Abstract
Single-nucleotide polymorphisms (SNPs) are the most common type of genetic variation in man. Genes containing one or more SNPs can give rise to two or more allelic forms of mRNAs. These mRNA variants may possess different biological functions as a result of differences in primary or higher order structures that interact with other cellular components. Here we report the observation of marked differences in mRNA secondary structure associated with SNPs in the coding regions of two human mRNAs: alanyl tRNA synthetase and replication protein A, 70-kDa subunit (RPA70). Enzymatic probing of SNP-containing allelic fragments of the mRNAs revealed pronounced allelic differences in cleavage pattern at sites 14 or 18 nt away from the SNP, suggesting that a single-nucleotide variation can give rise to different mRNA folds. By using phosphorothioate oligodeoxyribonucleotides complementary to the region of different allelic structures in the RPA70 mRNA, but not extending to the SNP itself, we find that the SNP exerts an allele-specific effect on the accessibility of its flanking site in the endogenous human RPA70 mRNA. This further supports the allele-specific structural features identified by enzymatic probing. These results demonstrate the contribution of common genetic variation to structural diversity of mRNA and suggest a broader role than previously thought for the effects of SNPs on mRNA structure and, ultimately, biological function.
Single-nucleotide polymorphisms (SNPs) are single base-pair substitutions that occur within and outside genes (1–3). SNPs account for many well characterized human phenotypes, including disease susceptibility and resistance (4, 5) and drug response (6, 7). As a result, SNP discovery efforts have markedly expanded over the past 2 years. The frequency of SNPs varies between genomic regions and between coding and noncoding sequences. The extent of nucleotide diversity ranges from 0.0003 to 0.005; in other words, there are from 3 to 50 SNPs per 10 kilobases when two chromosomes are compared (for review, see ref. 2).
Genes containing one or more SNPs can give rise to two or more allelic forms of mRNAs. mRNAs containing different bases at SNP sites may vary in their interactions with cellular components involved in mRNA synthesis, maturation, transport, translation, or degradation. It has been documented that a number of single base-pair substitutions alter or create essential sequence elements for splicing, processing, or translation of human mRNA (1). These SNPs are associated with altered length and/or steady-state level of cytoplasmic mRNA. On the other hand, SNPs that do not affect RNA consensus and protein sequences have not been analyzed in detail. It is conceivable that such SNPs could also lead to phenotypic effects, most likely through non-consensus-dependent mechanisms.
A growing body of evidence shows that the folding of mRNA influences a diverse range of biological events such as mRNA splicing (8, 9) and processing (10–12), and translational control (13–16) and regulation (17–19). Because the structure of mRNA is determined by its nucleotide sequence and its environment, we were interested in examining whether the folding of mRNA could be influenced by the presence of SNPs. In this report, we describe the analysis of SNPs in the coding regions of two essential genes: a U/C transition at nucleotide 1,013 of human alanyl tRNA synthetase (AARS, GenBank Accession no. D32050) and a U/C transition at nucleotide 1,674 of human replication protein A, 70-kDa subunit (RPA70, GenBank Accession no. M63488). Neither SNP affects the amino acid sequence of the respective protein. An initial screen of lymphoblast cDNA from 36 unrelated individuals (V.P.S., unpublished results) revealed allele frequencies of 0.49 and 0.51 for the AARS U and C alleles (57% heterozygotes observed) and allele frequencies of 0.85 and 0.15 for the RPA70 U and C alleles (31% heterozygotes observed). Using structural mapping and structure-based targeting strategies, we show that the SNPs have marked effects on the structural folds of the mRNAs. The contribution of common genetic variation to structural diversity of mRNA has not been described previously. These results suggest that phenotypic consequences of SNPs could arise from mechanisms that involve allele-specific structural motifs in mRNA.
MATERIALS AND METHODS
In Vitro Synthesis and Purification of mRNA.
Human cDNAs for PCR synthesis of transcription templates were prepared from total RNA of Epstein–Barr virus transformed lymphoblasts (Coriell Cell Repositories, Camden, NJ) or tumor cell line NCI-H292 (American Type Culture Collection). The cDNAs were genotyped for SNPs in human AARS and RPA70 either by sequencing or by restriction fragment length polymorphism analysis. A second round of cDNA sequencing was performed to confirm the nucleotide sequence of AARS and RPA70 cDNAs. Fragments of human mRNA alleles (113- to 1,000-mers) varying in a single base were obtained by using in vitro transcription from PCR-derived DNA templates with MEGAshortscript or MEGAscript T7 kits (Ambion, Austin, TX). Crude transcripts were either purified on an 8% denaturing polyacrylamide gel or on a G-50 Sephadex spin column (Boehringer Mannheim), and the purified mRNA was subsequently dephosphorylated by using calf intestinal alkaline phosphatase (Amersham Pharmacia) and 5′-end-labeled with 32P by using [γ-32P]ATP (DuPont/NEN) and T4 polynucleotide kinase (Amersham Pharmacia).
