundles in interphase cls1+ and cls1-36 cells (n=100 cells for each strain).
(D) Quantification of interphase MT plus end behavior. Plus end growth/shrinkage rates and dwell times at cell tips (n=24 for cls1+; n=23 for cls1-36), and the number of MTs contacting cell tips at one time (n>70 cell tips for each strain) were quantified from time-lapse images of cells at 30ºC. MTs did not curl around cell tips in 36 wild-type and 41 cls1-36 cells imaged for >6 min.
Grallert et al. (2006) recently reported that the loss of cls1p/peg1p function causes a decrease in catastrophe of interphase MT plus ends at cell tips in a dynein-dependent manner. However, we were unable to detect any significant defect in catastrophe regulation of MT plus ends in cls1-36, peg1.1, or in a dynein heavy chain mutant dhc1Δ(Figures 3D and S6). In cls1-36, dwell times of MT plus ends at cell tips and the number of MTs contacting a single cell tip at any one time were normal. MT plus ends maintained contact with the cortex at cell tips and did not curl around cell tips. Occasional buckling of bundles that contacted both cell tips occurred at similar rates and extents in both wild-type and cls1-36 cells (Figure 3A and data not shown). The +TIPs tip1p (CLIP170) and mal3p (EB1) localized normally on MT plus ends, and cell polarity regulation, which depends on MT plus end factors, was not affected (data not shown; Grallert et al., 2006). Nuclear positioning was also normal (data not shown) suggesting that attachment of MT bundles to the nuclear envelope was not affected. One reason for the differences between our data and those of Grallert et al. may be their use of a GFP-α-tubulin construct that replaces one of the endogenous α-tubulin genes (see Supplemental Discussion). Our analysis of cls1p phenotypes and localization show that cls1p does not affect general MT plus end dynamics at cell tips.
Cls1p Affects Stability of Interphase MTs within Overlap Zones
As cls1p marks regions of MT overlap stable to MBC, we next examined the role of cls1p in regulating MT stability in the overlap zones of interphase MT bundles. In contrast to wild-type cells, in which stable MT remnants were maintained in the presence of MBC, such remnants were either entirely missing or very weak in cls1 and peg1.1 mutant cells at the restrictive temperature (Figures 4A and S6). Thus, cls1p is required for stabilizing a zone within MT overlapping regions.
Figure 4. Cls1p is required for interphase MT stability at overlap zones.
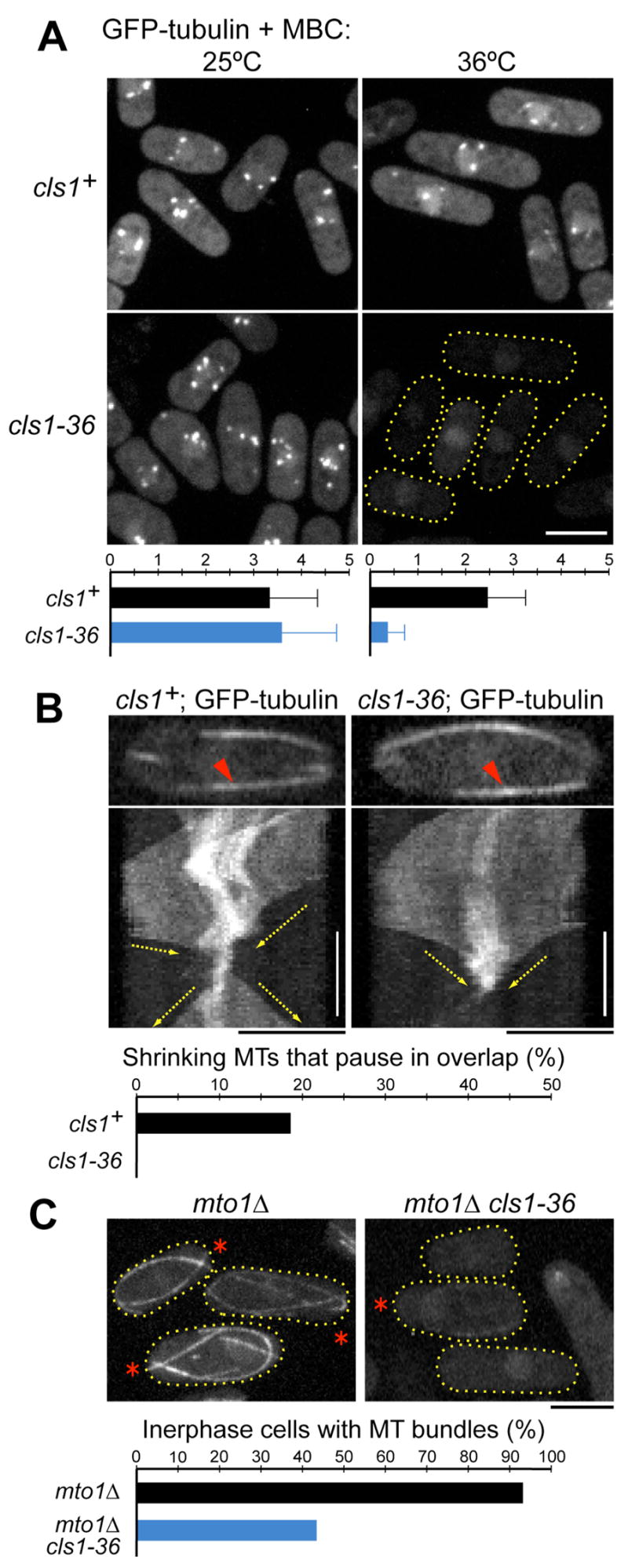
(A) Wild-type and cls1-36 cells expressing GFP-tubulin were grown at 25ºC, and then treated with MBC for 5 min at 25ºC. Cells were then shifted to 36ºC for 5 min or left at 25ºC. Maximum projection confocal images are shown. Scale bar: 5μm. Below: the number of stable MT remnants per cell (n>50 cells for each strain).
(B) MT regrowth from a short overlap zone. Cells expressing GFP-tubulin were imaged at 30ºC. Kymographs of the indicated bundle (red arrowheads) were constructed from single-plane confocal time-lapse images (0.5 fps). In wild-type, MT plus ends (yellow arrows) shrink from both sides to a short stable stub, and after a short pause MTs grow back toward cell tips. In cls1-36, the bundle disassembles completely. Vertical scale bar: 1 min. Below: the fraction of MTs that pause within an overlap zone during depolymerization (n>60 MTs for each strain).
