Abstract
Selection of novel molecular markers is an important goal of cancer genomics studies. The aim of our analysis was to apply the multivariate bioinformatical tools to rank the genes – potential markers of papillary thyroid cancer (PTC) according to their diagnostic usefulness. We also assessed the accuracy of benign/malignant classification, based on gene expression profiling, for PTC. We analyzed a 180-array dataset (90 HG-U95A and 90 HG-U133A oligonucleotide arrays), which included a collection of 57 PTCs, 61 benign thyroid tumors, and 62 apparently normal tissues. Gene selection was carried out by the support vector machines method with bootstrapping, which allowed us 1) ranking the genes that were most important for classification quality and appeared most frequently in the classifiers (bootstrap-based feature ranking, BBFR); 2) ranking the samples, and thus detecting cases that were most difficult to classify (bootstrap-based outlier detection). The accuracy of PTC diagnosis was 98.5% for a 20-gene classifier, its 95% confidence interval (CI) was 95.9–100%, with the lower limit of CI exceeding 95% already for five genes. Only 5 of 180 samples (2.8%) were misclassified in more than 10% of bootstrap iterations. We specified 43 genes which are most suitable as molecular markers of PTC, among them some well-known PTC markers (MET, fibronectin 1, dipeptidylpeptidase 4, or adenosine A1 receptor) and potential new ones (UDP-galactose-4-epimerase, cadherin 16, gap junction protein 3, sushi, nidogen, and EGF-like domains 1, inhibitor of DNA binding 3, RUNX1, leiomodin 1, F-box protein 9, and tripartite motif-containing 58). The highest ranking gene, metallophosphoesterase domain-containing protein 2, achieved 96.7% of the maximum BBFR score.
Introduction
Discrimination between benign thyroid nodules and cancer is an important aspect of determining the optimal extent of thyroid surgery. Currently, this is achieved by routine morphologic assessment of cytopathology samples. However, this method does not allow proper classification of all thyroid tumors (Baloch & Livolsi 2002, Franc et al. 2003). At several institutions, genomic studies have been undertaken which besides focusing on basic biological issues (Huang et al. 2001, Giordano et al. 2005), also explore potential diagnostic applications (Aldred et al. 2004, Chevillard et al. 2004, Finley et al. 2004a,b). Our recent microarray-based analysis brought a 20-gene classifier to differentiate between papillary thyroid cancer (PTC) and normal thyroid tissue (Jarzab et al. 2005), further verified using three independent datasets (Eszlinger et al. 2006). Very large and easily distinguishable differences between the molecular profiles of PTC and normal thyroid have clearly demonstrated the applicability of gene expression findings to diagnostic purposes. However, even more desirable for the clinician would be genomic profiling-based capability to discriminate between malignant tumors and various benign lesions. Therefore, we decided to use a balanced mixture of samples from malignant and benign tumors and normal thyroid tissue to mimic the clinical situation, where the material from any of these may be obtained and shall be properly classified. This large 180-array dataset is derived respectively from de novo studies (n=40), previously published own microarray data (n=124; Eszlinger et al. 2001, 2004, Jarzab et al. 2005), and accessible datasets published by other authors (n=16; Huang et al. 2001).
We set the following goals for the study:
To assess accuracy of benign/malignant classification of thyroid specimens in relation to gene set size, in the context of PTC and
To optimize the list of diagnostically relevant genes in PTC.
To answer both questions, we used the support vector machines (SVMs) method with bootstrapping. This approach relies on iterative construction of SVM classifiers based on randomly selected sets of specimens (bootstrap samples) and testing the classifiers on remaining samples. We applied bootstrap to obtain both gene (feature) ranking and outlier detection. The ranking of the genes that are most important for classification quality was based on the frequency of their occurrence in the classifiers of different size (bootstrap-based feature ranking, BBFR). The ranking of the misclassified samples allowed to detect outliers (bootstrap-based outlier detection, BBOD) and to obtain a reliable estimate of classification accuracy with appropriate confidence intervals (CI) for gene sets of different size.
Material and methods
Microarray data used in the study
Microarray datasets from three sources were included in the analysis:
Dataset obtained in Gliwice, Poland; in total, 90 specimens analyzed with GeneChip HG-U133A microarrays. The specimens were collected from 71 patients with PTC (9 males and 40 females; mean age 36 years, range 6–71 years) and 22 with other thyroid diseases, 6 with follicular adenoma, 13 with nodular or colloid goiter and 3 with chronic thyroiditis (9 males and 13 females; mean age 45 years, range 11–71 years). The thyroid tissue specimens included 49 PTC tumors and 41 normal/benign thyroid tissue samples. The latter samples were from patients with PTC (n=17) or other benign thyroid lesions (n=24), among them six follicular adenomas, four nodular goiters, nine colloid goiters, and five cases of thyroiditis, two of them taken from the contralateral lobe from patients with PTC. Fifty microarrays were included in our previously published study and publicly available at www.genomika.pl/thyroidcancer (Jarzab et al. 2005); 40 microarrays were from de novo studies. All new samples were processed according to description given in Jarzab et al. (2005).
Dataset obtained in Leipzig, Germany; 74 specimens analyzed with GeneChip HG-U95Av2 microarrays. The specimens included 15 autonomously functioning thyroid nodules, 22 cold thyroid nodules, and 37 samples of their respective surrounding thyroid tissues. The analysis of these datasets was published previously (Eszlinger et al. 2001, 2004) and the datasets are available at http://www.uni-leipzig.de/innere/_forschung/schwerpunkte/etiology.html.
Dataset obtained in Columbus, OH, USA; 16 specimens analyzed with GeneChip HG-U95A microarrays. The specimens were derived from eight patients and included both PTC tumors and their surrounding thyroid tissues. The dataset (Huang et al. 2001) is publicly available at http://thinker.med.ohio-state.edu.
