Abstract
Background
The Mitotic Spindle Assembly Checkpoint (MSAC) is an evolutionary conserved mechanism that ensures the correct segregation of chromosomes by restraining cell cycle progression from entering anaphase until all chromosomes have made proper bipolar attachments to the mitotic spindle. Its malfunction can lead to cancer.
Principle Findings
We have constructed and validated for the human MSAC mechanism an in silico dynamical model, integrating 11 proteins and complexes. The model incorporates the perspectives of three central control pathways, namely Mad1/Mad2 induced Cdc20 sequestering based on the Template Model, MCC formation, and APC inhibition. Originating from the biochemical reactions for the underlying molecular processes, non-linear ordinary differential equations for the concentrations of 11 proteins and complexes of the MSAC are derived. Most of the kinetic constants are taken from literature, the remaining four unknown parameters are derived by an evolutionary optimization procedure for an objective function describing the dynamics of the APC:Cdc20 complex. MCC:APC dissociation is described by two alternatives, namely the “Dissociation” and the “Convey” model variants. The attachment of the kinetochore to microtubuli is simulated by a switching parameter silencing those reactions which are stopped by the attachment. For both, the Dissociation and the Convey variants, we compare two different scenarios concerning the microtubule attachment dependent control of the dissociation reaction. Our model is validated by simulation of ten perturbation experiments.
Conclusion
Only in the controlled case, our models show MSAC behaviour at meta- to anaphase transition in agreement with experimental observations. Our simulations revealed that for MSAC activation, Cdc20 is not fully sequestered; instead APC is inhibited by MCC binding.
Introduction
The growth of all organisms requires that the genome is accurately replicated and equally partitioned between two cellular progenies. In eukaryotes, the duplication of chromosomes, the separation of sister chromatids, and their segregation to opposite poles of the cell prior to cytokinesis are features of the cell cycle and grant maintenance of genomic integrity [1]. Eukaryotic cells have evolved a surveillance mechanism for DNA segregation, the MSAC. This checkpoint blocks anaphase onset and prevents exit from mitosis until all chromosomes are properly attached and have aligned on the mitotic spindle. Its malfunction leads to cell death [2]–[4], generates aneuploidy [5]–[7] (deviation from euploidy is seen in 70–80% of all types of human cancers [8]), might facilitate tumorgenesis [9], [10] and aging [11], and might contribute to cancer [12]–[14] (reviewed in [9], [15]–[19]).
Current models of the MSAC
Despite considerable experimental knowledge, the MSAC has not yet been modeled at a detailed molecular level. Doncic et al. [20] compared several mechanisms that could account for the inhibition of the APC:Cdc20 complex in yeast. They noticed that the design of the MSAC network is limited by physical constraints imposed by realistic diffusion constants and the relevant spatial and temporal dimensions in the yeast cell. Designing a simplified model of radial symmetry, they observed that amplifying the signal through the release of a diffusible inhibitory complex can describe checkpoint function. Nevertheless, their model does not fully take into account the molecular complexity. A similar approach was presented by Sear et al. [21]. They investigated two mechanisms for MSAC in metazoan cells: one involves free diffusion and sequestration of cell cycle regulators requiring a two-stage signal amplification cascade. The second mechanism involves spatial gradients of a short-lived inhibitory signal that propagates by diffusion and primarily by active transport along spindle microtubules. Both mechanisms might act in parallel. Mathematical modeling of cell cycle control in budding yeast was analyzed in more details in [22], however not focusing on MSAC. A model for the exit from mitosis [23] describes the control of the checkpoint, however not considering BubR1 (Mad3 in yeast) nor MCC.
Here, we suggest a kinetic model based on a set of time dependent nonlinear ordinary differential equations for protein concentrations. The model describes the MSAC on the molecular level. It focuses on MSAC control in mitosis at metaphase to anaphase transition; it does not include exit from mitosis (e.g., Cdh1). The Mad1/Mad2 action and Cdc20 inhibition is described by a recently developed mathematical model [24] based on the biochemical Template Model [25], [26]. The description of MCC formation and APC inhibition is based on results from biochemical experiments [15], [27]–[31]. We present the chemical basis of the reactions and explain the chemical reaction equations in detail. Then, we describe the corresponding ordinary differential equations and their mathematical treatment.
It is still unclear how the MCC:APC complex falls apart and how the APC:Cdc20 complex is formed afterwards. Therefore we consider here two alternative pathways in our MSAC Model, the “Dissociation” and the “Convey” variants, differing in one reaction: either the MCC:APC complex dissociates into the MCC and the APC (“Dissociation variant”), or, alternatively, Cdc20 being a member of the MCC remains at the APC and only the other MCC complex members leave the MCC:APC (“Convey variant”). We noticed that checkpoint behavior requires that the dissociation of the MCC:APC is regulated by microtubule attachment. For this purpose we introduced a factor for the attachment dependent control of the associated reactions. We compared the controlled versus the uncontrolled case. Those resulting model variants that describe checkpoint function properly are validated by comparison to ten different deletion and over-expression experiments taken from literature. From our model calculations we conclude that the meta- to anaphase transition and the APC are not inhibited by Cdc20 sequestering but instead the APC is bound and blocked by the MCC.
Methods
Biochemical background
Our model incorporates three MSAC-related mechanisms: the Template Model, the (kinetochore dependent) MCC formation, and the APC inhibition. Their biochemical details will be explained in the following.
Mad2 Template Model
DeAntoni et al. [25] proposed the “Template Model” explaining the mechanism of Mad2 recruitment to the kinetochore during checkpoint activation and subsequent transfer to sequester Cdc20. Recent work by Vink et al. [27] and Mapelli et al. [26] provide additional support for the Template Model. Moreover, this model has been confirmed by Nezi et al. [32], and is entirely consistent with recent Fluorescence Recovery After Photobleaching (FRAP) data [27], [28]. The Template Model [25] is superior and more solid than the Exchange Model [33], which we confirmed in a recent in silico study [24] (for comparison and details see [25]–[27], [32], [34], for reviews see [35]–[39]).
The Mad2 Template Model is described by the reaction equations Eqs. (1)–(3) (see chemical reaction scheme, below). It is assumed that Mad1 and C-Mad2 form a stable core complex Mad1:C-Mad2 at unattached kinetochores [25]. In our nonlinear ordinary differential equations (ODEs) model, we assume that this process has already been completed. Therefore, there is no free Mad1. Equation (1) describes how the Mad1:C-Mad2 core complex binds additional molecules of O-Mad2 through formation of conformational heterodimers between the C- Mad2 subunit of the Mad1:C-Mad2 complex and O-Mad2. Upon Mad1:C-Mad2 binding, O-Mad2 adopts an intermediate conformation (O-Mad2*), which can quickly and efficiently bind Cdc20 and switch to the C-conformation. This process is documented by Eq. (2): Cdc20 binding to the complex Mad1:C-Mad2:O-Mad2* leads to the conversion of O-Mad2* to C-Mad2 forming together with Cdc20 the complex Cdc20:C-Mad2; Cdc20:C-Mad2 is assumed then to dissociate off Mad1:C-Mad2 [40]. Finally, we assume that the Cdc20:C-Mad2 complex can dissociate into Cdc20 and O-Mad2 (Eq. (3)).