(C) Synthetic effect of cls1 and mto1 mutations on maintenance of interphase MT bundles. Cells expressing GFP-tubulin were shifted to 30ºC for 5 min. Below: the fraction of cells (n>60 for each strain) that contain cytoplasmic MTs in interphase cells (asterisks).
The contribution of cls1p and this stable zone was more subtle in the context of the whole MT bundle. In cls1 mutants, the dynamic organization of the bundles was grossly normal, but in general, the placement and length of the overlap zones seemed more variable over time (also Grallert et al., 2006). Furthermore, we detected a quantitative defect in the stability of very short MT bundles, which occasionally form when MTs from both sides of a bundle shrink simultaneously toward the overlap zone, leaving only a short bundle of <1μm (Figures 4B and S7A). In wild-type cells these short bundles did not completely shrink away and generally regrew in a bipolar manner (13/14 events). In contrast, in cls1-36 cells about half of these short bundles (10/19 cases) depolymerized completely. Thus, these findings, which are consistent with effects in MBC-treated cells, show that cls1p is needed to stabilize short MT bundles.
Another indication of cls1p function was the pattern of MT growth from the overlap zone (Figures S7B and S7C). In wild-type cells, upon shrinkage of a MT plus end to the overlap zone, another MT grew back to replace it almost immediately. In cls1-36 cells, the time between MT shrinkage to the overlap region and MT regrowth was erratic and often delayed (51% of regrowth events in cls1-36 were delayed >30 sec, vs. 25% in wild-type).
Kymographs allowed us to analyze whether individual MTs are normally stabilized within bundles (Figure S7D). Recent data indicate that wild-type bundles are dynamic structures in which new MTs are nucleating off of pre-existing MTs, suggesting that to maintain a steady state, individual MTs must also be lost from bundles. We found that in wild-type cells, the majority of depolymerizing MTs appear to shrink thprough the overlap zone without any rescue or change in depolymerization rate. These findings disprove a simple model that cls1p stabilizes every MT at the overlap zone. However, we did note that in wild-type cells 19% of MTs shrank down to a short fragment that then paused. In the cls1-36 mutants MTs depolymerized past the overlap zone, and pause/rescue events were not observed. Together these findings show that cls1p prevents the complete depolymerization of a subset of MTs within overlap zones.
MT Bundles are Maintained by Nucleation and Stabilization
As CLASP has been proposed to affect MT nucleation (Efimov et al., 2007; Hannak and Heald, 2006), we tested the effects of cls1p on MT nucleation. First, we examined MT regrowth after MBC washout (Figure S8A). Even without starting with stable remnants, MTs quickly regrew in cls1 mutant cells, indicating that MT nucleation activity was still robust. Second, kymographs of intact MT bundles in cls1-36 mutants showed that nucleation and movement of short MTs along existing interphase MT bundles also occurred normally (Figure S7D, blue outlines). We further noted that particles of γ-tubulin complexes, as marked by mto1-GFP (Sawin et al., 2004; Zimmerman and Chang, 2005; Zimmerman et al., 2004), were localized normally on MT bundles in cls1-36 cells (Figure S8B). Thus, cls1p does not affect MT nucleation.
We reasoned that MT bundles may be maintained both by continpuous nucleation of new MTs on the bundle and by cls1p-dependent MT stabilization. We thus tested the role of cls1p in mto1 mutants in which MT nucleation is deficient (Figure 4C). Under our growth conditions, most (92%, n=65) mto1Δinterphase cells exhibited MT bundles. Strikingly, upon 5 min of shifting to 30ºC, all cytoplasmic MTs bundles were lost in the majority (56%, n=62) of cls1-36 mto1Δcells. Therefore, cls1p is critical for the maintenance of cytoplasmic MTs in the absence of MT nucleation.
Overexpression of Cls1p Promotes MT Rescue Independent of other MAPs
Cls1p overexpression led to MT stabilization in a highly specific manner. We overexpressed cls1 from its chromosomal locus by insertion of an inducible promoter upstream of cls1+. Imaging of cells overexpressing GFP-cls1p suggested that at these levels, cls1p localized to the MT lattice over much of the bundle (Figure 5A). Fluorescence intensity measurements indicated that cls1p was expressed 200–300 times higher than normal. In the cls1p-overexpressing cells, interphase MT bundles and spindles were in a grossly normal configuration and organization. However, time-lapse microscopy showed that the interphase bundles seemed largely frozen over significant lengths of time (Figure 5C and Movie 2). While in some cells MTs were entirely frozen, in others, a segment of the MTs at the plus ends remained dynamic. These MTs still grew to the cell tip, and then exhibited catastrophe (Figures 5D and 5E). But instead of shrinking back to the medial region of the cell, MT growth was promptly rescued so that MTs showed only short cycles of growth and shrinkage at their tips. The length of MT bundles stable to MBC was correlated with the level of cls1p expression (Figure 5B). Together with the loss-of-function experiments, these observations provide strong evidence that cls1p stabilizes MTs by preventing MT disassembly.
Figure 5. Overexpressed cls1p stabilizes MTs by promoting rescue.