In total, the three analyzed datasets comprised 57 PTCs, 61 benign thyroid lesions, and 62 apparently normal thyroid tissues analyzed on 180 GeneChips of two different generations. Half of them were U133A and the rest U95A platforms.
Data pre-processing and generation of datasets
Each dataset was pre-processed by the MAS5 algorithm. To compare the expression data generated using the U95A GeneChips (12 625 probe sets) with those from the U133A GeneChips (22 283 probe sets), we used the ‘Human Genome U95 to Human Genome U133 Best Match Comparison Spreadsheet’ (www.affymetrix.com/support/technical/comparison_spreadsheets.affx) which yielded an intersection of 9530 probe sets. The obtained data were log2 transformed.
Neighborhood analysis and recursive elimination in gene selection
For selection of gene sets with diagnostic potential, we applied here the recursive feature elimination (RFE) algorithm (Guyon et al. 2002) which is computationally less demanding than recursive feature replacement used in our previous studies (Jarzab et al. 2005, Eszlinger et al. 2006). The introductory gene selection was performed using neighborhood analysis (200 genes; Golub et al. 1999, Slonim et al. 2000), further selection of the 100 best genes set was carried out by RFE.
SVMs and classification
The linear SVM (Boser et al. 1992, Vapnik 1995) was used for developing the classification rule. As mentioned earlier, the classifier was independently trained for different numbers of selected genes (from 1 to 100).
Bootstrap for estimation of classifier accuracy and its CI
In order to determine the accuracy of the developed classifier, we performed classical bootstrap procedure in 500 resampling iterations (selection with equal probability and return of samples; Efron 1979). Iterations of all stages of the classifier construction (i.e. gene preselection, gene selection, and classifier learning) were performed in each bootstrap, as suggested previously (Simon et al. 2003). The accuracy of the classifier was calculated using the 0.632 bootstrap estimator (Efron 1983). The distribution of the misclassification rate obtained during all bootstrap runs was used to estimate the 95% CI. The accuracy of the classifier and the CI were calculated for different numbers of selected genes (up to 100).
Bootstrap based feature ranking (BBFR) and outlier detection (BBOD)
The primary purpose of the bootstrap used in this study was to estimate the accuracy of the molecular classifier for different sizes of gene subsets with appropriate CIs. However, the computational effort for the bootstrap technique may also be exploited to derive some additional information. We apply two methods that use the information collected during bootstrapping: BBFR and BBOD. They are similar to the methods of statistical learning based on resampling, such as bagging and boosting. In both techniques, an ensemble of many base classifiers is created. Each base classifier is trained on different bootstrap subsamples. The final decision is based on decisions of all base classifiers. The simplest approach is bagging (bootstrap aggregating) originally proposed by Breiman (1996). In bagging, the subsamples are randomly drawn as in classical bootstrapping where each observation is picked with the same probability 1/m, where m is the number of all observations. The final decision is the decision of most base classifiers. In boosting, different observations may be picked with different probability and the final decision is weighted sum of decisions of base classifiers. The well-known boosting algorithm is AdaBoost (Freund & Schapire 1996).
In our approach, we do not create an ensemble (committee) of many base classifiers but we use the information collected during bootstrap-based validation step of the SVM classifier.
Let the data contain m instances (observations). One instance is a vector of Nmax features (gene expression values) with a corresponding class label specified by an expert. Let LB be the number of bootstrap iterations. In each run, we select (with equal probability and return of samples) m instances from the dataset (bootstrap sample). Then, the bootstrap sample is used for feature selection and classifier learning. Finally, the classifier is tested on the test set containing all instances not belonging to the bootstrap sample.
To find the optimal size for the feature set, we select N feature sets Ω1,Ω2,…,ΩN of sizes 1,2,…,N respectively. In general, selected sets may not overlap, but in most commonly used feature selection methods, based on feature ranking or backward/forward searching, feature subsets satisfy the relation
| (1) |
BBFR
Let rj(i) be a number of subsets Ωi, i=1,2,…,N where the gene j belongs to. For gene selection methods satisfying equation (1), we have
| (2) |
The BBFR score Rj of the feature j is defined as a sum of rj(i) over all bootstrap runs as follows:
| (3) |
The maximum possible value of the BBFR score is LBN.
BBOD
Let qk be the number of bootstrap iterations where the observation k is chosen as a test instance (not a member of the bootstrap sample). Let qk true be the number of bootstrap iterations where the instance j is correctly classified at the test stage.
The BBOD score for k-th observation is
| (4) |
The value of Qk belongs to the interval 〈0,1〉 and the low value indicates outliers.
Comparison of different class prediction methods
We used BRB ArrayTools (developed by Dr Richard Simon and Amy Peng Lam) to compare different class prediction algorithms (Compound Covariate Predictor, Linear Diagonal Discriminant Analysis, Nearest Centroid, 1-Nearest Neighbor, 3-Nearest Neighbors and SVMs). To compute misclassification rate, 0.632 bootstrap cross-validation method was used. All genes with univariate misclassification rate below 0.2 were used for this analysis.
Results
Accuracy of malignant/benign classification and redundancy of PTC gene classifiers
The huge difference in gene expression between PTC and benign/normal thyroid tissues implies that many multi-gene classifiers with similar classification ability may be created. For preliminary assessment of accuracy of the differentiation between PTC and benign lesions or normal thyroid, we randomly divided the 180-array dataset into two subgroups, according to sample number: A (odd numbers) and B (even numbers). Each subgroup contained data from similar number of benign and malignant tumor specimens analyzed with U133A or U95A GeneChips. We used set A to obtain a 20-gene classifier; this classifier was tested on set B and the procedure was repeated, using set B as a training set and testing the classifier on set A. Using the classifier obtained from set A, we were able to correctly predict 86 out of 90 samples (95.6%) within set B, while using the classifier obtained from set B, we accurately diagnosed 88 out of 90 samples in set A (97.8%). Both classifiers differed partly from our previous 20-gene classifier (37) obtained on a smaller dataset.