MCC formation
Equations (4) and (5) describe the formation of the MCC, which contains Mad2, Bub3, BubR1 and Cdc20 in apparently equal stoichiometries [41]–[44]. Bub3 associates quite stably with BubR1 [18], [41], [45]. This interaction is constitutive and is required for the localization of BubR1 to the kinetochores during mitosis. Like for the Mad1:C-Mad2 complex, we do not model the dynamics of the formation of the BubR1:Bub3 complex. BubR1 cannot bind Mad2 directly [40]. Moreover, BubR1 does not form a ternary complex with Mad2 and Cdc20. Two Cdc20 binding sites were identified on BubR1 [46], [47]. Binding of the N-terminal region of BubR1 to Cdc20 requires prior binding of Mad2 to Cdc20 [47]. Consistently, the Bub3:BubR1 complex can bind to Cdc20:C-Mad2 in order to form the MCC (Eq. (4), rate constants k4 and k-4). The other site of BubR1 (between residues 490 and 560) can bind Cdc20 tightly regardless of Mad2 being bound to Cdc20 [47]. Thus, BubR1 can form a ternary complex with Bub3 and Cdc20 (Eq. (5)) which however has no inhibitory activity at the APC (unpublished data [41]). Equation (6) and its low rate were mentioned by Musacchio & Salmon [15].
APC inhibition
The MCC is considered to be essential for MSAC function, because it binds and inhibits the APC [41]–[43], [48]–[55]. However, MCC inhibits only the mitotic, and not the interphase APC [56]. The interaction between APC and MCC is quite labile in the absence of unattached kinetochores [41]. How the MCC inhibits APC activity is poorly understood [15]. The MCC might bind to the APC as a pseudosubstrate due to a KEN-box motif in BubR1 [40], [53], [57], [58]. This indicates that the MCC needs to disassemble from the APC at metaphase to elicit anaphase [40], [53]. Bub1 and Aurora-B kinase contribute directly to the formation of a complex of the MCC with the APC [53] (represented by k7 in Eq. (7)). Unattached kinetochores might sensitize the APC for inhibition by the MCC [20], [21], [38], [41] (represented by u in Eq. (7)). In addition to kinetochore attachment, tension is important for MSAC inactivation [59], [60]: if both sister kinetochores attach to microtubules from the same pole, not enough tension is generated and microtubules kinetochore attachment is destabilized to correct the problem [15]. This destabilization depends on Aurora-B kinase [56], [61]–[63]. Again, these effects are subsumed by the switching parameter u. For complex dissociation we consider two model variants:
In the “Dissociation variant”, we assume that MCC binds to APC and that this binding is reversible (Eq. (7a)). Free Cdc20 has to bind reversibly to APC (Eq. (8)), effectively competing with MCC.
In the “Convey variant”, we do not assume that the APC:MCC complex simply dissociates into APC and MCC, but that the MCC complex falls apart so that the Cdc20 contained in the MCC complex can bind to the APC (Eq. (7b)).
Control by attachment
Several reactions in the reaction scheme are controlled by the attachment of microtubules to the kinetochore which is realized by the factor u present in several reaction equations [64]. Factor u represents the function of proteins like p31comet, UbcH10, and Dynein (and its activator Spindly [65]). p31comet prevents further Mad2 turnover on Mad1 and neutralizes the inhibitory activity of Cdc20-bound Mad2 [26], [66]–[68]. Catalytically active UbcH10 can promote the release of checkpoint proteins from APC [69]. Dynein [70] removes the Mad1:C-Mad2 2:2 complex from the kinetochore site after microtubule attachment. Thus, p31comet, UbcH10, and Dynein work in concert during checkpoint inactivation.
Also the MCC:APC complex dissociation might be attachment controlled. We therefore introduced the factor u′ in Eq. (7a) and Eq. (7b), allowing us to compare the uncontrolled (u′ = 1) with the controlled (u′ = 1−u, i.e., u′ = 0 before and u′ = 1 after attachment) case. The switching parameter u′ might represent the protein function of Usp44, which deubiquitinates the APC co-activator Cdc20 both in vitro and in vivo, and thereby directly counteracts the APC-driven disassembly of Mad2:Cdc20 complexes [71], [72].
Chemical reaction scheme
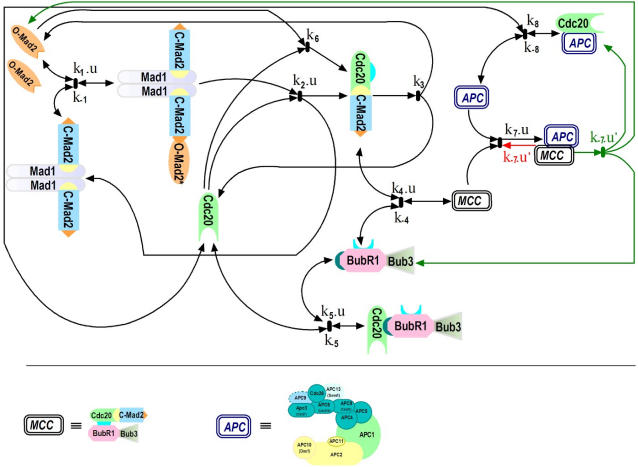
In our model of the MSAC mechanism, 9 biochemical reaction equations describe the dynamics of the following 11 species: Mad1:C-Mad2, O-Mad2, Mad1:C-Mad2:O-Mad2*, Cdc20, Cdc20:C-Mad2, Bub3:BubR1, MCC, Bub3:BubR1:Cdc20, APC,MCC:APC, and APC:Cdc20. Because the dissociation of the MCC:APC complex is not known in detail, we introduce two variants for the reaction equation for MCC:APC dissociation.
The Dissociation variant is defined by the following reaction rules (Figure 1, red lines):
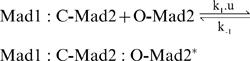 |
(1) |
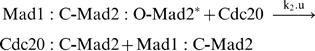 |
(2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (7a) |
| (8) |
Figure 1. Schematic network of the MSAC model.