Enzymatic Mapping of Allelic mRNA Structures.
Partial digestion of 1 pmol of 5′-32P-labeled mRNA by nuclease S1 or RNase T1 (Amersham Pharmacia) was carried out at 37°C for 5 min in 10 μl of buffer containing 10 mM Tris⋅HCl, pH 7.5/100 mM KCl/5 mM MgCl2/5 μg of phenol-extracted glycogen. Before the addition of S1 or T1, the RNA was renatured in the assaying buffer by heating for 45 sec at 85°C and cooling for 5 min at room temperature followed by 10 min at 37°C. The digestion products were fractionated on either a 4, 6, or 8% denaturing polyacrylamide gel, and the cleavage patterns were visualized by using autoradiography. RNase T1 digestion at 55°C and alkaline hydrolysis at 85°C were done to generate sequence ladders for cleavage site identification. RNase T1 digestion at 55°C was done for 1 min in 15 μl of buffer containing 25 mM sodium acetate, pH 5.2/25 mM KCl/3 μg of yeast tRNA. Alkaline hydrolysis was done for 15 min in 15 μl of buffer containing 50 mM sodium bicarbonate, pH 9.2/1 mM EDTA/10 μg of phenol-extracted glycogen.
Escherichia coli RNase H Digestion Assay.
Phosphorothioate oligodeoxyribonucleotides (PS-oligos) were used in our studies because of their enhanced stability against nucleases (20). PS-oligos were chemically synthesized and desalted on C-18 cartridges by Synthetic Genetics (San Diego). One picomole of 5′-32P-labeled mRNA was renatured in 12 μl of RNase H buffer (20 mM Tris⋅HCl, pH 7.5/10 mM KCl/5 mM MgCl2/0.1 mM DTT/5% glycerol/10 μg of phenol-extracted glycogen) by heating for 1 min at 85 °C, and then cooling for 5 min at room temperature followed by 10 min at 37°C. E. coli RNase H (0.6 units, Amersham Pharmacia) was then added to the renatured mRNA and incubated for 5 min at 37°C. PS-oligos (5 pmol in RNase H buffer) were subsequently added, and the reaction mixture (15 μl total) was incubated for 20 min at 37 °C. The digestion products were fractionated on a 4% denaturing polyacrylamide gel, and the cleavage patterns were visualized by autoradiography. RNA levels were quantitated by using a Fluorescent Image Analyzer (Fujifilm Model FLA-2000).
Oligodeoxyribonucleotide-Directed Cleavage of Endogenous RPA70 mRNA in Human Cell Extracts.
Human tumor cell lines HeLa and Calu-1 (American Type Culture Collection) were genotyped for SNPs in RPA70 cDNA by restriction fragment length polymorphism analysis and by sequencing. The genotyping results showed that HeLa and Calu-1 express only the U or the C base at the 1,674 U/C polymorphism, respectively. HeLa and Calu-1 cells were cultured in monolayers in minimum essential medium (GIBCO/BRL) and McCoy’s 5A medium (Sigma), respectively, under 5%CO2/95% air in a humidified incubator at 37°C. Both media were supplemented with 10% heat-inactivated fetal bovine serum (JRH Biosciences, Lenexa, KS), 100 units/ml penicillin and 100 μg/ml streptomycin (GIBCO/BRL), and 2 mM l-glutamine (GIBCO/BRL). For preparation of cell extracts, the cells were grown to 80–90% confluence, trypsinized, and washed twice with the complete medium as specified above and twice with PBS (GIBCO/BRL). Approximately 5 × 107 cells were lysed in a 15-ml tissue grinder with a Teflon pestle (Wheaton) in the presence of 1.5 ml of lysis buffer (20 mM Tris⋅HCl, pH 7.5/420 mM NaCl/2 mM MgCl2/0.2 mM EDTA/1 mM DTT/0.5 mM PMSF/25% glycerol). The sodium concentration was chosen to facilitate the disruption of the nuclear membrane to release RNase H from nuclei (21). Up to 200 strokes were needed to completely lyse the cells, as checked by using a hemacytometer. The cell lysate was centrifuged at 16,000 × g for 10 min at 4°C, and cell extracts