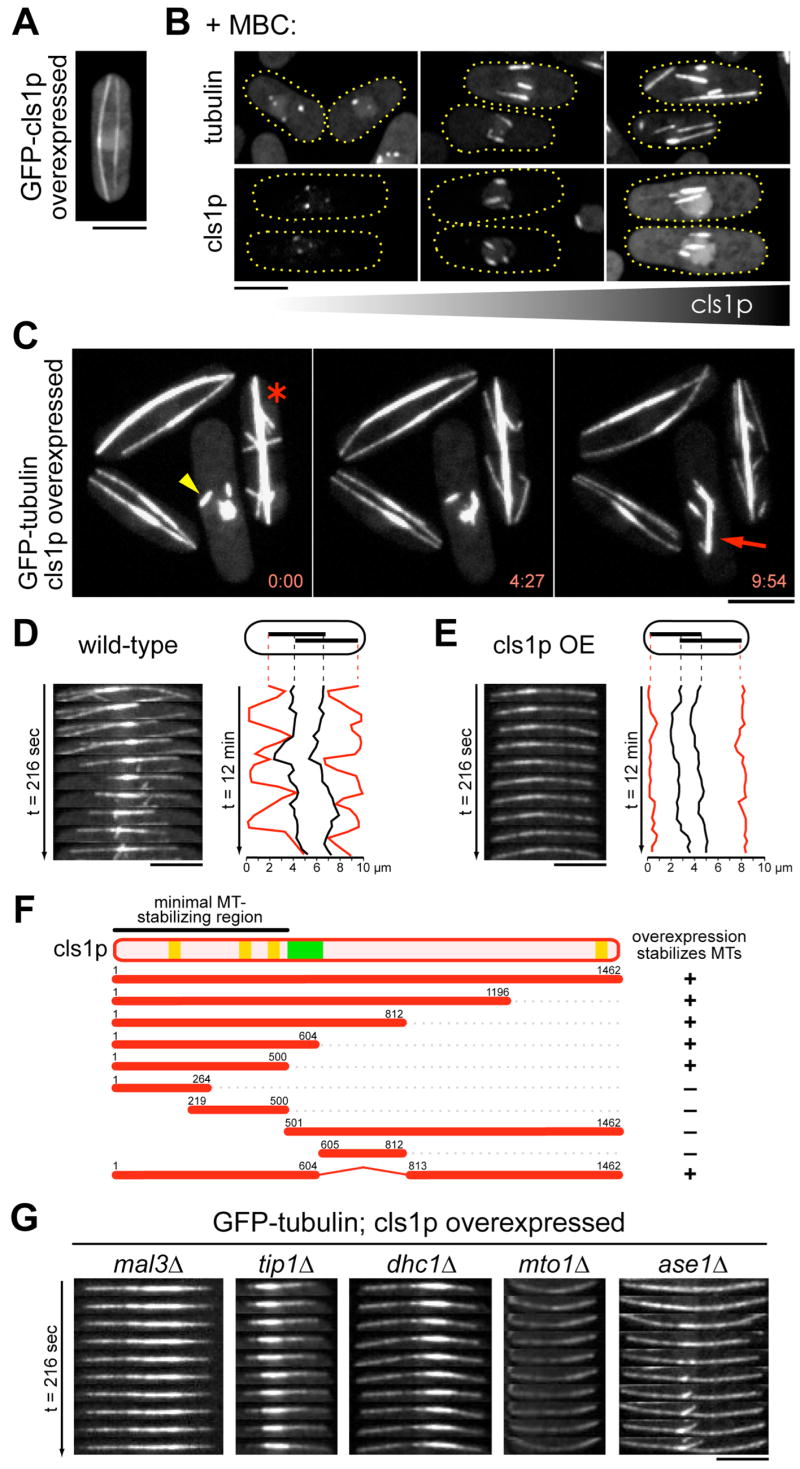
(A) Localization of GFP-cls1p overexpressed from a full strength nmt1 promoter induced by the removal of thiamine.
(B) Cls1p overexpression stabilizes MTs to MBC. Cells expressed GFP-tubulin (top panels), cls1-3GFP (bottom left), or GFP-cls1p (bottom middle, bottom right). Different levels of expression of untagged (top panels) or GFP-tagged cls1p (bottom panels) were driven by the endogenous cls1 promoter (left panels), or an integrated medium-strength (middle panels) or full-strength nmt1 promoter (right panels) in the presence of thiamine. (C,E,G) Cells expressing GFP-tubulin were induced for cls1p overexpression from an integrated full-strength nmt1 promoter by the removal of thiamine.
(C) Images of MT structures in cells overexpressing cls1p. A remnant of an interphase MT bundle persists in mitosis (yellow arrowhead). A spindle elongates (red arrow), while another fails to disassemble during septation (red asterisk). Time is in min:sec.
(D,E) Single interphase MT bundles from a wild-type cell (D) or a cls1p-overexpressing cell (E). Traces represent positions of plus ends (red) and boundaries of the overlap zone (black).
(F) An N-terminal region of cls1p is necessary and sufficient for MT stabilization. Cells were induced to express the indicated cls1 fragments from a full-strength nmt1 promoter by removal of thiamine. HEAT repeats (orange boxes) and a basic serine-rich stretch (green box) are indicated in the cls1p schematic. Cells were scored for interphase MT stability by time-lapse imaging of GFP-tubulin.
(G) Single interphase MT bundles in the indicated mutant backgrounds overexpressing cls1p. All images are maximum projections of confocal stacks. Scale bars: 5μm.
We mapped the region of cls1p responsible for the MT-stabilizing activity by overexpressing cls1 fragments (Figure 5F). An N-terminal cls1 fragment (aa 1-500), which contains a series of HEAT repeats, was necessary and sufficient for this activity.
To further examine the mechanism of cls1p function, we tested whether cls1p stabilizes MTs through other proteins implicated in MT stability. It has been proposed that CLASPs may regulate MT dynamics through interactions with CLIP170, EB1, or dynein (Akhmanova et al., 2001; Grallert et al., 2006; Mimori-Kiyosue et al., 2005). The effects of cls1p overexpression were monitored in various mutants (Figure 5G). MT stabilization was still apparent in the absence of tip1p (CLIP170), mal3p (EB1), and dhc1p (dynein heavy chain). Importantly, stabilization also occurred in ase1 (PRC1/MAP65) mutants, in which MT bundling is reduced. Thus, cls1p can function on single unbundled MTs, and under these conditions of overexpression, is not dependent on ase1p.
Cls1p overexpression also appeared to stabilize MT minus ends. The γ-tubulin complex can function as a cap that stabilizes the MT minus end (Wiese and Zheng, 2000); mutants such as mto1Δ, which are defective in γ-tubulin complex function, exhibit shrinkage from MT minus ends (Zimmerman and Chang, 2005). Cls1p overexpression appeared to stabilize all MT ends in mto1Δcells. In addition, cls1p also stabilized MT fragments generated by laser microsurgery (Khodjakov, Magidson, and Chang, unpublished observations). Although overexpression studies of MT-associated proteins do carry caveats, these striking phenotypes, which are the opposite of cls1p loss-of-function phenotypes, provide further support for a role of cls1p in preventing MT disassembly and promoting MT rescue.