The arrows describe the interactions between the proteins and complexes. Red lines represent the Dissociation variant, green lines represents the Convey variant, while the black arrows are common to both. The switching parameter u models the effect of the attachment. We set u = 1 for the unattached case and u = 0 for the attached case. We set u′ = 1 for the uncontrolled scenario and u′ = 1−u for the controlled scenario (Table 1). The model incorporates three central control mechanisms, namely Mad1/Mad2 induced Cdc20 sequestering based on the Template Model, MCC formation, and APC inhibition. These sub-systems can be red from left to right. Nine biochemical reaction equations describe the interactions of 11 species: Mad1:C-Mad2, O-Mad2, Mad1:C-Mad2:O-Mad2*, Cdc20, Cdc20:C-Mad2, Bub3:BubR1, MCC, Bub3:BubR1:Cdc20, APC, MCC:APC, and APC:Cdc20. Below the network, the subunits of MCC as well as APC are depicted.
The reaction rules defining the second variant, the Convey variant, are different from this set by replacing the back reaction Eq. (7a) by Eq. (7b) (Figure 1, green lines):
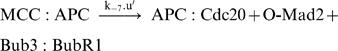 |
(7b) |
Both variants are controlled by the switching parameters u and u′. They represent a signal generated by the unattached and attached kinetochores, respectively. If the kinetochore is unattached, we set u = 1, otherwise u = 0. For instance, formation of Mad1:C-Mad2:O-Mad2* (Eq. (2)) can only take place as long as the kinetochores are unattached [25].
The switching parameter u′ represents an additional hypothetical control, whose biochemical realization is described above. For each of the two dissociation variants, we therefore considered two scenarios: In the first, we assume that this control does not exist by setting u′ = 1. In the second, we assume that there is a control by setting u′ = 1−u. This is summarized in Table 1:
Table 1. Model Variants.
| Scenario | Model variants | Reaction rules | Control of MCC:APC Dissociation |
| Uncontrolled | Dissociation | Eqs. (1)–(7), (7a), (8) | u′ = 1 |
| Controlled | Dissociation | Eqs. (1)–(7), (7a), (8) | u′ = 1−u |
| Uncontrolled | Convey | Eqs. (1)–(7), (7b), (8) | u′ = 1 |
| Controlled | Convey | Eqs. (1)–(7), (7b), (8) | u′ = 1 − u |
Mathematical treatment and simulation
By applying general principles of mass-action kinetics, we converted the reaction rules into sets of time dependent nonlinear ordinary differential equations (ODEs) for the Dissociation variant (Text S1, Eqs. (D.1)–(D.11)) and for the Convey variant (Text S1, Eqs. (C.1)–(C.11)).
For the rate constants ki, we selected experimentally determined values, if available (Table 2). In the other cases, we selected representative values exemplifying their whole physiologically possible range. We also fitted unspecified parameters by minimizing an APC:Cdc20 concentration dependent objective functional (Text S1, C), taking into account the range of parameter values from experiments [15], [27], [40], [55], [73].
Table 2. Model parameters.
| Parameters | Comments and References |
| Species initial concentration | |
| [Cdc20] = 2.2* 10−7M | [40], [55], [71] |
| [Mad2]total = 2* 10−7M | [40], [55], [71] |
| [BubR1:Bub3] = 1.3* 10−7M | [40], [55] |
| [APC] = 0.9*10−7M | [69] |
| Other species are zero | |
| Species concentration ratios | |
| 25% of [Mad2]total associated with Mad1, [Mad1:C-Mad2] = 25%[Mad2]total | [33], [40] |
| [O-Mad2] = 75%[Mad2]total | [40], [71] |
| Model–Parameters | |
| k1 = 2*105 M−1s−1 | [31] |
| k-1 = 2*10−1 s−1 | [31] |
| K2 = 108 M−1s−1 | [24] |
| K3 = 10−2 s−1 | [24] |
| K4 = 107 M−1s−1 | [62] |
| k-4 = 10−2 s−1 | [62] |
| K5 = 104 M−1s−1 | [62] |
| k-5 = 10−1 s−1 | [62] |
| K6 = 103 M−1s−1 | [15] |
| K7 = 108 M−1s−1 | This study |
| k-7 = 8*10−2 s−1 | This study |
| K8 = 5*106 M−1s−1 | This study |
| k-8 = 8*10−2 s−1 | This study |
In a typical simulation, we initialized all reaction partners according to Table 2 and numerically integrated the ODEs until steady state was reached using u = 1 (denoting unattached kinetochores). Then we set u = 0 (kinetochores attached) and continued integrating the ODEs until we again reached a steady state.
The minimum concentration of APC:Cdc20 before attachment and the speed of recovery after attachment (recovery time) are criteria for MSAC function and were analyzed to compare the models. Deduced from the biochemical data (see above), the APC:Cdc20 concentration must be low before and the recovery must be fast after attachment.
Results
We developed a theoretical model of the human biochemical mitotic checkpoint at meta- to anaphase transition. As described in the literature, many proteins contribute to checkpoint function. The key players and their interactions are captured by the reaction equations introduced in the previous section. We transformed these equations into ODEs and selected specific values for the initial concentrations and rate constants from the literature and our previous publications (summarized by Table 2). For only four values we could not identify specific data in the literature. We obtained these values by optimizing the properties of the model according to the APC:Cdc20 level: this complex level should be low in metaphase and high in anaphase; furthermore, the switching should be fast (see Text S1 for details). We found good behavior of the model network for the values k7 = 108M−1s−1, k−7 = 8*10−2s−1, k8 = 5*106M−1s−1, and k−8 = 8*10−2s−1.
MSAC Model behavior
We analyzed the dynamics of the model integrating 11 proteins and complexes of the MSAC. The literature does not provide a clear view, yet, about how the MCC:APC complex dissociates resulting in APC activation. Therefore, we introduced two alternative reaction pathways: In the first variant, we assume that the MCC:APC complex dissociate into MCC and APC (reaction Eq. (7a)), subsequently allowing the MCC to disassemble into its parts according to reaction Eq. (4) (Dissociation variant). In the second variant, the MCC component Cdc20 may stay in the complex with APC and only the further MCC complex members dissociate according to reaction Eq. (7b)(Convey variant).