were obtained by transferring supernatant to clean microcentrifuge tubes and freezing immediately at −80 °C. Total protein concentration of cell extracts was determined by using the Bradford assay (Bio-Rad). For PS-oligo-directed cleavage of endogenous mRNA reactions, cell extracts were diluted 3- to 4-fold with the lysis buffer to normalize the total protein concentration to 3 mg/ml for the two cell types. A 10× PS-oligo solution was subsequently added to the normalized cell extracts, and the reaction mixture was incubated for 40 min at 37°C. The incubation time and oligo concentration range were chosen to maintain the extent of mRNA cleavage below 100% for subsequent quantitation of intact mRNA. The level of intact RPA70 mRNA was determined by total RNA isolation and Northern blot hybridization by using procedures described previously (22, 23). Blots were analyzed with a Fluorescent Image Analyzer (Fujifilm Model FLA-2000). Northern probes specific for RPA70 mRNA were synthesized by random primed [α-32P]dCTP labeling of a cDNA probe corresponding to nucleotides 1,519–2,080 of human RPA70 mRNA. To normalize the amount of total RNA loaded into individual gel wells for Northern analysis, the level of glyceraldehyde 3-phosphate dehydrogenase mRNA was also measured by first stripping the RPA70 mRNA probe and then reprobing the same Northern blot with a random-primed [α-32P]dCTP-labeled cDNA probe corresponding to a 600-nt sequence of human glyceraldehyde 3-phosphate dehydrogenase (Stratagene).
RESULTS AND DISCUSSION
Characterization of SNP-Containing mRNA Structures.
Two SNPs in the coding regions of two human mRNAs were analyzed in this study: a U/C transition at nucleotide 1,013 of AARS (GenBank Accession no. D32050) and a U/C transition at nucleotide 1,674 of RPA70 (GenBank Accession no. M63488). To investigate effects of these SNPs on mRNA structure, we first synthesized both allelic SNP-containing fragments of the two mRNAs by using in vitro transcription and then performed mRNA structural mapping by using structure-specific nuclease S1 and RNase T1.
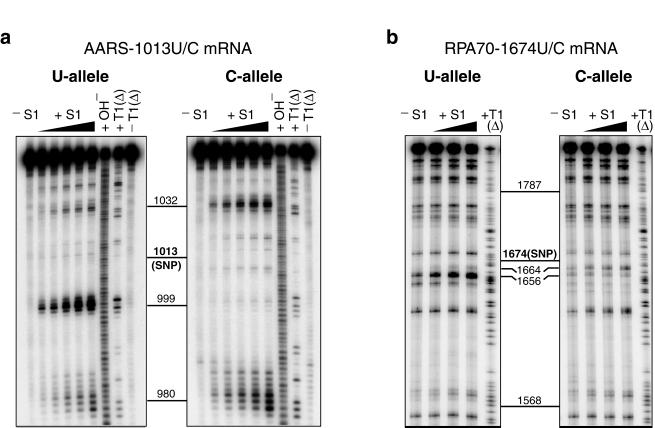
Nuclease S1 cleaves both DNA and RNA in single-stranded regions (24). It is a useful structural probe for identifying large unpaired accessible regions (>4 nt) in RNA. Fig. 1a shows nuclease S1 mapping of a 113-nt fragment of AARS mRNA (nucleotides 951–1,063). Differences in S1 cleavage patterns were observed between the U and C alleles. Strong cleavages centered at nucleotide 999 of the U allele revealed the presence of a single-stranded region ≈14 nt upstream of the SNP. The majority of the species of the C allele lack such a single-stranded region, as shown by the absence of strong cleavage bands in the same region. Allelic differences in S1 accessibility were also observed at several other sites, including regions 980 and 1,032 (Fig. 1a). At higher S1 concentrations, both alleles exhibited similar cleavage patterns at sites 980 and 1,032, suggesting that the stronger cleavage intensities at these sites in the C allele may be due partly to the absence of a large unpaired region at position 999. Structural mapping by RNase T1 (data not shown), which identifies unpaired guanosine residues (24), corroborated the finding of substantial allelic structural differences within ±14 nt of the SNP. The enzymatic mapping experiments were also done on a 1,000-nt fragment (nucleotides 529–1,528) of AARS mRNA. Differential S1 digestion patterns of the two alleles identical to those at sites 980, 999, and 1,032 in Fig. 1a were observed with the 1,000-mer mRNA sequence (data not shown).