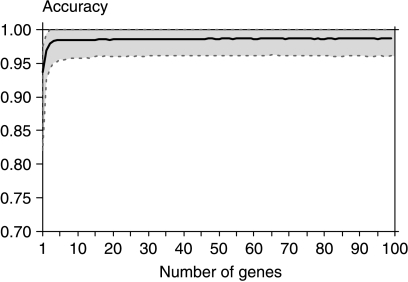
To avoid a bias in gene selection and accuracy estimation, related to the arbitrary selection of the training set, we carried out the procedure of accuracy estimation by bootstrapping, i.e. randomly selecting large numbers of slightly different training sets and validating them on the remaining samples. This procedure allows using sufficiently large training sets while simultaneously obtaining a reliable estimation of classification accuracy. By applying this method, we estimated the accuracy of discrimination between benign and malignant samples to be 98.6%, with a rather narrow CI (see Fig. 1). For small gene sets, the accuracy was a bit lower (93.7% for one-gene set, 96.9% for two-gene set, 97.9 for three-gene set, and from 98.3 to 98.6 for larger sets, up to n=100). For the 20-gene classifier, the accuracy was 98.5% and the estimated 95% CI was 95.9–100% for the classifiers built from more than five genes.
Figure 1.
Accuracy of bootstrapping-estimated benign–malignant classification for different gene set sizes. The 95% confidence interval is marked by dashed lines.
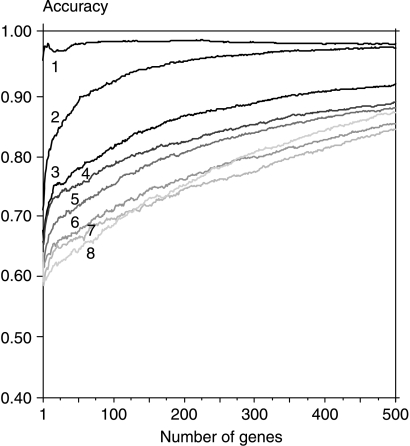
We compared the results of classification by the best 500 genes (Fig. 2) with the classification by consecutive 500-gene sets (i.e. first 500, 500–1000, 1000–1500, etc). We noted that only the first 500 genes allow accurately classifying samples by single genes or small gene sets. Genes ranked 500–1000 achieved 90% accuracy only for classifiers larger than 50 genes, while genes beyond the first 1000 hardly achieve this limit of accuracy. When we excluded all genes analyzed in Fig. 2 (8×500=4000), the accuracy obtained for small sets was only ∼60%, close to random. However, the accuracy rose with gene set size, and for classifier sets larger than 700 genes it achieved 90% (data not shown). These results support the conclusion that the PTC transcriptome differs from the normal one in thousands of genes; they also provide evidence that optimizing a diagnostic gene set is a necessary step of analysis in order to make this set useful for molecular PTC classification.
Figure 2.
Accuracy of classification obtained by successive gene set reduction. The accuracy of the best 500 genes was evaluated in one iteration using the bootstrap technique, then the selected 500-gene set was removed from the whole dataset, and the next 500 genes were selected in the following iteration. This procedure was repeated seven times, thus 3500 genes were excluded (line no. 8). To speed up the procedure, only neighbourhood analysis (NA) was used for gene selection.
Ranking of PTC genes for their classification ability
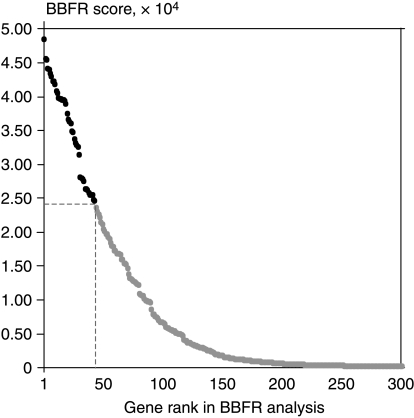
To obtain the ranking of genes based on their usefulness in the diagnostic context, we performed subsequent repetitive gene selection process by bootstrapping of the whole dataset. We ranked all genes according to the frequency of appearance within the selected gene sets (BBFR). Genes important for the majority of diagnostic datasets were highly ranked, while less importance was given to complementing transcripts, which exhibited higher variability (Fig. 3). During the selection process, 365 transcripts occurred at least once within the obtained classifiers and some of them were present in nearly all classifiers. The maximum theoretical score to be obtained by a gene was 5×104 and the gene with the best rank, encoding metallophosphoesterase domain-containing protein 2 (MPPED2), had a score of 4.84×104, i.e. 96.7% of the maximum one. The first 20 genes were given scores >3.74×104 (>77% of the maximum score), only slightly lower than the top gene, and the first 100 transcripts were characterized by scores >0.64×104, which is >13.2% of the maximum score obtained. In total, 43 transcripts representing 41 genes scored higher than half of the value for the top gene (>2.42×104, Fig. 3). Among them, there were both genes known for their changed expression in PTC or described in previous microarray studies, some used already as single markers, as well as new genes, not considered previously for their diagnostic potential (Table 1).
Figure 3.
Result of bootstrap-based feature ranking (BBFR). Each dot represents one gene, dashed lines define the subset of 43 genes with BBFR score larger than half of the maximum one (black dots).
Table 1.