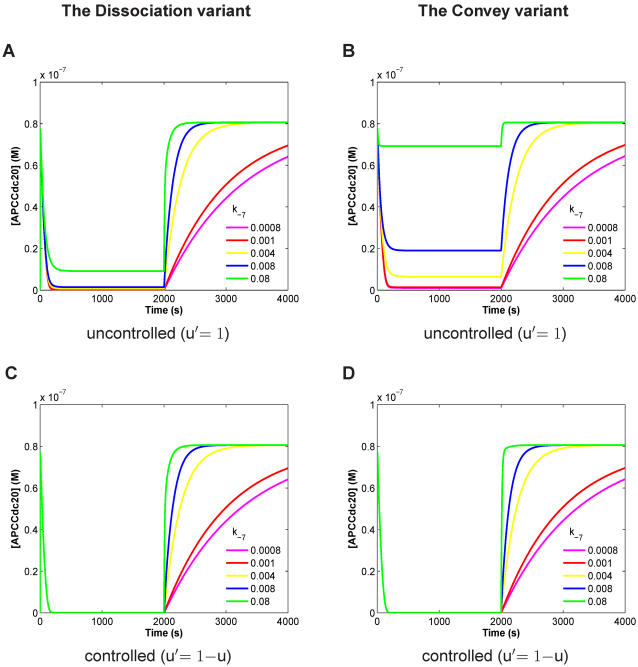
Figure 2 displays the APC:Cdc20 concentrations over time. For both, Dissociation and Convey variant, we have selected the time range such that each concentration can reach steady state. For all calculations, the concentrations and rates of Table 2 were chosen including those for k7, k8, and k−8. We varied the rate of k−7 (dissociation of MCC:APC) between 0.0008 and 0.08, because k−7 is unknown and crucial for model behavior. For both model variants, we distinguished 2 scenarios: in one scenario reaction Eq. (7a) (or Eq. (7b)) of the checkpoint is valid all the time (“uncontrolled”), while in the other case this reaction is silenced until it is activated by microtubule attachment to the kinetochore (“controlled”). This property of the controlled case is realized by introducing the factor u′ for reaction Eq. (7a) and Eq. (7b).
Figure 2. Dynamical behavior of APC:Cdc20 concentration versus time for the Dissociation variant (A, C) and the Convey variant (B, D) each in the uncontrolled (A, B) and the controlled (C, D) case.
Calculation results are presented for different values of the rate k−7 in [s−1 >(dissociation of MCC:APC) between 0.0008 and 0.08, because k−7 is unknown and crucial for model behavior, as indicated. The APC:Cdc20 concentration should be close to zero before attachment and should rise quickly after attachment. Spindle attachment occurs at t = 2000s (switching parameter u from 1 to 0). For the uncontrolled case (A, B), both variants cannot explain the checkpoint behavior; and the Convey variant is even less satisfying compared to the Dissociation variant. In the controlled case (C, D), both variants fully inhibit APC:Cdc20 before attachment and both show fast switching recovery for high k−7 values. The controlled Convey variant (D) is slightly faster (by about 5 mins) in switching compared to the controlled Dissociation (C) variant. Parameters setting according to Table 1 and Table 2.
In the uncontrolled case, our model cannot explain the checkpoint behavior, independently of which pathway is chosen (Figure 2A–B). For the Dissociation variant, the APC:Cdc20 concentration is low for low values of k−7, however, in this case the switching recovery is unrealistically slow. On the other hand, for fast switching, k−7 must be high resulting in an increased APC:Cdc20 concentration before attachment (Figure 2A–B). This behavior is even worse for the Convey variant in the uncontrolled case (Figure 2B). For low values of k−7, both pathways behave rather similarly; for higher values of k−7 the Convey variant is even less satisfying compared to the Dissociation variant.
In the controlled case, we introduced the factor u′ (see above) regulating reaction Eq. (7a) and Eq. (7b), and re-calculated the model. Both pathways fully inhibit APC:Cdc20 before attachment and both show very fast switching recovery for high k−7 values (Figure 2C–D). Thus, a distinction between the two pathways in the controlled case is not possible based on our theoretical results. We observed that the controlled Convey variant is slightly faster (by about 5 mins) in switching compared to the controlled Dissociation variant. This makes the Convey variant slightly superior, however, we think that this difference is too small for a clear preference between the two pathways. Experimental measurements have to distinguish between these cases. Such experiments are in progress in our laboratory.
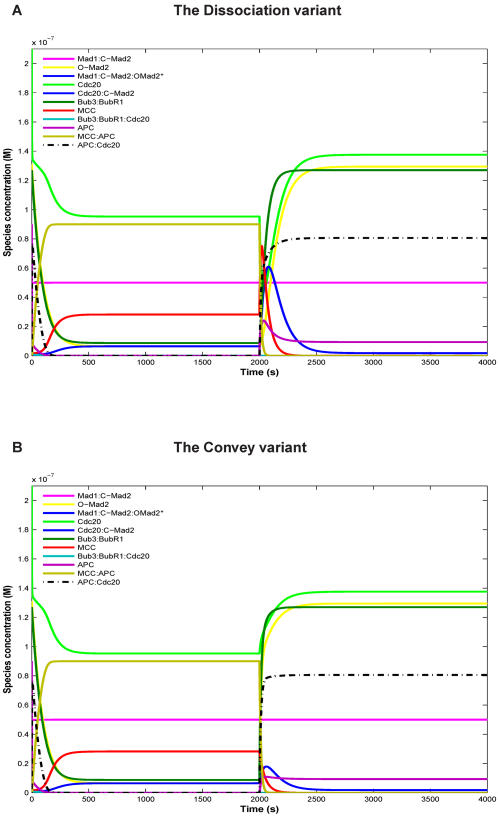
In addition to the APC:Cdc20 concentration values, we also analyzed the time-dependent concentrations of all reaction components. We observed differences between the two pathways in the controlled case for sub-complexes like Cdc20:C-Mad2 and MCC (Figure 3).
Figure 3. Species concentration over time for the controlled Dissociation variant (A) and the controlled Convey variant (B).
Spindle attachment occurs at t = 2000s (switching parameter u from 1 to 0 and u' from 0 to 1). Both variants show similar qualitative dynamics. However, quantitative differences can be observed for species like Cdc20:C-Mad2 and MCC. Parameters setting according to Table 2 (“wild type”).
In our simulations, the MCC completely sequesters the APC so that no free APC is available until the microtubules are attached. Thus, Cdc20 has a dual function: until kinetochore attachment, Cdc20 contributes to MCC formation and thus APC inhibition, while after attachment Cdc20 acts as the APC activator.
Model validation by mutation experiments
In order to validate our model, we tested different mutations (deletion and over-expression) of the proteins and complexes involved, measured in different organisms (Table 3).
Table 3. In-silico mutation experiments for validation.