Figure 1.
Nuclease S1 mapping of allelic mRNA structures. −S1, control reaction without S1 added; +S1, partial digestion by S1 at 37°C with increasing S1 concentration; +OH−, alkaline hydrolysis reaction; +T1(Δ), partial digestion by T1 at 55°C to create a G ladder to allow determination of cleavage sites; −T1(Δ), control reaction without T1 added. (a) Mapping of accessible single-stranded regions of the U and C alleles of a 113-mer fragment (nucleotides 951–1,063) of AARS mRNA. (b) Mapping of accessible single-stranded regions of the U and C alleles of a 544-mer fragment (nucleotides 1,378–1,921) of RPA70 mRNA. Polymorphic positions are indicated in boldface and labeled SNP.
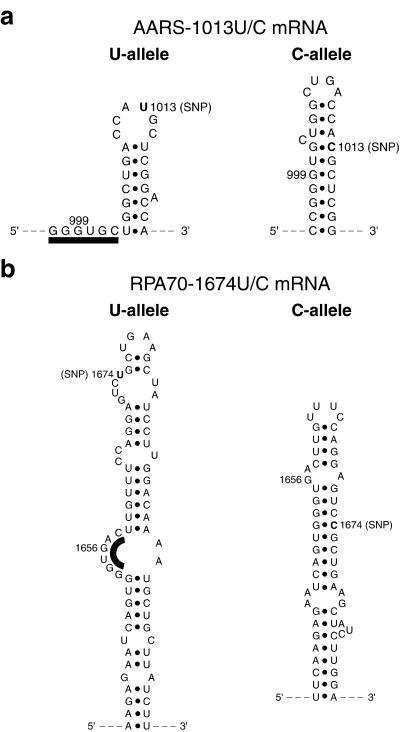
Pronounced allelic structural differences caused by a single-nucleotide variation in the mRNA sequence were also detected in a 544-nt fragment of RPA70 mRNA (Fig. 1b). The fragment, consisting of nucleotides 1,378–1,921, was centered about the 1,674 U/C polymorphism. Intense S1 cleavages were detected in the U allele around nucleotide 1,656, indicating the presence of a single-stranded region ≈18 nt upstream of the SNP. In contrast, no cleavage above background intensity was detected at the same site of the C allele. The allelic difference at this site was confirmed by structural probing with RNase T1 (data not shown). Differential S1 cleavage between the two alleles was also observed at nucleotide 1,664, where the U allele was more weakly cleaved than the C allele. These data thus defined a differential-structure motif between the two allelic forms of RPA70 mRNA, with a maximum allelic difference in structural accessibility in the region of nucleotide 1,656. In addition, the structural mapping data on both the RPA70 and the AARS mRNAs revealed that the polymorphic sites were relatively inaccessible by single-strand-specific nuclease S1. Proposed secondary-structure models of the polymorphic regions of the two pairs of mRNA alleles are shown in Fig. 2.
Figure 2.
Proposed secondary-structure models of the polymorphic regions of the pairs of mRNA alleles. The polymorphic bases are shown boldfaced and labeled SNP. The model building was assisted by the use of computer program mfold (M. Zuker, Washington University, St. Louis, MO). Only modeled regions supported by S1 and T1 mapping data are shown. (a) Proposed allelic structures of the polymorphic region of AARS mRNA. (b) Proposed allelic structures of the polymorphic region of RPA70 mRNA. The black bars highlight the highly accessible single-stranded regions.
Because the SNP-dependent allelic structural differences described above were identified by using mRNA fragments in a purified system, it is critical to examine whether our findings of allele-specific structural features exist in the context of full-length mRNA in an environment better mimicking the physiological conditions in cells. To address this issue, we used a structure-based mRNA targeting strategy to monitor allelic structural accessibility of RPA70 mRNA in two different assay systems: a purified system containing mRNA fragments and a cell-mimicking system containing endogenous human RPA70 mRNA. We designed a series of PS-oligos complementary to the 1,656 region of RPA70 mRNA that contains the maximum allelic difference in structural accessibility as identified by enzymatic mapping. The target site at the region of 1,656 was chosen to span 20 or 15 nt that do not include the 1,674 U/C polymorphism. Thus, the target site was identical in nucleotide sequence between the U and C alleles and was predicted based on enzymatic mapping results to be accessible for PS-oligo binding in the U allele, and inaccessible in the C allele. Exogenous or endogenous RNase H, which cleaves RNA in RNA·DNA hybrids (25), was used to monitor the targeting efficiency and allele specificity of PS-oligos for the two alleles of RPA70 mRNA.