Ase1p Targets Cls1p to Overlap Zones
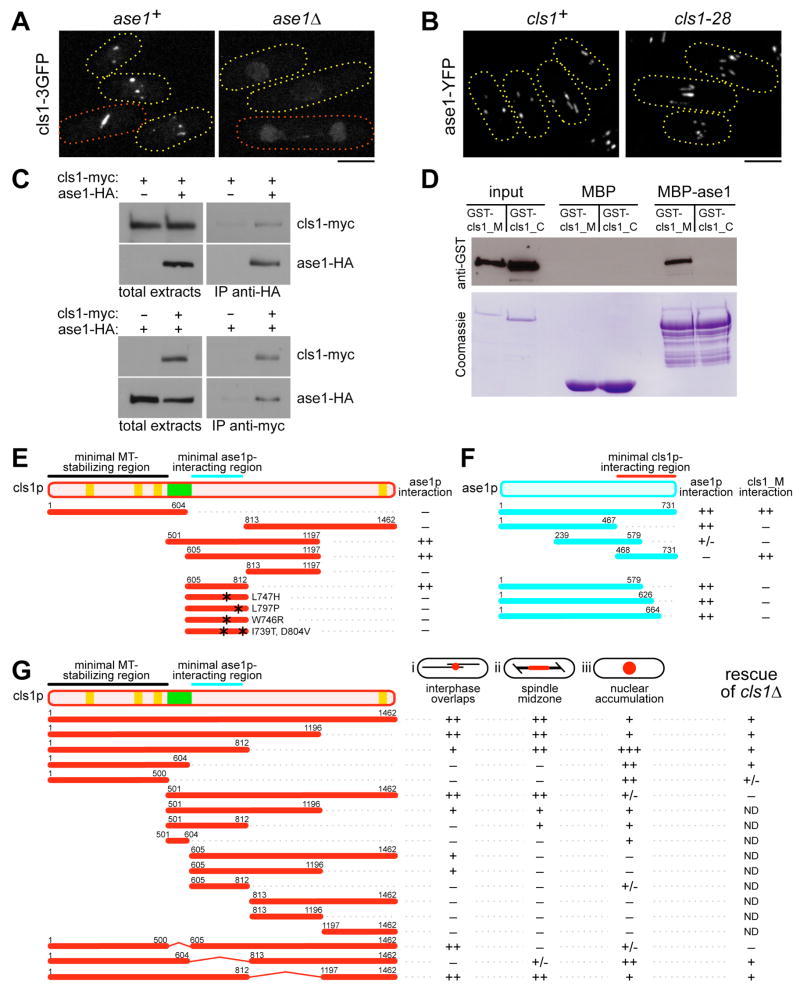
We next determined how cls1p is targeted to specific regions on the MT. The localization of cls1p (expressed at endogenous levels) was unaffected by the loss of the +TIPs tip1p (CLIP170), mal3p (EB1), tea1p (kelch repeat protein), and tea2p (kinesin) (Figure S1C; Grallert et al., 2006). In contrast to Grallert et al (2006), we also detected no localization defects in a dhc1 (dynein heavy chain) mutant. Consistent with its localization to the medial overlap zone, cls1p localization was dependent on ase1p (PRC1/MAP65) (Figures 6A and S1C). In an ase1 background, cls1-3GFP localization was severely disrupted in interphase and post-anaphase MT arrays. Cls1p localization was also significantly reduced at the spindle, although it was detected at low levels all along the length of anaphase spindles. In contrast, ase1-YFP localized normally to discrete regions along interphase MT bundles in cls1ts cells (Figure 6B). Thus ase1p is needed to recruit cls1p to regions of MT overlap.
Figure 6. Ase1p recruits cls1p to overlapping MTs by direct binding.
(A) Cls1-3GFP in wild-type and ase1Δcells. Cls1p localization is defective in ase1Δ. Cells in interphase (yellow) and anaphase (red) are shown. Maximum projection confocal images are shown. Scale bar: 5μm.
(B) Ase1-YFP localization in wild-type and cls1-28 interphase cells at 33ºC.
(C) Coimmunoprecipation. Extracts from yeast cells expressing ase1-HA3 and/or cls1-myc13 at endogenous levels were immunoprecipitated and probed by immunoblotting with anti-HA or anti-myc antibodies.
(D) In vitro binding. GST-cls1_M (aa 501-1197) and GST-cls1_C (aa 813-1462) were expressed in bacteria, purified, and tested for binding to columns containing MBP-ase1 (full-length) or MBP. GST-tagged proteins were detected by immunoblotting. Coommassie staining shows loading control.
(E) Two-hybrid assays between GAD-cls1 fragments and GBD-ase1 (full-length). The minimal ase1p-interacting region (aa 605-812) is indicated in blue. This interaction was disrupted by point mutations identified in cls1ts alleles (asterisks).
(F) Two-hybrid assays between GBD-ase1 fragments and GAD-ase1 (full-length) or GAD-cls1_M. The minimal cls1p-interacting region (aa 468-731) is indicated in red. “++” denotes growth on −HIS and on −ADE, and “+/−” denotes growth only on −HIS.
(G) Localization and function of exogenously expressed cls1 fragments fused to mCherry. For localization, fragments were expressed in haploid cls1+ cells co-expressing GFP-tubulin and semi-quantitatively scored for localization to the indicated cellular locales. For (i) and (ii), “++” indicates wild-type levels, and for (iii), “+” indicates wild-type levels. For functional assays, fragments were expressed in diploid cls1+/cls1Δcells, and their ability to rescue viability in haploid cls1Δspores was determined. “+” indicates full rescue. “ND”, not determined.
Given these findings, we tested whether ase1p and cls1p interact. Using epitope-tagged proteins expressed at endogenous levels in fission yeast, we found that ase1p and cls1p coimmunoprecipitated (Figure 6C). Two-hybrid assays also revealed the interaction. We then demonstrated that this interaction was direct by the binding of bacterially expressed proteins in vitro (Figure 6D).
Using two-hybrid and direct binding assays, we mapped the minimal ase1p interaction domain on cls1p to a ~200 aa region in the middle of the protein (Figures 6D and 6E). This region was responsible for localization of cls1p at MT overlap zones (Figure S9) and was distinct from the N-terminal MT stabilization domain. Interestingly, this ase1p-interacting region encompasses the ~60 aa stretch that contains the bulk of the mutated residues in our randomly generated cls1ts alleles (Figure S5C). The ase1p interaction domains from four of these mutant alleles failed to bind ase1p. These results suggest that the ase1p interaction domain of cls1p is important for its function in vivo.