| No. | Species | Organisms | Exp. | Experimental effects | Effects in our models |
| 1. | Mad2 | H. s. | D | -Impaired MSAC [32] | - MSAC fails to arrest |
| Mad2 | M. | D | -Unable to arrest [72] | & no Cdc20 sequestering. | |
| Mad2 | H. s. & M. | D | -Defective MSAC [73] | - [APC:Cdc20] very high. | |
| Mad2 | H. s. | D | -Unable to bind Cdc20 or Mad1 [74]. More refs.: [31], [40], [108] [1], [34], [109]-[111]. | ||
| 2. | Mad2 | H.s. | O | -Activates the MSAC [25] | -Activates the MSAC |
| Mad2 | S.p. | O | -Blocks mitosis [90] | & full Cdc20 sequestering. -[APC:Cdc20] very low. | |
| 3. | Mad1 | H.s. | D | - MSAC inactivation & aneuploidy [75]. | - MSAC fails to arrest & no Cdc20 sequestering. |
| Mad1 | S.p. | D | - cell death [76]. More refs.: [77]-[79]. | -[APC:Cdc20] very high. | |
| 4. | BubR1 | H.s. | D | -Reduced MSAC function, Reduced MSAC binding to Cdc20:C-Mad2 [47]. | -MSAC fails to arrest. |
| -[APC:Cdc20] very high. | |||||
| BubR1 | M. | D | -Increased polyploidy [82]. More refs.: [83], [84]. | ||
| 5. | Bub3 | M. | D | -Fails to arrest [80], [81]. | -MSAC fails to arrest. |
| -[APC:Cdc20] very high. | |||||
| 6. | Cdc20 | S.c. | O | -Allows cells with a depolymerized spindle or damaged DNA to leave mitosis [91]. | -MSAC fails to arrest. |
| -[APC:Cdc20] very high. | |||||
| -Impairment MSAC and aneuploidization in oral cancer [112]. | |||||
| 7. | Cdc20 | H.s. | D | -Reduced binding to Mad2, selective disruption from Mad2 [85]. | - blocks mitosis. |
| -[APC:Cdc20] very low. | |||||
| Cdc20 | S.p. | D | -Arrest in metaphase [113]. | ||
| 8. | Bub1 | Drosophila | Inh. | -Chromosome missegregation [86]. | -MSAC fails to arrest. |
| -[APC:Cdc20] very high. | |||||
| Bub1 | H.s. | Inh. | -Disruption of Bub3 localization, disruption of Bub3 binding to BubR1 [81]. | ||
| 9. | Aurora B | Xenopus | Inh. | -Overriding the MSAC function, perturbs MTs dynamics [87]. | -MSAC fails to arrest. |
| -[APC:Cdc20] very high. | |||||
| Aurora B | S.c. | Inh. | -Unregulated MTs, | ||
| - MSAC fails to arrest [88], [89] | |||||
| 10. | APC (units) Cdc26, apc9 Cdc6,doc1 | S.p. | D | -Disruption of complex association [92], [93], More refs.: [114]. | -Activates the MSAC. |
| -[APC:Cdc20] very low. |
Abbreviations: D for deletion or knockdown experiments, O for over-expression experiments, and Inh. for inhibition; S.c., Saccharomyces cerevisiae; S.p., Schizosaccharomyces pombe; H. s., Homo sapiens(Human); and M., Murine.
Recent experimental studies report that deletion in different organisms of any ofMad2 [32], [74]–[76], Mad1 [77]–[81], Bub3 [82], [83], BubR1 [47], [84]–[86], Cdc20 [87], Bub1 [83], [88], or Aurora B [89]–[91] resulted in MSAC defects like premature sister-chromatid separation, no mitotic arrest, reduced partner binding, increase of polyploidy, or death. Experimental details and our model predictions are in qualitative agreement as summarized in Table 3: For example, over-expression of Mad2 [25], [92] activates the MSAC resulting in mitotic arrest, while over-expression of Cdc20 [93] allows cells to exit from mitosis, however with a depolymerized spindle or damaged DNA. Deletion of any of the APC subunits Cdc26, Apc9, Cdc6 or Doc1 disrupts complex association [94], [95] with no anaphase initiation. We observed that deletions and/or over-expression of proteins, realized experimentally or in our model, change checkpoint function in the same way.
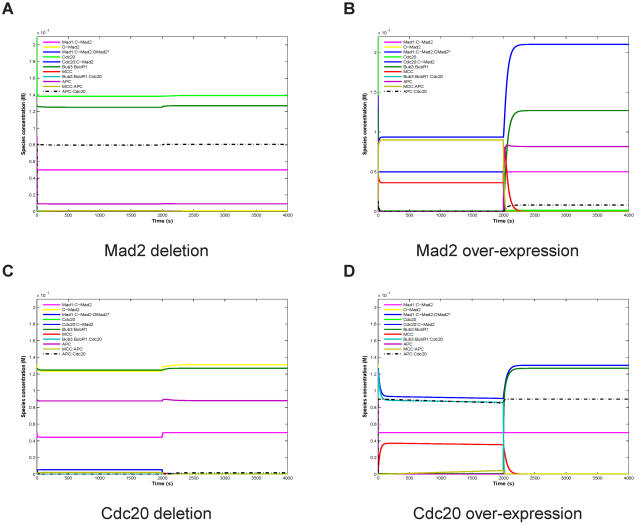
For the essential checkpoint proteins, Mad2 and Cdc20, we present the mutation effect on our model in detail in Figure 4 for the Convey variant. The effect is basically the same for the Dissociation variant (Figure S1). For Mad2 or Cdc20 deletion, the concentrations of all model components are rather stable, that is, they are almost not affected by microtubule attachment. However, in the case of Mad2 deletion, the APC:Cdc20 concentration is high (Figure 4A) while for Cdc20 deletion this concentration is zero by definition (Figure 4C). In the case of Mad2 or Cdc20 over-expression, many component concentrations were affected. In particular, for Mad2 over-expression the APC:Cdc20 concentration remains low before and significantly lower than in the wild type after attachment, explaining mitotic arrest and the delay of exit from mitosis (Figure 4B). In contrast, for Cdc20 over-expression, the APC:Cdc20 concentration is high before and after attachment (Figure 4D) resulting in total checkpoint failure. Thus, our MSAC Model is able to explain the presented mutation phenotypes (Table 3, Figures 4, S1, S2, and S3).
Figure 4. Simulation of Mad2 and Cdc20 mutations for the controlled Convey variant (cf. Table 3).
For deletion we set the respective initial concentration 100 times lower, and for over-expression 10000 times higher. For proper functioning, APC:Cdc20 concentration should be very low(zero) before the attachment, and should increase quickly after attachment. Deletion of Mad2 (A) or Cdc20 (C) destroys the switching behavior, that is, the concentrations of all model species are rather constant. Mad2 deletion (A) causes high APC:Cdc20 concentration right from the beginning, while for Cdc20 deletion (C) APC:Cdc20 concentration is zero, by definition. For Mad2 over-expression (B) or Cdc20 over-expression (D), many species concentrations are affected. Particularly, for Mad2 over-expression (B) the APC:Cdc20 concentration remains low before attachment and, after attachment, stays significantly lower than in the wild type (meaning mitotic arrest). In contrast, for Cdc20 over-expression (D), the APC:Cdc20 concentration is high before attachment and also after attachment (meaning checkpoint failure). Spindle attachment occurs at t = 2000s (switching parameter u from 1 to 0 and u' from 0 to 1). Further setting as in Figure 2.