Allele-Specific Cleavage of RPA70 mRNA in a Purified System.
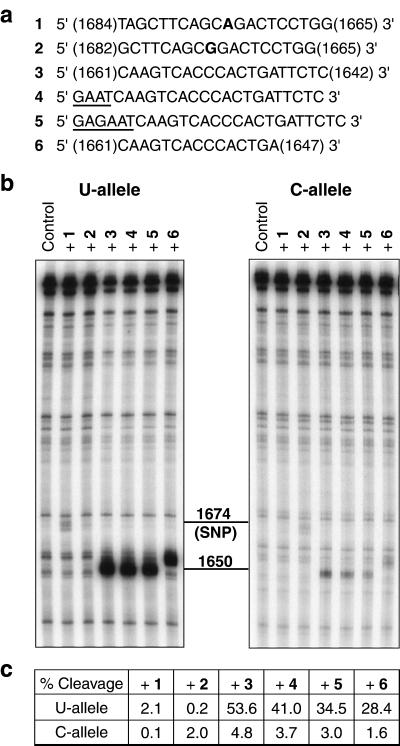
We first used an in vitro assay with E. coli RNase H and the in vitro-transcribed 544-mer RPA70 mRNA fragments to examine the efficiency and allele specificity of structure-based targeting as compared with directly targeting the 1,674 U/C polymorphism. Fig. 3a shows nucleotide sequences of the PS-oligos designed to target either the RPA70 polymorphism (oligo 1, U allele-specific; oligo 2, C allele-specific) or the site of maximum differential structural accessibility in the 1,656 region (oligos 3–6). SNP-targeting oligos 1 and 2 were previously optimized for allele discrimination in a cell transfection assay (31). Each of the SNP-targeting oligos is perfectly matched only with one of the two alleles; it is singly mismatched with the other allele. Single-base mismatches formed between the SNP-targeting oligos and their respective nontargeted alleles contribute to the allele discrimination observed previously. Structure-targeting oligos 3 and 6 match perfectly with both alleles, and were designed to hybridize maximally with the single-stranded 1,656 region of the U allele, while avoiding the 1,664 region, which is slightly accessible in both alleles (Fig. 1b). Four and six additional nucleotides were added to the 5′-end of oligo 3 to produce oligos 4 and 5, respectively, to facilitate folding into hairpin structures for enhanced allele discrimination (26).
Figure 3.
E. coli RNase H digestion of both alleles of the 544-mer fragment of the RPA70–1,674 U/C mRNA. (a) Nucleotide sequence of the PS-oligos used in the assay. Oligos 1 and 2 target the polymorphic site; the bases opposing the polymorphism in the mRNA are shown in boldface. Oligos 3–6 target the site of different allelic structures around nucleotide 1,656. The terminal residues of oligos 1, 2, 3, and 6 are labeled according to the positions of the complementary nucleotides in RPA70 mRNA. Oligos 4 and 5 share the same target hybridization sequence as 3, but with additional nucleotides (shown underlined) added to facilitate folding into hairpin structures for intended enhancement in allele discrimination. (b) E. coli RNase H digestion patterns of the 5′-32P-labeled U and C alleles hybridized to the oligos in a. Cleavage sites were confirmed by running on the same gel RNase T1 digestion ladders for the two alleles (data not shown): they are centered at position 1,674 for oligos 1 and 2, 1,650 for oligos 3–5, and 1,654 for oligo 6. (c) Comparison of percent cleavages of the U and C alleles calculated from b. Oligo-dependent cleavage bands were integrated as a percentage of total RNA in each lane with the overlapping background-cleavage bands subtracted. Variations in sample loading to each lane were internally corrected by measuring percent cleavage instead of absolute band intensity.
Results of oligo-directed, site-specific cleavage of the 544-mer U and C alleles by E. coli RNase H are shown in Fig. 3 b–c. As predicted, the U allele that was more sensitive to S1 cleavage than the C allele in the 1,656 region was over 10-fold more efficiently cleaved by E. coli RNase H in the presence of structure-targeting PS-oligos 3–6. Although the cleavage efficiencies with PS-oligos 1 and 2 that target the SNP at position 1,674 are greater for their respective matched alleles than for the mismatched alleles, allele discrimination cannot be accurately assessed because of uncertainties in the measurement of cleavage efficiencies below 1%. The polymorphic site targeted by oligos 1 and 2 was much less efficiently cleaved than the single-stranded 1,656 region in the U-allele targeted by oligos 3–6. This is consistent with the difference in accessibility between the two sites as identified by S1 mapping.