Ranking of papillary thyroid cancer (PTC) genes as assessed by bootstrap-based feature ranking (BBFR) approach. For each transcript selected, rank and score obtained by the BBFR method are given, together with basic univariate statistics (log2 mean and log2 ratio)
| Gene symbol | Gene name | Affy_ID (U133) | Rank | Score | PTC mean log2 | Benign mean log2 | Log ratio | Log ratio U133 | Log ratio U95 | References of microarray or other high throughput studiesa | Referred to in single studies of thyroid cancer | Other data relevant for functional role in thyroid cancer | Gene functionb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MPPED2 | Metallophosphoesterase domain-containing protein 2 | 205413_at | 1 | 48 449 | 4.66 | 7.82 | −3.16 | −3.46 | −3.26 | Aldred et al. (2003, 2004), Mazzanti et al. (2004) and Griffith et al. (2006) | Fetal brain protein of unknown function | ||
| H/HBA2 | Hemoglobin, α-1/hemoglobin, α-2 | 209458_x_at | 2 | 45 521 | 9.79 | 12.04 | −2.25 | −2.28 | −1.87 | Griffith et al. (2006) | Onda et al. (2005) | Oxygen transport | |
| MET | Met proto-oncogene (hepatocyte growth factor receptor) | 213807_x_at | 3 | 45 363 | 8.15 | 5.22 | 2.93 | 1.73 | 2.60 | Barden et al. (2003), Wasenius et al. (2003), Finley et al. (2004a,b), Prasad et al. (2004), Zou et al. (2004) and Giordano et al. (2005) | cBelfiore et al. (1997) and cIppolito et al. (2001) | Ramirez et al. (2000), Ruco et al. (2001) and Scarpino et al. (2004) | Membrane tyrosine kinase receptor enhances cell motility, invasiveness, and chemokine production (Ruco et al. (2001)) |
| FN1 | Fibronectin 1 | 210495_x_at | 4 | 44 017 | 12.24 | 8.72 | 3.52 | 2.75 | 3.91 | Chen et al. (2001), Barden et al. (2003), Wasenius et al. (2003),Finley et al. (2004a,b), Prasad et al. (2004), Giordano et al. (2005), Hamada et al. (2005) and Griffith et al. (2006) | Takano et al. (1998), 1999 and cPrasad et al. (2005) | Ghinea et al. (2002) and Liu et al. (2005) | Extracellular matrix glycoprotein participates in cell adhesion, regulates proliferation and survival of thyroid cells via integrin receptors (Illario et al. (2003)) |
| GALE | UDP-galactose-4-epimerase | 202528_at | 5 | 43 974 | 7.12 | 3.70 | 3.42 | 2.41 | 3.50 | Converts glucose to galactose and N-acetylglucosamine to its UDP-derivatives | |||
| QPCT | Glutaminyl-peptide cyclotransferase (glutaminyl cyclase) | 205174_s_at | 6 | 43 317 | 7.60 | 4.99 | 2.61 | 3.14 | 2.56 | Barden et al. (2003), Chevillard et al. (2004), Finley et al. (2004a,b) and Griffith et al. (2006) | Converts glutaminyl peptides to cyclic pyroglutamyl ones | ||
| NELL2 | NEL-like 2 (chicken) | 203413_at | 7 | 42 953 | 9.68 | 7.56 | 2.12 | 2.02 | 2.53 | Barden et al. (2003) and Finley et al. (2004a,b) | Brain protein with six EGF-like repeats | ||
| PGCP | Plasma glutamate carboxypeptidase | 203501_at | 8 | 42 153 | 7.70 | 9.23 | −1.53 | −1.05 | −1.33 | Aldred et al. (2003, 2004), Barden et al. (2003), Finley et al. (2004a,b), Weber et al. (2005) and Sarquis et al. (2006) | Breakdown of secreted peptides, homologous to prostate membrane-specific antigen (Gingras et al. (1999)) | ||
| DPP4 | Dipeptidylpeptidase 4 (CD26, adenosine deaminase complexing protein 2) | 203717_at | 9 | 42 115 | 7.87 | 3.77 | 4.11 | 3.21 | 3.81 | Huang et al. (2001), Takano et al. (2002, 2004), Prasad et al. (2004) and Griffith et al. (2006) | Kehlen et al. (2003), cKholova et al. (2003a,b) and Ozog et al. (2006) | Aratake et al. (2006) and Schagdarsurengin et al. (2006) | Membrane enzyme, participates in breakdown of secreted peptides |
| ADORA1 | Adenosine A1 receptor | 205481_at | 10 | 41 699 | 7.16 | 4.85 | 2.30 | 2.00 | 2.81 | Aldred et al. (2003, 2004) and Prasad et al. (2004) | Lelievre et al. (1998), Woodhouse et al. (1998) and Schnurr et al. (2004) | Membrane receptor, stimulates motility and modulates proliferation | |
| HMGA2 | High-mobility group AT-hook 2 | 208025_s_at | 11 | 40 713 | 7.90 | 4.66 | 3.24 | 3.58 | 2.62 | Baris et al. (2004, 2005) and Jacques et al. (2005) | Fedele et al. (2001), Berlingieri et al. (2002) and Musholt et al. (2006) | Architectural transcription factor (Noro et al. (2003)) | |
| RYR1 | Ryanodine receptor 1 (skeletal) | 205485_at | 12 | 40 473 | 6.96 | 4.70 | 2.27 | 2.53 | 1.92 | Barden et al. (2003) and Finley et al. (2004a,b) | Present mainly in excitable cells | Calcium release channel of the sarcoplasmic reticulum | |