Domain Analysis of Cls1p Function and Localization
In a complementary line of experiments, we carried out a truncation analysis of cls1p to determine the regions of cls1p responsible for its localization and function in vivo (Figures 6G, S9, and S10). Cls1 fragments were fused to mCherry and exogenously expressed from the repressed nmt1 promoter in cls1+ cells co-expressing GFP-tubulin. Functionality of the fragments in these studies was measured by the ability to rescue cls1Δmutants for growth of colonies.
Notably, the N-terminus of cls1p was sufficient and necessary for the essential function of cls1p. This region includes the MT stabilization domain, which contains HEAT repeats and has predicted structural similarities to dis1/TOG proteins (Figure S11). The MT stabilization domain alone provided partial rescue, while a fragment containing the MT stabilization domain and the basic, serine-rich domain provided full rescue of viability. These fragments localized to the nucleus, consistent with an essential function in mitosis, but did not appear to accumulate on MT structures. The basic domain, which has been implicated in MT binding in other CLASPs (Mimori-Kiyosue et al., 2005; Wittmann and Waterman-Storer, 2005; Yin et al., 2002), appeared to mediate efficient nuclear localization; this region was also required for rescue activity, and for association with spindle MTs but not with interphase MTs. These data suggest that the MT stabilization activity and nuclear accumulation are critical for cls1p function.
The ase1p-binding domain in the middle of the protein was critical for localization to MT overlap zones. Deletion of this region caused the protein to be localized poorly along the entire anaphase spindle, but as in ase1Δcells, did not affect the ability of cls1p to support viability. Although the ase1p-binding domain did not localize by itself to regions of MT overlap, it targeted neighboring sequences to these sites.
The C-terminus of CLASP is predicted to interact with CLIP170 and mediate targeting to kinetochores (Akhmanova et al., 2001; Maiato et al., 2003). Interestingly, we found that deletions of this region did not affect cls1p rescue activity or localization to overlapping MTs. These results were confirmed with a truncated cls1p expressed from the endogenous locus (data not shown).
In summary, these structure-function analyses demonstrate that the MT stabilization and basic domains are sufficient for the essential cls1p function in vivo, and that the medial ase1p-interacting domain, while non-essential, is important for enriched localization to overlap zones.
Separating Roles of Ase1p in MT Stabilization from MT Bundling
We further mapped functional domains of ase1p. Two-hybrid studies showed that the C-terminups of ase1p was necessary and sufficient for the interaction with cls1p (Figure 6F). An N-terminal portion of ase1p, which has been shown to mediate MT binding, also binds to itself, presumably for ase1p dimerization and MT bundling (Janson et al., 2007; Schuyler et al., 2003; Smertenko et al., 2004).
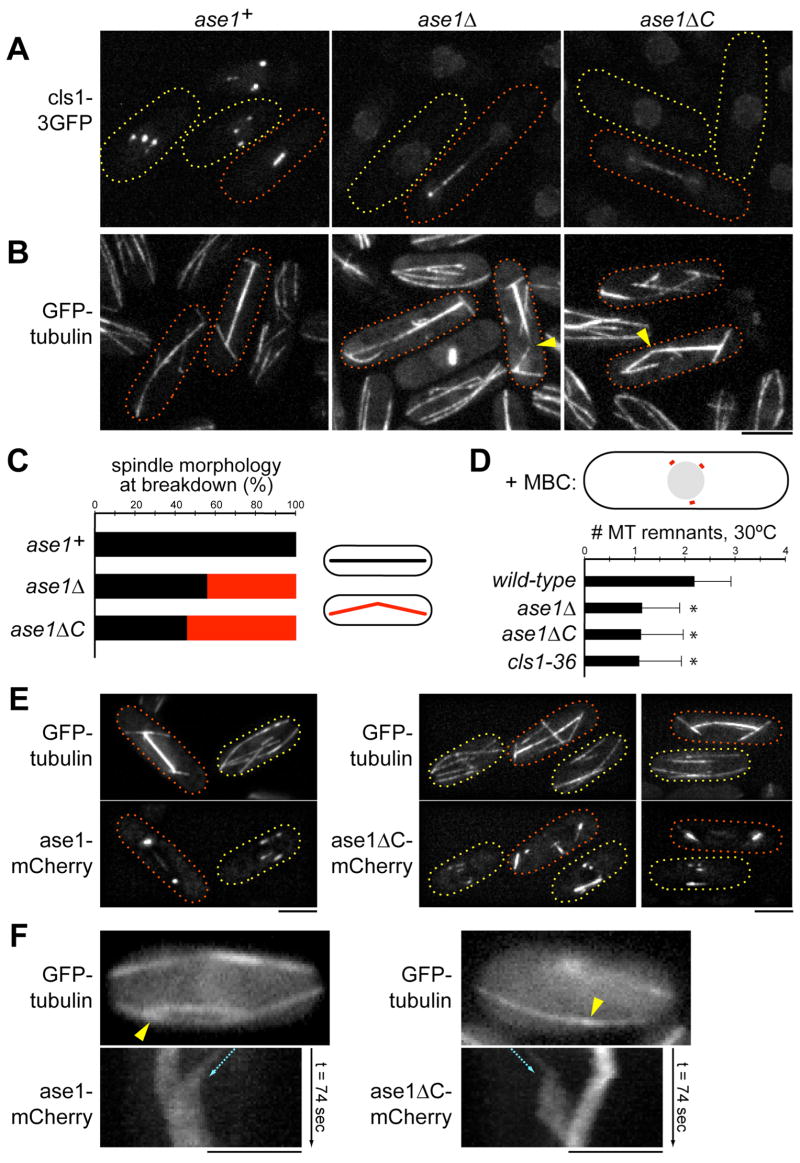
These findings suggested that ase1p has distinct domains, one to dimerize and bundle MTs, and another to stabilize MTs through cls1p. To test this model, we generated an ase1 mutant lacking the cls1p-binding site by creating a C-terminal truncation of the chromosomal ase1 gene (ase1ΔC). This mutant ase1p, which lacks the C-terminal 67 aa, was capable of binding and bundling MTs in vivo (Figures 7E and 7F). However, it was defective in targeting cls1p to overlap zones (Figure 7A) and exhibited similar defects as ase1Δcells in anaphase spindle stability (Figures 7B and 7C; Movies 3–5) and stabilization of interphase MT stubs within overlap zones (Figure 7D). Thus, the formation of bundles that lack stability in ase1ΔC and cls1ts cells demonstrates that stability is not an intrinsic property of overlapping MTs, but relies on the recruitment of cls1p.
Figure 7. Ase1p C-terminus is necessary for cls1p targeting but not MT bundling.