Oligos 4 and 5 with predicted hairpin structures exhibited a 23% or 36% reduction, respectively, in the cleavage of both alleles as compared with their parent oligo 3. Therefore, no enhancement in allele discrimination was obtained with oligos 4 and 5 under the experimental conditions.
Allele-Specific Cleavage of Endogenous RPA70 mRNA in Human Cell Extracts.
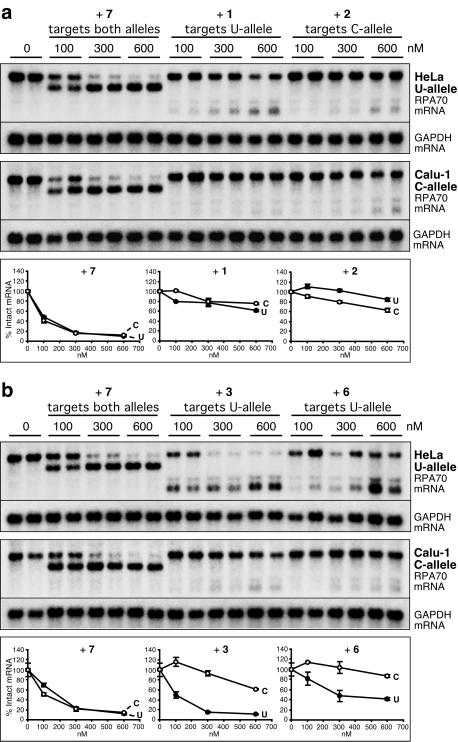
We next performed oligo-directed cleavage of the full-length, endogenous RPA70 mRNA in whole-cell extracts prepared from two genotyped human cell lines, HeLa (1,674 U) and Calu-1 (1,674 C). Cell lysis conditions were chosen to allow the disruption of both the plasma and the nuclear membranes in the absence of denaturants deleterious to the integrity of cellular proteins and nucleic acids. To control for cellular component-associated, nonspecific effects of PS-oligos in the two different cell types, we introduced an RPA70-specific but non-allele-discriminating 20-mer PS-oligo 7 to the assay to serve as a positive control, in addition to the use of SNP-targeting oligos 1 and 2, which serve as reciprocal negative controls. Oligo 7 is complementary to the 2,230–2,249 sequence region of the 3′-untranslated region of human RPA70 mRNA; it was previously shown to target the 1,674 U and 1,674 C alleles with equally high efficiency in a cell transfection assay (31).
Fig. 4 compares the extent of site-specific cleavage of the 1,674 U and 1,674 C alleles of the endogenous human RPA70 mRNA in the presence of 100, 300, and 600 nM of either U or C allele-specific oligos 1, 2, 3, and 6, or oligo 7, which targets both alleles. Both the full-length and one or two cleavage products were detected by using Northern blot hybridization. Because no E. coli RNase H was added to the reaction mixture, the site-specific cleavage of the mRNA is attributable to the presence of endogenous human RNase H in the cell extracts. Quantitative analysis of the extent of mRNA cleavage was done by measuring the full-length mRNA signal instead of the cleavage products. The latter did not hybridize with probes to the same extent because of probe design and were also likely more susceptible to inhomogeneous degradation by RNases in the cell extracts.
Figure 4.
Northern blot analysis of PS-oligo-directed cleavage of endogenous RPA70 mRNA in human cell extracts. The extracts were prepared from HeLa and Calu-1 cells that express only the 1,674 U or the 1,674 C allele of RPA70 mRNA, respectively. In addition to PS-oligos 1, 2, 3, and 6 as described in Fig. 3, PS-oligo 7 (5′-TGGTCTGCAGTTAGGGTCAG-3′) was used in the assay as a non-allele-discriminating control oligo that targets nucleotides 2,230–2,249 of the 3′-untranslated region of RPA70 mRNA. (a) Targeting the 1,674 U/C polymorphic site by using oligos 1 and 2. (b) Targeting the site of different allelic structures around nucleotide 1,656 by using oligos 3 and 6. Reactions were done in duplicate. The averaged data points of the duplicates are plotted with standard deviations as error bars and are shown below the corresponding Northern blots. Intact RPA70 mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels and expressed as a percentage of no-oligo control for each experiment.