| CDH16 | Cadherin 16, KSP-cadherin | 206517_at | 13 | 39 770 | 3.47 | 8.07 | −4.60 | −4.68 | −1.43 | Thought to be kidney specific (Thomson et al. (1995)) | Calcium-dependent, membrane-associated glycoprotein, participates in cell adhesion | ||
| GJB3 | Gap junction protein β-3, 31 kDa (connexin 31) | 205490_x_at | 14 | 39 526 | 6.49 | 4.04 | 2.44 | 2.71 | 0.62 | Does not normally appear in thyroid, in adult mouse becomes restricted to epidermis, testis and placenta (Tonoli et al. (2000), Plum et al. (2002) and Green et al. (2005) | Forms incompatible hemichannels with thyroidal connexin 43 (Dahl et al. (1996)) | ||
| EMID1 | EMI domain containing 1 | 213779_at | 15 | 39 505 | 6.44 | 8.12 | −1.68 | −1.09 | −0.76 | Barden et al. (2003), Cerutti et al. (2004) and Finley et al. (2004a,b) | Extracellular matrix protein, able to promote cell movements (Spessotto et al. (2003)) | ||
| NRIP1 | Nuclear receptor-interacting protein 1 | 202599_s_at | 16 | 39 358 | 8.31 | 6.33 | 1.98 | 1.36 | 2.06 | Barden et al. (2003) and Finley et al. (2004a,b) | Interacts with nuclear receptors | ||
| MET | Met proto-oncogene (hepatocyte growth factor receptor) | 211599_x_at | 17 | 39 348 | 8.44 | 5.68 | 2.76 | 1.53 | 2.54 | Barden et al. (2003), Wasenius et al. (2003), Finley et al. (2004a,b), Prasad et al. (2004), Zou et al. (2004) and Giordano et al. (2005) | See the information given above for another probeset of the same gene | ||
| DTX4 | Deltex 4 homolog (Drosophila) | 212611_at | 18 | 39 298 | 10.24 | 8.24 | 2.00 | 2.07 | 1.45 | Prasad et al. (2004) | Participates in protein ubiquination | ||
| RAB27A | RAB27A, member RAS oncogene family | 210951_x_at | 19 | 38 913 | 8.62 | 5.62 | 3.00 | 1.60 | 1.24 | Barden et al. (2003), Finley et al. (2004a,b), Weber et al. (2005), Musholt et al. (2006) and Sarquis et al. (2006) | Prenylated membrane bound protein with GTP-ase function | ||
| – | CDNA clone IMAGE:4152983 | 214803_at | 20 | 37 397 | 7.30 | 5.39 | 1.90 | 1.94 | 1.23 | Not identified | |||
| BCL2 | B-cell CLL/lymphoma 2 | 203684_s_at | 21 | 36 483 | 2.92 | 5.88 | −2.95 | −2.74 | −1.39 | Hoos et al. (2002), Baris et al. (2004, 2005), Prasad et al. (2004), Wreesmann et al. (2004), Giordano et al. (2005) and Jacques et al. (2005) | Mitselou et al. (2004), cAksoy et al. (2005) and cLetsas et al. (2005) | Stassi et al. (2003) and Basolo et al. (1999) | Anti-apoptotic protein |
| TACSTD2 | Tumor-associated calcium signal transducer 2 | 202286_s_at | 22 | 36 170 | 10.42 | 6.29 | 4.13 | 4.02 | 4.02 | Giordano et al. (2005) | May serve as cell surface receptor | ||
| DIO1 | Deiodinase, iodothyronine, type I | 206457_s_at | 23 | 35 971 | 6.19 | 9.94 | −3.75 | −3.79 | −4.33 | Eszlinger et al. (2001, 2004), Huang et al. (2001), Barden et al. (2003), Finley et al. (2004a,b), Prasad et al. (2004), Wreesmann et al. (2004), Giordano et al. (2005) and Griffith et al. (2006) | cDe Micco et al. (1999), cCzarnocka et al. (2001), cLe Fourn et al. (2004), cAmbroziak et al. (2005) and cArnaldi et al. (2005) | Kohrle (1999) | 5′ Deiodination of thyroxine |
| ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 | 203710_at | 24 | 34 804 | 6.75 | 8.84 | −2.09 | −2.06 | −1.83 | Barden et al. (2003), Finley et al. (2004a,b), Prasad et al. (2004), Wreesmann et al. (2004) and Hamada et al. (2005) | Signal transducer coupled with calcium channels, participates in apoptosis (Sedlak & Snyder (2006)) | ||
| HBB | Hemoglobin β | 209116_x_at | 25 | 34 591 | 9.62 | 12.13 | −2.51 | −2.48 | −1.24 | Aldred et al. (2003, 2004) and Onda et al. (2005) | See above the HBA gene | ||
| SNED1 | Sushi, nidogen, and EGF-like domains 1 | 213493_at | 26 | 33 625 | 2.87 | 5.88 | −3.01 | −2.14 | −2.06 | Participates in cell–matrix adhesion, contains sushi, nidogen-and calcium-binding domains | |||
| AHR | Aryl hydrocarbon receptor | 202820_at | 27 | 33 003 | 7.52 | 6.02 | 1.50 | 1.20 | 1.59 | Barden et al. (2003), Wasenius et al. (2003) and Finley et al. (2004a,b) | A ligand-activated transcription factor able to form complexes with other nuclear receptors (Widerak et al. (2005) | ||
| HGD | Homogentisate 1,2-dioxygenase (homogentisate oxidase) | 205221_at | 28 | 32 816 | 4.57 | 7.83 | −3.26 | −3.17 | −3.92 | Huang et al. (2001), Aldred et al. (2003), Barden et al. (2003), Aldred et al. (2004), Finley et al. (2004a,b), Prasad et al. (2004) and Giordano et al. (2005) | Fe(II)-dependent enzyme responsible for aromatic ring cleavage | ||