(A) Defective localization of cls1-3GFP in ase1Δand ase1ΔC (truncation of aa 665-731). Maximum projection confocal images of cells in interphase (yellow) and anaphase (red) are shown. Scale bar: 5μm.
(B) Anaphase spindle elongation defects in ase1Δand ase1ΔC. Maximum projection confocal images of cells expressing GFP-tubulin show similarly broken (arrowheads) and asymmetric spindles in ase1 mutants.
(C) Quantification of anaphase spindles that displayed broken morphology, as seen in (B). For each strain, >20 spindles were examined in time-lapse.
(D) Quantification of the number of stable MT remnants in MBC-treated cells at 30°C (n>120 cells for each strain). ase1 mutants are defective in stabilization of interphase MT bundle overlap zones (“*” P<10−4).
(E) Localization of ase1-mCherry and ase1ΔC-mCherry to MT overlap zones. Interphase (yellow) and anaphase cells (red) are shown. Images are maximum projections of deconvolved stacks.
(F) Ase1-mCherry and ase1 C-mCherry localization on single interphase MT bundles. Top, single-plane image of GFP-tubulin. Bottom, kymographs of the mCherry fusion proteins along the indicated bundle (yellow arrowheads)—constructed from single-plane time-lapse images (0.5 fps)—showing association with a newly-nucleated MT sliding toward the main overlap zone of the bundle (blue arrow).
Discussion
A Mechanism for Stabilization of Overlapping MTs
Here, we provide a new mechanism for how overlapping MT arrays are stabilized. Although it has been thought that ase1p or MT bundling itself stabilizes MTs, we show that cls1p is responsible for this activity. Ase1p, which binds and bundles MTs directly, also binds and recruits cls1p to zones of overlapping MTs in the spindle and interphase MT bundles. Inactivation of cls1p led to the rapid loss of mitotic spindle MTs and, in the absence of MT nucleation, of interphase MT bundles. Our data indicate that cls1p stabilizes MTs by preventing MT disassembly while still allowing for MT plus end growth; we find no measurable effects of cls1p on MT nucleation, polymerization, catastrophe, or bundling. Although CLASP has been primarily studied as a MT plus end protein, these studies demonstrate that CLASP can form a MT-stabilizing “patch” that can be directed to other parts of the MT to stabilize a particular subset of the MT cytoskeleton.
We note that these studies lead to different conclusions from those of a previous paper on this same gene by Grallert et al. (2006). A discussion of these differences and possible reasons is presented in the Supplement.
CLASP and Rescue of the Spindle
The mitotic spindle contains the most stable MTs in the fission yeast cell cycle. Many of the interpolar MTs that assemble in initial stages of spindle formation persist until the end of anaphase, and are resistant to MBC and cuts by laser microsurgery (Khodjakov et al., 2004; Mallavarapu et al., 1999). For most of anaphase, the plus ends of these MTs remain dynamic within the midzone but do not generally shrink past this region; thus, there has been a parodox of how these spindle MTs can be dynamic at their ends, but do not shrink away completely during catastrophe. Cls1p appears to be responsible for the maintenance of these overlapping spindle MTs. Our data indicate that it functions by increasing the rescue frequency at the midzone.
It is likely that this role of CLASP in stabilizing midzone MTs is conserved. Functional studies in Xenopus suggest that CLASP is needed for maintenance of the anaphase spindle, and there are clear effects of CLASP on the prominent spindle midzone MTs in Drosophila cells (Hannak and Heald, 2006; Inoue et al., 2004). Computational modeling of MT behavior in spindles has suggested that a catastrophe gradient exists at the poles (Cheerambathur et al., 2007; Pearson et al., 2006); we note that this is operationally equivalent to having a rescue gradient at the midzone. Thus, CLASP may be a key regulator of spindle MT dynamics.
CLASP and Rescue within Interphase MT Bundles
Our data provide new insight into the inner workings of the interphase MT bundle. These bundles are dynamic, in that MTs are continuously nucleated on the bundles, while others are lost to depolymerization. These bundles are also known to contain a medial “patch”, which is stable to MBC treatment; these stable zones were the original definition of the iMTOC (Tran et al., 2001). Cls1p, which decorates only a subset of the MT overlap zone, is the first protein identified that marks specifically this patch.
We propose that this cls1p patch is a fail-safe mechanism that prevents the complete disassembly of a MT bundle. Although continuous MT nucleation from γ-tubulin complexes appears to compensate for loss of MT stability normally, cls1p becomes critical for maintenance of bundles when nucleation is defective. Cls1p was also critical for maintenance of short bundles in wild-type cells, where there may be less chance of MT nucleation, and in conditions leading to MT depolymerization.
Each cls1p patch may stabilize only one or a small subset of MTs within these interphase bundles. In wild-type interphase bundles, kymographs reveal that most shrinking MTs are not rescued in the overlap zone but appear to depolymerize away completely. However, about 20% of MTs exhibited cls1p-dependent pause events within the overlap zone. Each bundle contains around 4–5 MTs in the overlap zone, and about half of these are <1μm in length (Hoog et al., 2007). Therefore, a cls1p patch may normally stabilize on average only one MT within the overlap zone. To maintain a bundle, the stabilization of 1–2 MTs may be sufficient, even advantageous: if every MT were stabilized within an interphase bundle, new nucleation would lead to over-proliferation of MTs. Thus, nucleation, depolymerization, and selective stabilization may maintain the number of MTs within bundles.
CLASP Mechanism
Our studies suggest that cls1p functions to halt MT disassembly and promote MT rescue. Very little is known about the mechanism of MT rescue, a multi-step process that involves stopping depolymerization and then promoting re-initiation of MT growth. The localization pattern of cls1p suggests that it acts as a brake or clamp on the MT lattice that anticipates the arrival of the shrinking MT end, and prevents the MT from shrinking past it. Cls1p patches consist of 10–13 molecules per stable MT in both interphase and spindle MTs. This number is intriguing, as it suggests that very small numbers of CLASP molecules (possibly 1 per protofilament) are sufficient for this activity. It will be interesting to see if CLASP acts by independently stabilizing individual protofilaments, by regulating the GTP/GDP state of tubulin subunits, or as a collar at the lattice to stop the splaying of depolymerizing protofilaments.