Discussion
Although our model is able to explain checkpoint function, this explanation does not contain details regarding the bio-molecular nature of the switching signal represented by the abstract factor u in our model. For a general explanation of mitosis it is desirable to replace the abstract factor “u” by chemical reactions of species like p31comet, Dynein, Usp44, and/or UbcH10. These species play a role in the signaling of the attachment to the MSAC control network we modeled here. When further biochemical details become available, we will replace “u” by a network model encompassing these species. Other additional proteins and complexes are involved in MSAC function implicitly. These species grant localization of outer kinetochore proteins as well as checkpoint proteins, which do not appear in our model explicitly. Examples are Bub1 (responsible for Bub3 and BubR1 localization [45], [96], [97]) and Mps1, an essential component of the MSAC [43], [98]–[101] required for kinetochore localization of Mad1 and Mad2 [102]–[109]. Considering these additional proteins and their spatial localization would be an important next step towards a systems level model of mitosis.
Supporting Information
Supplement: Differential equations, Materials, Methods, and Optimization
(0.03 MB PDF)
Simulation of Mad2 and Cdc20 mutations for the controlled Convey Dissociation (cf. Table 3). For deletion we set the respective initial concentration 100 times lower, and for over-expression 10000 times higher. For proper functioning, APC:Cdc20 concentration should be very low(zero) before the attachment, and should increase quickly after attachment. Deletion of Mad2 (A) or Cdc20 (C) destroys the switching behavior, that is, the concentrations of all model species are rather constant. Mad2 deletion (A) causes high APC:Cdc20 concentration right from the beginning, while for Cdc20 deletion (C) APC:Cdc20 concentration is zero, by definition. For Mad2 over-expression (B) or Cdc20 over-expression (D), many species concentrations are affected. Particularly, for Mad2 over-expression (B) the APC:Cdc20 concentration remains low before attachment and, after attachment, stays significantly lower than in the wild type (meaning mitotic arrest). In contrast, for Cdc20 over-expression (D), the APC:Cdc20 concentration is high before attachment and also after attachment (meaning checkpoint failure). Spindle attachment occurs at t = 2000s (switching parameter u from 1 to 0 and u' from 0 to 1). Further setting as in Figure 2.
(0.69 MB TIF)
Simulation of BubR1, Aurora B, Mad1, and APC (subunits) mutations for the controlled Dissociation variant (cf. Table 3). For deletion we set the respective initial concentration 100 times lower, and for APC subunit 10 times lower. Spindle attachment occurs at t = 2000s (switching parameter u from 1 to 0 and u' from 0 to 1). Note that Bub3 deletion has the same effect like BubR1 (data not shown), and Bub1 deletion has the same effect like Aurora B (data not shown). APC:Cdc20 for the wild type should be very low (zero) before the attachment and increase quickly after attachment. Deletion of any of BubR1, Mad1, or Aurora B (as well as Bub3 and Bub1) results in high concentration of APC:Cdc20 right from the beginning (meaning checkpoint failure). Deletion of APC subunits disrupts the complex and thus makes APC:Cdc20 unavailable, which implies mitotic arrest. Parameter setting according to Table 2.
(0.74 MB TIF)
Simulation of BubR1, Aurora B, Mad1, and APC (subunits) mutations for the controlled Convey variant (cf. Table 3). The qualitative effect of the mutations is the same as for the Dissociation variant shown in Figure S2. There are quantitative differences in some species concentrations (c.f. Panal B, deletion of Aurora B). Same parameter settings as in Figure S2.
(0.72 MB TIF)
Acknowledgments
We thank Michael Rape for corresponding information on USP44 component.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: We acknowledge funding by the DAAD (German Academic Exchange Service, grant A/04/31166) the DFG (German Research Foundation, grant Di 852/4-x), the BMBF (Federal Ministry of Education and Research of Germany, grant 0312704A), and the EU (ESIGNET, grant 12789).
References
- 1.Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–9. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver BAA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Dorer RK, Zhong S, Tallarico JA, Wong WH, Mitchison TJ, et al. A small- molecule inhibitor of Mps1 blocks the spindle-checkpoint response to a lack of tension on mitotic chromosomes. Curr Biol. 2005;15:1070–6. doi: 10.1016/j.cub.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Heald R. Cell biology. Serving up a plate of chromosomes. Science. 2006;311:343–4. doi: 10.1126/science.1123525. [DOI] [PubMed] [Google Scholar]
- 5.Kim M, Kao GD. Newly identified roles for an old guardian: profound deficiency of the mitotic spindle checkpoint protein BubR1 leads to early aging and infertility. Cancer Biol Ther. 2005;4:164–5. doi: 10.4161/cbt.4.2.1506. [DOI] [PubMed] [Google Scholar]
- 6.Steuerwald N. Meiotic spindle checkpoints for assessment of aneuploid oocytes. Cytogenet Genome Res. 2005;111:256–9. doi: 10.1159/000086897. [DOI] [PubMed] [Google Scholar]
- 7.Mondal G, Roychoudhury S. The spindle assembly checkpoint and its defects in human cancer. Int J Hum Genet. 2003;3:89–97. [Google Scholar]
- 8.Iwanaga Y, Kasai T, Kibler K, Jeang KT. Characterization of regions in hsMAD1 needed for binding hsMAD2. A polymorphic change in an hsMAD1 leucine zipper affects MAD1-MAD2 interaction and spindle checkpoint function. J Biol Chem. 2002;277:31005–13. doi: 10.1074/jbc.M110666200. [DOI] [PubMed] [Google Scholar]
- 9.Kops GJPL, Weaver BAA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–85. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 10.Michel L, Diaz-Rodriguez E, Narayan G, Hernando E, Murty VVVS, et al. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci U S A. 2004;101:4459–64. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker D, Jeganathan K, Cameron JD, Thompson M, Jueja S, et al. BubR1 insufficiency causes early onset of aging-associted phenotypes and infetility in mice. Natrure Genetics. 2004;36:744–49. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 12.Compton DA. Chromosomes walk the line. Nat Cell Biol. 2006;8:308–10. doi: 10.1038/ncb0406-308. [DOI] [PubMed] [Google Scholar]
- 13.Gupta A, Inaba S, Wong OK, Fang G, Liu J. Breast cancer-specific gene 1 interacts with the mitotic checkpoint kinase BubR1. Oncogene. 2003;22:7593–9. doi: 10.1038/sj.onc.1206880. [DOI] [PubMed] [Google Scholar]
- 14.Mondal G, Sengupta S, Panda CK, Gollin SM, Saunders WS, et al. Overexpres-sion of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidiza-tion in oral cancer. Carcinogenesis. 2007;28:81–92. doi: 10.1093/carcin/bgl100. [DOI] [PubMed] [Google Scholar]
- 15.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature Reviews Molecular Cell Biology. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 16.Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–99. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Taylor SS, Scott MIF, Holland AJ. The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 2004;12:599–616. doi: 10.1023/B:CHRO.0000036610.78380.51. [DOI] [PubMed] [Google Scholar]
- 19.Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol. 2002;3:731–41. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- 20.Doncic A, Ben-Jacob E, Barkai N. Evaluating putative mechanisms of the mitotic spindle checkpoint. Proc Natl Acad Sci U S A. 2005;102:6332–7. doi: 10.1073/pnas.0409142102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sear RP, Howard M. Modeling dual pathways for the metazoan spindle assembly checkpoint. Proc Natl Acad Sci U S A. 2006;103:16758–63. doi: 10.1073/pnas.0603174103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B, et al. Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell. 2004;15:3841–62. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciliberto A, Lukacs A, Toth A, Tyson JJ, Novak B. Rewiring the exit from mitosis. Cell Cycle. 2005;4:1107–12. [PubMed] [Google Scholar]