| RXRG | Retinoid X receptor, γ | 205954_at | 29 | 32 444 | 7.35 | 4.69 | 2.66 | 2.80 | 2.62 | Haugen et al. (2004) | Klopper et al. (2004), Schmutzler et al. (2004) and Frohlich et al. (2005) | Heterodimer partner of several nuclear receptors | |
| CA4 | Carbonic anhydrase IV | 206209_s_at | 30 | 31 332 | 6.33 | 8.51 | −2.18 | −2.62 | −1.41 | Barden et al. (2003), Finley et al. (2004a,b), Weber et al. (2005) and Sarquis et al. (2006) | An ancient isozyme | ||
| SDC4 | Syndecan 4 (amphiglycan, ryudocan) | 202071_at | 31 | 28 036 | 10.76 | 8.31 | 2.45 | 1.86 | 2.41 | Barden et al. (2003), Chevillard et al. (2004), Finley et al. (2004a,b), Prasad et al. (2004) and Griffith et al. (2006) | Transmembrane heparan sulfate proteoglycan involved in the organization of the actin cytoskeleton and in cell–matrix interactions, binds fibronectin, behaves as CXCL12 receptor (Lin et al. (2005)) | ||
| ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | 209473_at | 32 | 27 859 | 8.71 | 6.75 | 1.97 | 1.49 | 1.48 | Weber et al. (2005) and Sarquis et al. (2006) | Membrane bound enzyme converts adenine nucleotides to adenosine, interacts with caveolin 1 and 2 (Kittel et al. (2004)) | ||
| TPO | Thyroid peroxidase | 210342_s_at | 33 | 27 658 | 7.29 | 12.24 | −4.95 | −4.93 | −3.75 | Barden et al. (2003), Cerutti et al. (2004), Finley et al. (2004a,b) and Griffith et al. (2006) | Arturi et al. (1997), Lazar et al. (1999) and | Furuya et al. (2004) | Thyroid-specific enzyme crucial for organification of iodine and synthesis of thyroid hormones |
| KRT19 | Keratin 19 | 201650_at | 34 | 27 398 | 8.92 | 5.71 | 3.22 | 3.55 | 3.07 | Barden et al. (2003), Chevillard et al. (2004), Finley et al. (2004a,b), Prasad et al. (2004) and Griffith et al. (2006) | Schelfhout et al. (1989) | The smallest known keratin expressed in some types of cancer | |
| ID3 | Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 207826_s_at | 35 | 26 271 | 9.17 | 11.25 | −2.08 | −1.26 | −1.29 | Downstream target of pituitary tumor transforming gene (PTTG) | |||
| RUNX1 | Runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 209360_s_at | 36 | 26 202 | 7.37 | 4.80 | 2.58 | 3.50 | 2.01 | Kim et al. (2007) | Transcription factor may promote E-cadherin expression (Liu et al. (2005)) | ||
| LMOD1 | Leiomodin 1 (smooth muscle) | 203766_s_at | 37 | 26 044 | 5.60 | 7.80 | −2.20 | −2.77 | −0.95 | Present both in thyroid cells and eye muscle (Kromminga et al. (1998)) | 64 kDa antigen, considered for its role in thyroid autoimmunity | ||
| RAB27A | RAB27A, member RAS oncogene family | 209514_s_at | 38 | 25 684 | 8.57 | 6.29 | 2.28 | 1.43 | 1.53 | Barden et al. (2003), Finley et al. (2004a,b), Weber et al. (2005) and Sarquis et al. (2006) | See above information on the alternative probeset identifying the same gene | ||
| FBXO9 | F-box protein 9 | 212987_at | 39 | 25 331 | 8.47 | 9.29 | −0.83 | −0.50 | −0.57 | Members of this gene family in complexes may act as protein–ubiquitin ligases | |||
| TRIM58 | Tripartite motif-containing 58 | 215047_at | 40 | 25 304 | 3.91 | 6.99 | −3.08 | −2.27 | −1.74 | Not identified | |||
| – | – | 210524_x_at | 41 | 25 302 | 9.73 | 12.70 | −2.97 | −2.95 | −2.12 | Not identified | |||
| MT1G | Metallothionein 1G | 204745_x_at | 42 | 24 688 | 9.94 | 12.39 | −2.45 | −1.97 | −4.00 | Baris et al. (2004, 2005), Prasad et al. (2004), Jacques et al. (2005) and Griffith et al. (2006) | Cherian et al. (2003) | Low molecular weight, cysteine-rich, zinc-donating protein. Associated with protection against DNA damage, stress, and apoptosis (Theocharis et al. (2004)) | |
| ICAM1 | Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | 202638_s_at | 43 | 24 534 | 8.18 | 5.61 | 2.57 | 1.70 | 2.40 | Kawai et al. (1998) | Epithelial adhesion molecule plays a key role in lymphocyte infiltration into the thyroid |
The original papers (Eszlinger et al. 2001, 2004, Huang et al. 2001, Jarzab et al. 2005) containing datasets included in the present study were not cited here. RXRG was listed in our previous microarray-based analysis (Jarzab et al. 2005), together with FN1, MET, KRT19, DPP4, HBB, QPCT, GJB3, and DTX4, also occurring in this table.
OMIM-based information if not otherwise specified.
Denotes immunohistochemistry studies.