Structure-function analyses indicate that the N-terminal region is sufficient for the essential function of cls1p. The majority of this region is comprised of the MT stabilization domain, which is responsible for the effects of cls1p overexpression to increase rescue frequency. This activity was independent of numerous plus and minus end-stabilizing factors, as well as ase1p and MT bundling. This region includes HEAT repeats and has predicted structural similarity to TOG domains; a recent structure of a TOG domain reveals how these HEAT repeats may bind directly on tubulin (Al-Bassam et al., 2007). An analogous fragment from S. cerevisiae CLASP Stu1p binds MTs with low affinity in vitro (Yin et al., 2002). Therefore it is likely that this domain of cls1p can interact with tubulin directly. The basic region of cls1p, which has also been implicated in MT binding in other CLASPs (Mimori-Kiyosue et al., 2005; Wittmann and Waterman-Storer, 2005; Yin et al., 2002), also contributes to its essential function.
The ase1p-interacting region on CLASP is sufficient and necessary for concentrating cls1p to MT overlap regions. Conservation of this region suggests that other CLASPs may also interact with ase1p orthologs. This region binds to the C-terminus of ase1p, which is also functionally separable from other functions of ase1p. Interestingly, we note that this region on ase1p contains several putative cdc2p phosphorylation sites, which could potentially regulate this interaction in mitosis (Khmelinskii et al., 2007; Loiodice et al., 2005; Mollinari et al., 2002; Smertenko et al., 2006)
A Module for Building MT Structure
In developing concepts and models of cellular assembly, a toolbox of functional modules needs to be defined. This study advances the concept that a rescue factor, which can be attached to specific sites on MTs, can help to generate a MT structure that has both dynamic and stable components. Many MT assemblages need to be dynamic for their function, but also must be stable enough so that they do not fall apart completely during shrinkage events. Stabilizing MTs using stable caps that stop both polymerization and depolymerization are not suitable in this system, as the continued dynamics are critical for the function of these arrays. Ways to counter the complete disassembly of a dynamic bundle are threefold: nucleation of a new MT off a pre-existing MT, stabilization and rescue of shrinking MTs, or formation of an entire new bundle. In fission yeast, all three mechanisms can occur in interphase MT arrays, but MT stabilization acts alone to maintain anaphase spindles.
Mathematical modeling of fission yeast MT bundles now makes this a powerful simple system amenable to detailed mechanistic analysis (Janson et al., 2007). In addition to the mitotic spindle, antiparallel MT bundles are a prominent feature of arrays in fungal and plant cells (Wasteneys, 2002), as well as in differentiated animal cells such as neurons, myotubes, and megakaryocytes (Baas et al., 1988; Patel et al., 2005; Tassin et al., 1985). It is likely that the complex of CLASP with Ase1/PRC1/MAP65 will be an important element in building many types of MT structures.
Experimental Procedures
Yeast Strains, Media, and Genetic Methods
S. pombe strains used in this study are listed in Table S1. Standard methods for media and genetic manipulations were used. Deletion strains and strains with integrated tags and/or promoters were constructed by PCR and homologous recombination (see Supplemental Methods) and confirmed by PCR from genomic DNA. Tagged cls1 strains were functional as determined by viability, mitotic spindle behavior, interphase MT dynamics, and MT stability to MBC.
Microscopy
Microscopy was performed using either a wide-field fluorescence microscope or a spinning disk confocal microscope (Pelham and Chang, 2001). Images were acquired and analyzed with OpenLab software (Improvision) and ImageJ (NIH). 3-D wide-field stacks were processed where indicated by maximum entropy deconvolution, and kymographs were constructed with the “Volume Slicing” tool in OpenLab. Cells were imaged on agar pads or in liquid media under a coverslip. An objective heater (Bioptechs) was used for temperature control during imaging.
Generation of cls1ts Alleles
A C-terminal cls1 fragment was randomly mutagenized in vitro and incorporated into the endogenous cls1 locus by homologous recombination using a “marker switch” method (MacIver et al., 2003) (see Figure S5 and Supplemental Methods). Of thirteen recessive cls1ts alleles analyzed, all contained at least one mutation in a ~60 aa region near the middle of the protein.
Cytoskeletal Analysis
Plasmids used in this study are listed in Table S3. For colocalization studies, MTs were observed in cells transformed with either CFP-α-tubulin (pRL74) or mRFP-α-tubulin (pSB62; see Supplemental Methods). For MT kymographs (Figures 2D, 4B, and S7D; Movie 1), MTs were observed in cells expressing GFP-α-tubulin from pDQ105. Otherwise, GFP-α-tubulin was expressed from the SV40 early promoter (see Supplemental Methods).
For MT drug treatments, cells were added to agar pads containing 25μg/mL MBC (Aldrich) from a fresh 100X stock in DMSO. MT depolymerization and regrowth experiments in flow chambers were performed as described (Zimmerman et al., 2004). To control temperature during the washout of MBC in Figure S8, prewarmed media and a heated objective were used.
Coimmunoprecipitation, In Vitro Binding, and Two-Hybrid Analyses
Immunoprecipitations from yeast extracts and in vitro binding assays were performed as described (Martin et al., 2005) with modifications (see Supplemental Methods). For expression in E. coli or S. cerevisiae, ase1 and cls1 gene fragments were cloned by standard methods into pMal, pGEX, pGAD, or pGBD vectors. Introns were removed in PCR oligos. For two-hybrid analysis, GAD and GBD fusions were expressed in S. cerevisiae strain AH109 (Clontech). The assay for interaction was growth on medium lacking adenine or medium lacking histidine with 10mM 3′AT.