The two cell types responded similarly to the addition of control oligo 7 over a concentration range of 100–600 nM (Fig. 4). In contrast, oligos 1, 2, 3, and 6 displayed varying degrees of targeting efficiency and allele specificity qualitatively similar to the results obtained in the purified system. In the presence of 300 nM of 20-mer 1 or 18-mer 2 targeting the 1,674 U/C polymorphic site, the majority of the matched allele remained uncleaved, and the matched allele was equally or slightly more cleaved than the singly mismatched allele (Fig. 4a). The lack of efficient cleavage with 1 and 2 can be explained by the inaccessibility of the 1,674 U/C polymorphic site as identified by using in vitro structural mapping. On the other hand, in the presence of 300 nM 20-mer 3, which targets the single-stranded structural motif of the U allele, only 15% of the U allele remained uncleaved, whereas >90% of the nontarget C allele was intact (Fig. 4b). Such high efficiency and specificity of U allele targeting by 3 is consistent with an accessible single-stranded structure in the 1,656 region of the U allele and an inaccessible structure in the same region of the C allele (see Figs. 1b and 2b). Based on the data in Fig. 4, we conclude that under the experimental conditions, the allele discrimination obtained by targeting the different allelic structures of RPA70 mRNA with 20-mer 3 is 2- to 6-fold greater than that obtained by directly targeting the polymorphism with 20-mer 1 when the percentage of intact mRNA is evaluated. The 15-mer oligo 6 also showed better allele discrimination than SNP-targeting oligos 1 and 2 at all three oligo concentrations, albeit less prominent than that of 20-mer 3.
It is important to note that structure-targeting oligos 3 and 6 are completely complementary to their target site sequences, because the target sites do not contain the polymorphic bases. Thus, the pronounced allele discrimination by these structure-targeting oligos reflects a significant difference in target-site accessibility between the two alleles. This accessibility difference could be caused by either different structural folds of the two alleles of endogenous RPA70 mRNA, different contacts with cellular components, or both effects. The positive correlation we have observed between the results in the purified system and those in cell extracts strongly supports the argument that the pronounced allele discrimination obtained with the structure-targeting oligos is caused mainly by structural differences between the two endogenous RPA70 mRNA alleles.
SNPs are the most abundant type of human DNA sequence variation. The fraction of SNPs associated with detectable differences in mRNA folding is yet to be discovered. Statistical studies of RNA secondary structures by Fontana et al. (27) predicted that structures derived from GC- or AU-rich sequences are much more sensitive to point mutation than those from sequences with uniform base distributions. The GC content of the 100 nucleotides flanking the AARS and RPA70 SNPs is 63% and 40%, respectively. However, data from the two examples described here are insufficient to provide a statistically significant assessment of the determinants of SNP-associated structural changes.
In conclusion, we have demonstrated that SNPs can cause significant structural variation in allelic forms of human mRNA. The structural differences occur both at and in regions flanking the SNP. The structural differences allow allele-specific cleavage of mRNA with PS-oligos complementary to a site of maximum differential structural accessibility that is independent of single-base discrimination at the polymorphic site. These findings illustrate how SNPs can affect mRNA structure and suggest mRNA-structure-dependent mechanisms by which SNPs can cause allele-specific biological consequences. Further elucidation of the contribution of common genetic variation to structural and functional diversity of mRNA will provide insight into fundamental mechanisms of human phenotypic variation and facilitate studies of disease susceptibility and drug response (4–7). The results described here should also be heeded by researchers interested in RNA as a therapeutic target; the possible effect of genetic variation on target heterogeneity should be considered for target characterization and drug design. Finally, structure-based mRNA targeting may be applied therapeutically to diseases where allele-specific RNA down-regulation would provide a therapeutic benefit. Nucleic acid-based inhibitors could be used for such purposes, however, small-molecule inhibitors (28–30) have more attractive pharmaceutical properties, including enhanced cellular uptake and possibly improved allele discrimination.
Acknowledgments
We thank R. M. Adams, D. I. Chasman, D. R. Govindaraju, J. C. Olson, P. Rioux, A. R. Schievella, J. D. Stepp, and M. Zillmann for scientific and technical help; F. R. C. Lippincott for cDNA sequencing; Prof. C. Hélène, Muséum National d’Histoire Naturelle, for suggesting experiments in cell extracts; and Prof. D. E. Housman, Massachusetts Institute of Technology, for advice and helpful comments. We also gratefully acknowledge Prof. G. L. Verdine, Harvard University, for advice, support, and critical reading of the manuscript.