We analyzed fold-change differences between PTC and benign thyroid samples for the 43 selected transcripts to evaluate the potential influence of inter-platform differences on the obtained gene selection. Twenty of them showed more than fourfold increase (log ratio >2) and four transcripts were increased more than twice, whereas the remaining 19 transcripts were decreased. Generally, the consistency between fold-changes observed in subsets from U95 and U133 arrays was good, although for some genes (e.g. the well-known thyroid cancer markers fibronectin 1 (FN1) and MET or novel genes cadherin 16 (CDH16) or gap junction protein β-3 (GJB3)) there were inter-platform differences between the log ratios. However, 40 out of 43 selected genes exhibited more than twofold change in both the U133 and the U95 subsets. For all 43 genes, the PTC–benign difference was larger than the difference between fold-changes obtained with different GeneChip generation subsets. This confirms that the selection performed was robust to inter-array differences.
Misclassified thyroid samples
The algorithm with bootstrapping allows ranking the samples according to the frequency of their misclassification (Table 2). BBOF showed very frequent misclassifications for two samples. One of them was not properly classified by any gene set selected, and this was sample no. 154 from the U133 dataset no. 1, a small (10 mm in diameter) familial PTC found within a larger follicular adenoma. It was observed in an 18-year-old woman. A year later her mother, 43 years old, was diagnosed with 0.7 cm PTC (follicular variant). The other one, properly classified only in 8% of runs, was a benign follicular adenoma (diagnosed as atypical) from the same dataset (sample no. 97) which was derived from a 15-year-old boy of another family with familial PTC. In this family, there were two PTC cases (mother of the patient, diagnosed with pT2BNxM0 PTC and her aunt who died of a dissemination of PTC) and one follicular thyroid cancer case (pT2bNxM0, 11 years old, sister of the patient). These were the only two cases with a positive family history of thyroid cancer among 49 Polish patients included in the study. Two further samples were properly classified in 65–68% of runs (one from dataset no. 1 and one from dataset no. 3), again one benign adenoma and one PTC, respectively. For the fifth sample, the accuracy was much higher and it was properly classified in 88% of the runs. Thus, only 5 out of 180 samples (2.8%) were misclassified in more than 10% of the runs, while a total of 14 samples (7.8%) were misclassified in more than 1% of the runs. Seventy samples were classified with an excellent accuracy between 99 and 100%, and for further 64 cases no misclassification occurred during the bootstrapping process.
Table 2.
Ranking of thyroid samples by tumor–normal misclassification frequency, assessed by bootstrap-based outlier detection (BBOD) approach. The BBOD rank and score Qk, as defined in Material and methods, is given
| Sample number | Status | Array | Set | Rank | Score (%) |
|---|---|---|---|---|---|
| 154 | PTC | U133 | B | 1 | 0.04 |
| 97 | Benign | U133 | A | 2 | 7.23 |
| 148 | PTC | U133 | B | 3 | 65.34 |
| 95 | Benign | U133 | A | 4 | 68.25 |
| 88 | PTC | U95v1 | B | 5 | 88.28 |
| 166 | PTC | U133 | B | 6 | 90.02 |
| 84 | PTC | U95v1 | B | 7 | 93.11 |
| 161 | PTC | U133 | A | 8 | 95.96 |
| 94 | Benign | U133 | B | 9 | 97.26 |
| 116 | Normal | U133 | B | 10 | 97.30 |
| 120 | PTC | U133 | A | 11 | 97.98 |
| 77 | Normal | U95v1 | B | 12 | 98.30 |
| 100 | Benign | U133 | B | 13 | 98.70 |
| 139 | PTC | U133 | A | 14 | 98.91 |
| 90 | PTC | U95v1 | B | 15 | 99.09 |
| 42 | CTN | U95v2 | B | 16 | 99.22 |
| 3 | AFTN | U95v2 | A | 17 | 99.28 |
| 37 | CTN | U95v2 | A | 18 | 99.36 |
| 147 | PTC | U133 | A | 19 | 99.38 |
| 40 | CTN | U95v2 | B | 20 | 99.41 |
| 64 samples (28 PTCs, 36 benign/normal) | 21–84 | 99.46–99.98 | |||
| 96 samples (19 PTCs, 77 benign/normal) | 85–180 | 100 |
Comparison of classification accuracy by different class prediction methods
To evaluate our method, we compared the accuracy of prediction by different class prediction methods implemented in BRB-Array software. We based the class prediction on all genes that showed the univariate misclassification rate lower than 20%. We found out that the classification accuracy ranged from 89% (compound covariate predictor method) to 99% (SVM), and confirmed the best performance of SVM-based methods to analyze these data (Table 3).
Table 3.
Comparison of results obtained by different class prediction methods
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|
| Compound covariate predictor | 89 | 85 | 88 | 77 | 93 |
| Nearest centroid | 90 | 86 | 89 | 79 | 93 |
| Linear diagonal discriminant analysis | 92 | 87 | 92 | 83 | 94 |
| One-nearest neighbor | 98 | 94 | 99 | 98 | 97 |
| Three-nearest neighbors | 98 | 93 | 100 | 99 | 97 |
| Support vector machines | 99 | 95 | 99 | 98 | 98 |
PPV, positive predictive value; NPV, negative predictive value.