Supplementary Material
Acknowledgments
We are grateful to H. Maiato for encouraging us to initiate these studies, R. Daga and S. Salas for generation of the SV40:GFP-atb2 strain, and N. Minc for help with constructing flow chambers. We also thank A. Khodjakov and G. Gundersen for discussions and comments on the manuscript; I. Hagan, J. Millar, P. Nurse, T. Toda, P. Tran, and X. He, for strains and reagents; and S. Martin, N. Padte A. Yonetani, and other members of the Chang lab for advice and support. This work was supported by NIH grants T32-GM-07367 to S.B. and R01 GM069670 to F.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–362. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Baas PW, Black MM. Individual microtubules in the axon consist of domains that differ in both composition and stability. J Cell Biol. 1990;111:495–509. doi: 10.1083/jcb.111.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988;85:8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Antony C, Nurse P. The kinesin Klp2 mediates polarization of interphase microtubules in fission yeast. Science. 2005;309:297–300. doi: 10.1126/science.1113465. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Nurse P. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol. 2006;8:1102–1107. doi: 10.1038/ncb1479. [DOI] [PubMed] [Google Scholar]
- Cheerambathur DK, Civelekoglu-Scholey G, Brust-Mascher I, Sommi P, Mogilner A, Scholey JM. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J Cell Biol. 2007;177:995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga RR, Lee KG, Bratman S, Salas-Pino S, Chang F. Self-organization of microtubule bundles in anucleate fission yeast cells. Nat Cell Biol. 2006;8:1108–1113. doi: 10.1038/ncb1480. [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. Asymmetric CLASP-Dependent Nucleation of Noncentrosomal Microtubules at the trans-Golgi Network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, Ruthmann A. Evidence for three “classes” of microtubules in the interpolar space of the mitotic micronucleus of a ciliate and the participation of the nuclear envelope in conferring stability to microtubules. Chromosoma. 1982;85:687–706. doi: 10.1007/BF00330781. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grallert A, Beuter C, Craven RA, Bagley S, Wilks D, Fleig U, Hagan IM. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 2006;20:2421–2436. doi: 10.1101/gad.381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hannak E, Heald R. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J Cell Biol. 2006;172:19–25. doi: 10.1083/jcb.200508180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Hoog JL, Schwartz C, Noon AT, O’Toole ET, Mastronarde DN, McIntosh JR, Antony C. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev Cell. 2007;12:349–361. doi: 10.1016/j.devcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Inoue YH, do Carmo Avides M, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, Glover DM. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J Cell Biol. 2000;149:153–166. doi: 10.1083/jcb.149.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue YH, Savoian MS, Suzuki T, Mathe E, Yamamoto MT, Glover DM. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J Cell Biol. 2004;166:49–60. doi: 10.1083/jcb.200402052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Nedelec F, Surrey T. Modelling microtubule patterns. Nat Cell Biol. 2006;8:1204–1211. doi: 10.1038/ncb1498. [DOI] [PubMed] [Google Scholar]
- Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, La Terra S, Chang F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol. 2004;14:1330–1340. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Lemos CL, Sampaio P, Maiato H, Costa M, Omel’yanchuk LV, Liberal V, Sunkel CE. Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. Embo J. 2000;19:3668–3682. doi: 10.1093/emboj/19.14.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver FH, Glover DM, Hagan IM. A ‘marker switch’ approach for targeted mutagenesis of genes in Schizosaccharomyces pombe. Yeast. 2003;20:587–594. doi: 10.1002/yea.983. [DOI] [PubMed] [Google Scholar]
- Maiato H, Fairley EA, Rieder CL, Swedlow JR, Sunkel CE, Earnshaw WC. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–1426. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- Martin SG, McDonald WH, Yates JR, 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Sasaki H, Matsui C, Akhmanova A, Tsukita S, Vorobjev I. Mammalian CLASPs are required for mitotic spindle organization and kinetochore alignment. Genes Cells. 2006;11:845–857. doi: 10.1111/j.1365-2443.2006.00990.x. [DOI] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Nabeshima K, Kinoshita K, Yanagida M. Dissection of fission yeast microtubule associating protein p93Dis1: regions implicated in regulated localization and microtubule interaction. Genes Cells. 1996;1:633–644. doi: 10.1046/j.1365-2443.1996.00253.x. [DOI] [PubMed] [Google Scholar]
- Nedelec F, Surrey T, Karsenti E. Self-organisation and forces in the microtubule cytoskeleton. Curr Opin Cell Biol. 2003;15:118–124. doi: 10.1016/s0955-0674(02)00014-5. [DOI] [PubMed] [Google Scholar]
- Pasqualone D, Huffaker TC. STU1, a suppressor of a beta-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Richardson JL, Schulze H, Kahle E, Galjart N, Drabek K, Shivdasani RA, Hartwig JH, Italiano JE., Jr Differential roles of microtubule assembly and sliding in proplatelet formation by megakaryocytes. Blood. 2005;106:4076–4085. doi: 10.1182/blood-2005-06-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Gardner MK, Paliulis LV, Salmon ED, Odde DJ, Bloom K. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Mol Biol Cell. 2006;17:4069–4079. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Chang F. Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat Cell Biol. 2001;3:235–244. doi: 10.1038/35060020. [DOI] [PubMed] [Google Scholar]
- Pereira AL, Pereira AJ, Maia AR, Drabek K, Sayas CL, Hergert PJ, Lince-Faria M, Matos I, Duque C, Stepanova T, et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagolla MJ, Uzawa S, Cande WZ. Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J Cell Sci. 2003;116:4891–4903. doi: 10.1242/jcs.00796. [DOI] [PubMed] [Google Scholar]
- Salmon ED, Begg DA. Functional implications of cold-stable microtubules in kinetochore fibers of insect spermatocytes during anaphase. J Cell Biol. 1980;85:853–865. doi: 10.1083/jcb.85.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Lourenco PC, Snaith HA. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr Biol. 2004;14:763–775. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Tran PT. Cytoplasmic microtubule organization in fission yeast. Yeast. 2006;23:1001–1014. doi: 10.1002/yea.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Sonobe S, Fenyk SI, Weingartner M, Bogre L, Hussey PJ. Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J Cell Sci. 2006;119:3227–3237. doi: 10.1242/jcs.03051. [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Wagner V, Kaloriti D, Fenyk S, Sonobe S, Lloyd C, Hauser MT, Hussey PJ. The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell. 2004;16:2035–2047. doi: 10.1105/tpc.104.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Maro B, Bornens M. Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic-Norrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol. 2004;14:1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Wasteneys GO. Microtubule organization in the green kingdom: chaos or self-order? J Cell Sci. 2002;115:1345–1354. doi: 10.1242/jcs.115.7.1345. [DOI] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, You L, Pasqualone D, Kopski KM, Huffaker TC. Stu1p is physically associated with beta-tubulin and is required for structural integrity of the mitotic spindle. Mol Biol Cell. 2002;13:1881–1892. doi: 10.1091/mbc.01-09-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S, Chang F. Effects of {gamma}-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol Biol Cell. 2005;16:2719–2733. doi: 10.1091/mbc.E04-08-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S, Tran PT, Daga RR, Niwa O, Chang F. Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev Cell. 2004;6:497–509. doi: 10.1016/s1534-5807(04)00096-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.