ABBREVIATIONS
- SNP
single nucleotide polymorphism
- AARS
alanyl tRNA synthetase
- RPA70
replication protein A, 70-kDa subunit
- oligo
oligodeoxyribonucleotide
- PS-oligos
phosphorothioate oligos
References
- 1.Cooper D N, Krawczak M, Antonarakis S E. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. Vol. 1. New York: McGraw–Hill; 1995. pp. 259–291. [Google Scholar]
- 2.Chakravarti, A. (1999) Nat. Genet.21, Suppl., 56–60. [DOI] [PubMed]
- 3.Wang D G, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 4.Lander E S. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 5.Collins F S, Guyer M S, Chakravarti A. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 6.Weber W W. Pharmacogenetics. New York: Oxford Univ. Press; 1997. [Google Scholar]
- 7.Kleyn P W, Vesell E S. Science. 1998;281:1820–1821. doi: 10.1126/science.281.5384.1820. [DOI] [PubMed] [Google Scholar]
- 8.Coleman T P, Roesser J R. Biochemistry. 1998;37:15941–15950. doi: 10.1021/bi9808058. [DOI] [PubMed] [Google Scholar]
- 9.Côté J, Chabot B. RNA. 1997;3:1248–1261. [PMC free article] [PubMed] [Google Scholar]
- 10.van Gelder C W G, Gunderson S I, Jansen E J R, Boelens W C, Polycarpou-Schwarz M, Mattaj I W, van Venrooij W J. EMBO J. 1993;12:5191–5200. doi: 10.1002/j.1460-2075.1993.tb06214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allain F H-T, Gubser C C, Howe P W A, Nagai K, Neuhaus D, Varani G. Nature (London) 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 12.Vasserot A P, Schaufele F J, Birnstiel M L. Proc Natl Acad Sci USA. 1989;86:4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelletier J, Sonenberg N. Biochem Cell Biol. 1987;65:576–581. doi: 10.1139/o87-074. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Sarnow P, Siddiqui A. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ten Dam E B, Pleij C W A, Bosch L. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L X, Tinoco I., Jr J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 17.Hentze M W, Caughman S W, Rouault T A, Barriocanal J G, Dancis A, Harford J B, Klausner R D. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- 18.Casey J L, Koeller D M, Ramin V C, Klausner R D, Harford J B. EMBO J. 1989;8:3693–3699. doi: 10.1002/j.1460-2075.1989.tb08544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Addess K J, Basilion J P, Klausner R D, Rouault T A, Pardi A. J Mol Biol. 1997;274:72–83. doi: 10.1006/jmbi.1997.1377. [DOI] [PubMed] [Google Scholar]
- 20.Shaw J-P, Kent K, Bird J, Fishback J, Froehler B. Nucleic Acids Res. 1991;19:747–750. doi: 10.1093/nar/19.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peppel K, Baglioni C. BioTechniques. 1990;9:711–713. [PubMed] [Google Scholar]
- 23.Brown T, Mackey K. In: Current Protocols in Molecular Biology. Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Vol. 1. New York: Wiley; 1997. pp. 4.9.1–4.9.16. [Google Scholar]
- 24.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hostomsky Z, Hostomska Z, Matthews D A. In: Nucleases. Linn S M, Lloyd R S, Roberts R J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 341–376. [Google Scholar]
- 26.Tyagi S, Bratu D P, Kramer F R. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 27.Fontana W, Konings D A M, Stadler P F, Schuster P. Biopolymers. 1993;33:1389–1404. doi: 10.1002/bip.360330909. [DOI] [PubMed] [Google Scholar]
- 28.Werstuck G, Green M R. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 29.Mei H-Y, Cui M, Heldsinger A, Lemrow S M, Loo J A, Sannes-Lowery K A, Sharmeen L, Czarnik A W. Biochemistry. 1998;37:14204–14212. doi: 10.1021/bi981308u. [DOI] [PubMed] [Google Scholar]
- 30.Hamy F, Felder E R, Heizmann G, Lazdins J, Aboul-ela F, Varani G, Karn J, Klimkait T. Proc Natl Acad Sci USA. 1997;94:3548–3553. doi: 10.1073/pnas.94.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basilion, J. P., Schievella, A. R., Burns, E., Rioux, P., Olson, J. C., Monia, B. P., Lemonidis, K. M., Stanton, V. P., Jr., & Housman, D. E. (1999) Mol. Pharmacol., in press. [DOI] [PubMed]