- 24.Ibrahim B, Diekmann S, Schmitt E, Dittrich P. Mad2 binding is not sufficient for complete cdc20 sequestering (an in-silico study). Biophys Chem, submitted. 2007 doi: 10.1016/j.bpc.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 25.DeAntoni A, Pearson CG, Cimini D, Canman JC, Sala V, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–25. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 26.Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–84. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vink M, Simonetta M, Transidico P, Ferrari K, Mapelli M, et al. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr Biol. 2006;16:755–66. doi: 10.1016/j.cub.2006.03.057. [DOI] [PubMed] [Google Scholar]
- 28.Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, et al. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–52.10. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 29.Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–64. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 30.Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule- independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol. 2002;158:841–7. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–50. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nezi L, Rancati G, DeAntoni A, Pasqualato S, Piatti S, et al. Accumulation of Mad2-Cdc20 complex during spindle checkpoint activation requires binding of open and closed conformers of Mad2 in Saccharomyces cerevisiae. J Cell Biol. 2006;174:39–51. doi: 10.1083/jcb.200602109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, et al. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–45. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 34.DeAntoni A, Sala V, Musacchio A. Explaining the oligomerization properties of the spindle assembly checkpoint protein Mad2. Philos Trans R Soc Lond B Biol Sci. 2005;360:637–47, discussion 447–8. doi: 10.1098/rstb.2004.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lnrt P, Peters JM. Checkpoint activation: Donot get mad too much. Curr Biol. 2006;16:R412–4. doi: 10.1016/j.cub.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Nasmyth K. How do so few control so many? Cell. 2005;120:739–46. doi: 10.1016/j.cell.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Hardwick KG. Checkpoint signalling: Mad2 conformers and signal propagation. Curr Biol. 2005;15:R122–4. doi: 10.1016/j.cub.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Chan GK, Liu ST, Yen TJ. Kinetochore structure and function. Trends Cell Biol. 2005;15:589–98. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Hagan RS, Sorger PK. Cell biology: the more MAD, the merrier. Nature. 2005;434:575–7. doi: 10.1038/434575a. [DOI] [PubMed] [Google Scholar]
- 40.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–66. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in hela cells is mediated by a complex of BubR1, Bub3, Cdc20, and Mad2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. The Journal of Cell Biology. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millband DN, Hardwick KG. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol Cell Biol. 2002;22:2728–42. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung E, Chen RH. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nature Cell Biology. 2003;5:748–753. doi: 10.1038/ncb1022. [DOI] [PubMed] [Google Scholar]
- 45.Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolanos-Garcia VM, Beaufils S, Renault A, Grossmann JG, Brewerton S, et al. The conserved N-terminal region of the mitotic checkpoint protein BUBR1: a putative TPR motif of high surface activity. Biophys J. 2005;89:2640–9. doi: 10.1529/biophysj.105.063511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp Cell Res. 2006;312:1831–42. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 48.Fang G, Yu H, Kirschner MW. The checkpoint protein Mad2 and the mitotic regulator Cdc20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon KB, Canman JC, Salmon ED. Mad2 and BubR1 function in a single checkpoint pathway that responds to a loss of tension. Mol Biol Cell. 2002;13:3706–19. doi: 10.1091/mbc.E02-03-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, et al. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:664859. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poddar A, Stukenberg PT, Burke DJ. Two complexes of spindle checkpoint proteins containing Cdc20 and Mad2 assemble during mitosis independently of the kinetochore in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:867–78. doi: 10.1128/EC.4.5.867-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acquaviva C, Herzog F, Kraft C, Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat Cell Biol. 2004;6:892–8. doi: 10.1038/ncb1167. [DOI] [PubMed] [Google Scholar]
- 53.Morrow CJ, Tighe A, Johnson VL, Scott MIF, Ditchfield C, et al. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J Cell Sci. 2005;118:3639–52. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- 54.D'Angiolella V, Mari C, Nocera D, Rametti L, Grieco D. The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev. 2003;17:2520–5. doi: 10.1101/gad.267603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–37. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 56.Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–93. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–67. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.King EMJ, van der Sar SJA, Hardwick KG. Mad3 KEN Boxes Mediate both Cdc20 and Mad3 Turnover, and Are Critical for the Spindle Checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicklas R, Ward S, Gorbsky G. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J Cell Biol. 1995;130:929–39. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 61.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, et al. The small molecule hesperadin reveals a role for aurora b in correcting kinetochoremicrotubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, et al. Evidence that the Ipl1-Sli15 (Aurora Kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 63.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nature Cell Bio. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 64.Ibrahim B, Schmitt E, Dittrich P, Diekmann S. MCC assembly is not combined with full Cdc20 sequestering Submitted Paper. 2007 [Google Scholar]
- 65.Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Habu T, Kim SH, Weinstein J, Matsumoto T. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 2002;21:6419–28. doi: 10.1093/emboj/cdf659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia G, Luo X, Habu T, Rizo J, Matsumoto T, et al. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–43. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang M, Li B, Tomchick DR, Machius M, Rizo J, et al. p31(comet) Blocks Mad2 Activation through Structural Mimicry. Cell. 2007;131:744–55. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 70.Howell B, McEwen B, Canman J, Hoffman D, Farrar E, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–81. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 72.Diaz-Martinez LA, Yu H. Running on a treadmill: dynamic inhibition of APC/C by the spindle checkpoint. Cell Div. 2007;2:23. doi: 10.1186/1747-1028-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–50. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–45. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 75.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, et al. MAD2 haplo insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–9. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 76.Fang G, Yu H, Kirschner MW. The checkpoint protein mad2 and the mitotic regulator cdc20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kienitz A, Vogel C, Morales I, Muller R, Bastians H. Partial downregulation of MAD1 causes spindle checkpoint inactivation and aneuploidy, but does not confer resistance towards taxol. Oncogene. 2005;24:4301–10. doi: 10.1038/sj.onc.1208589. [DOI] [PubMed] [Google Scholar]
- 78.Lee MS, Spencer FA. Bipolar orientation of chromosomes in Saccharomyces cerevisiae is monitored by Mad1 and Mad2, but not by Mad3. Proc Natl Acad Sci U S A. 2004;101:10655–60. doi: 10.1073/pnas.0404102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–6. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 80.Hardwick KG, Murray AW. Madlp, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–20. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle check- point of budding yeast depends on a tight complex between theMad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–18. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kalitsis P, Earle E, Fowler KJ, Choo KH. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–82. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. The Journal of Cell Biology. 1998;142:111. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Q, Liu T, Fang Y, Xie S, Huang X, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–85. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 85.Schmidt M, Medema RH. Exploiting the compromised spindle assembly check-point function of tumor cells: dawn on the horizon? Cell Cycle. 2006;5:159–63. doi: 10.4161/cc.5.2.2309. [DOI] [PubMed] [Google Scholar]
- 86.Hanks S, Coleman K, Reid S, Plaja A, Firth H, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–9. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, et al. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol. 2002;12:900–5. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- 90.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–29. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheeseman IM, Anderson S, Jwa M, Green EM, seog Kang J, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–72. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 92.He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci U S A. 1997;94:7965–70. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, et al. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–4. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 94.Passmore LA. The anaphase-promoting complex (APC): the sum of its parts? Biochem Soc Trans. 2004;32:724–7. doi: 10.1042/BST0320724. [DOI] [PubMed] [Google Scholar]
- 95.Boronat S, Campbell JL. Mitotic Cdc6 stabilizes anaphase-promoting complex substrates by a partially Cdc28-independent mechanism, and this stabilization is sup-pressed by deletion of Cdc55. Mol Cell Biol. 2007;27:1158–71. doi: 10.1128/MCB.01745-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen RH. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J Cell Biol. 2002;158:487–96. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Larsen NA, Al-Bassam J, Wei RR, Harrison SC. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc Natl Acad Sci U S A. 2007;104:1201–6. doi: 10.1073/pnas.0610358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–6. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 99.Stucke VM, Sillje HHW, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–32. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Palframan WJ, Meehl JB, Jaspersen SL, Winey M, Murray AW. Anaphase inactivation of the spindle checkpoint. Science. 2006;313:680–4. doi: 10.1126/science.1127205. [DOI] [PubMed] [Google Scholar]
- 101.Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 102.Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, et al. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell. 2004;15:4584–96. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu ST, Chan GKT, Hittle JC, Fujii G, Lees E, et al. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–51. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stucke VM, Baumann C, Nigg EA. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 2004;113:1–15. doi: 10.1007/s00412-004-0288-2. [DOI] [PubMed] [Google Scholar]
- 105.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule- dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fisk HA, Mattison CP, Winey M. A field guide to the Mps1 family of protein kinases. Cell Cycle. 2004;3:439–42. [PubMed] [Google Scholar]
- 107.Fisk HA, Winey M. Spindle regulation: Mps1 flies into new areas. Curr Biol. 2004;14:R1058–60. doi: 10.1016/j.cub.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 108.Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 109.Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell. 2004;16:387–97. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 110.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between theMad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–18. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Luo X, Fang G, Coldiron M, Lin Y, Yu H, et al. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–9. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 112.Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, et al. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–82. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between theMad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–18. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mondal G, Sengupta S, Panda CK, Gollin SM, Saunders WS, et al. Overexpression of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidization in oral cancer. Carcinogenesis. 2007;28:81–92. doi: 10.1093/carcin/bgl100. [DOI] [PubMed] [Google Scholar]
- 115.Shirayama M, Toth A, Galova M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–7. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 116.Chang L, Morrell JL, Feoktistova A, Gould KL. Study of cyclin proteolysis in anaphase-promoting complex (APC) mutant cells reveals the requirement for APC function in the final steps of the fission yeast septation initiation network. Mol Cell Biol. 2001;21:6681–94. doi: 10.1128/MCB.21.19.6681-6694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hansen N, Kern S. Eighth International Conference on Parallel Problem Solving from Nature PPSN VIII. Springer; 2004. Evaluating the cma evolution strategy on multimodal test functions. pp. 282–291. [Google Scholar]
- 118.Hansen N, Muller SD, Koumoutsakos P. Reducing the time complexity of the derandomized evolution strategy with covariance matrix adaptation (CMA-ES). Evol Comput. 2003;11:1–18. doi: 10.1162/106365603321828970. [DOI] [PubMed] [Google Scholar]
- 119.Ibrahim B, Dittrich P, Diekmann S, Schmitt E. Stochastic effects in a compartmental model for mitotic checkpoint regulation. Journal of Integrative Bioinformatics. 2007;4 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement: Differential equations, Materials, Methods, and Optimization
(0.03 MB PDF)
Simulation of Mad2 and Cdc20 mutations for the controlled Convey Dissociation (cf. Table 3). For deletion we set the respective initial concentration 100 times lower, and for over-expression 10000 times higher. For proper functioning, APC:Cdc20 concentration should be very low(zero) before the attachment, and should increase quickly after attachment. Deletion of Mad2 (A) or Cdc20 (C) destroys the switching behavior, that is, the concentrations of all model species are rather constant. Mad2 deletion (A) causes high APC:Cdc20 concentration right from the beginning, while for Cdc20 deletion (C) APC:Cdc20 concentration is zero, by definition. For Mad2 over-expression (B) or Cdc20 over-expression (D), many species concentrations are affected. Particularly, for Mad2 over-expression (B) the APC:Cdc20 concentration remains low before attachment and, after attachment, stays significantly lower than in the wild type (meaning mitotic arrest). In contrast, for Cdc20 over-expression (D), the APC:Cdc20 concentration is high before attachment and also after attachment (meaning checkpoint failure). Spindle attachment occurs at t = 2000s (switching parameter u from 1 to 0 and u' from 0 to 1). Further setting as in Figure 2.
(0.69 MB TIF)
Simulation of BubR1, Aurora B, Mad1, and APC (subunits) mutations for the controlled Dissociation variant (cf. Table 3). For deletion we set the respective initial concentration 100 times lower, and for APC subunit 10 times lower. Spindle attachment occurs at t = 2000s (switching parameter u from 1 to 0 and u' from 0 to 1). Note that Bub3 deletion has the same effect like BubR1 (data not shown), and Bub1 deletion has the same effect like Aurora B (data not shown). APC:Cdc20 for the wild type should be very low (zero) before the attachment and increase quickly after attachment. Deletion of any of BubR1, Mad1, or Aurora B (as well as Bub3 and Bub1) results in high concentration of APC:Cdc20 right from the beginning (meaning checkpoint failure). Deletion of APC subunits disrupts the complex and thus makes APC:Cdc20 unavailable, which implies mitotic arrest. Parameter setting according to Table 2.
(0.74 MB TIF)
Simulation of BubR1, Aurora B, Mad1, and APC (subunits) mutations for the controlled Convey variant (cf. Table 3). The qualitative effect of the mutations is the same as for the Dissociation variant shown in Figure S2. There are quantitative differences in some species concentrations (c.f. Panal B, deletion of Aurora B). Same parameter settings as in Figure S2.
(0.72 MB TIF)