Discussion
Transcripts important for discriminating PTC from benign and normal thyroid samples
In the study, we performed an advanced optimization of putative PTC markers using a large group of benign thyroid lesions and normal thyroid tissues and proposed a list of 43 transcripts, selected by their most frequent appearance in the classifiers. An additional proof of their efficacy was obtained by hierarchical clustering (all samples clustered correctly, data shown in the web appendix to this article, www.genomika.pl/thyroidcancer). Forty-one of them (95.3%) could be attributed to 39 known genes, 32 well-defined ones, and 7 of unknown or not well-defined function. There were 12 genes which had never before been related to the thyroid gland nor mentioned in genomic studies of thyroid cancer, while 29 genes (74%) were identified in previous thyroid microarray studies. However, only ten of them were discussed in the original papers for their putative role in thyroid carcinoma. Within the list of the well-known genes which received high scores by BBFR, one should mention gene encoding FN1, met proto-oncogene (MET; both scored 4.4×104), dipeptidylpeptidase 4 (DPP4), adenosine A1 receptor (ADORA1), keratin 19, and B-cell CLL (BCL2) genes (Huang et al. 2001, Wasenius et al. 2003, Baris et al. 2004, Chevillard et al. 2004, Finley et al. 2004a, Wreesmann et al. 2004, Giordano et al. 2005), all up-regulated with the exception of BCL2. Their inclusion in our classifier positively validates the applied criteria. All these genes except ADORA1 were previously found by single gene studies (see Table 1) and later confirmed by microarray approaches. Moreover, in the recent meta-analysis of thyroid cancer gene expression profile, MET and FN1 were included into top 12 candidates for consistent gene expression markers (Griffith et al. 2006). Similarly, thyroid-specific (down-regulated) genes, deiodinase, iodothyronine, type I and thyroid peroxidase, were widely recognized previously for their diagnostic significance both in microarray-based (Eszlinger et al. 2001, Huang et al. 2001, Baris et al. 2004, Cerutti et al. 2004, Finley et al. 2004a, Wreesmann et al. 2004) and single gene studies (Arturi et al. 1997, Lazar et al. 1999, De Micco et al. 1999, Czarnocka et al. 2001, Le Fourn et al. 2004, Ambroziak et al. 2005, Arnaldi et al. 2005). Nevertheless, neither our approach nor the meta-analysis mentioned earlier indicated other thyroid-specific genes, confirming the lesser diagnostic potency of sodium iodide symporter, thyroglobulin, thyrotrophin receptor, or thyroid-specific transcription factors, shown to be down-regulated in previous single gene studies (Arturi et al. 1997, Lazar et al. 1999, Shimura et al. 2001, Scouten et al. 2004, Ambroziak et al. 2005, Wagner et al. 2005).
The top gene identified by our effort, MPPED2, which is lost in PTC, was not previously considered for its role in PTC, although it was previously listed by Aldred et al. (2004, in the context of FTC) and by Mazzanti et al. (2004). It is an ancient gene highly conserved from Caenorhabditis elegans to mammals and expressed in fetal brain. Its function is unknown.
Already the first microarray-based analysis of a PTC gene expression profile (Huang et al. 2001) indicated the dominant position of genes controlling cell–matrix adhesion and cell–cell communication. Besides, FN1 mentioned earlier, and intercellular adhesion molecule 1 (ICAM-1; Kawai et al. 1998), it seems important to mention syndecan 4 (SDC4), a transmembrane heparan sulfate proteoglycan known to bind FN1 and functioning also as CXCL12 receptor in signal transduction (Huang et al. 2001, Chevillard et al. 2004, Finley et al. 2004a). Loss of CDH16 (kidney-specific cadherin; Thomson et al. 1998) was indicated for the first time in our study, a gene closely related to cadherin E (CDH1), which is well known to be lost in a subgroup of PTCs with negative prognostic significance (Rocha et al. 2003), while cadherin P (CDH3) is up-regulated in PTC (Jarzab et al. 2005). Other genes involved in cell adhesion and present in our list comprise ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1) (up-regulated) and less known genes such as NEL-like 2 (up-regulated) and sushi, nidogen, and EGF-like domains 1 (down-regulated), both exhibiting EGF-like repeats (Watanabe et al. 1996). The GJB3 gene (connexin 31) encodes the protein subunit of gap junctions, essential for cell–cell communication.
DPP4 (CD26), ICAM1, and ENTPD1 (CD39) may be considered as immune-related genes, although their expression is not confined to immune or endothelial cells. ICAM1 was shown to be present in thyroid cancer cells (Kawai et al. 1998). ENTPD1 (ecto-ATPase), in turn, has not been described before for the thyroid gland; its expression was shown in some other organs like salivary glands or exocrine pancreas (Kittel et al. 2004). It converts adenine nucleotides to adenosine, thus participating in the control of signal transduction. DPP4, another membrane-bound enzyme which hydrolyzes peptides engaged in paracrine and autocrine regulation, is up-regulated in PTCs both on RNA and protein level (Huang et al. 2001, Kholova et al. 2003). The contribution of various enzymes to our list is striking: others, not described previously in the context of thyroid gland, comprise UDP-galactose epimerase (GALE) and glutaminyl-peptide cyclotransferase (QPCT), both with virtually unknown expression patterns. The latter was also indicated by the meta-analysis of Griffith et al. Among gene encoding enzymes lost in PTC are plasma glutamate carboxypeptidase, plasma glutamate carboxypeptidase (Gingras et al. 1999), not mentioned in any thyroid-related study before; carbonic anhydrase 4 (CA4), and even the well-known homogentisate oxidase (encoding HGD), not previously related to the thyroid in any context, although listed in many microarray-based reports (Table 1).
Underexpression of hemoglobin transcripts (HBA1/A2 and HBB scored at positions 2 and 25 respectively) was already discussed in our papers as a very characteristic feature of PTC gene expression profile (Jarzab et al. 2005). We believe that the down-regulation of hemoglobin gene could be associated with tumor hypoxia; HBA has also been considered a tumor suppressor since transduction of this gene in an anaplastic thyroid cancer cell line induces an anti-proliferative effect (Onda et al. 2005).
Many of the genes listed in Table 1 participate in signal transduction; among them are MET, ADORA1,RAB27A as well as tumor-associated calcium signal transducer 2, inositol 1,4,5-triphosphate receptor, type 1 (ITPR1), ryanodine receptor 1, all up-regulated in PTC except for ITPR1. Some enzymes mentioned above (DPP4, ENTPD1, and QPCT) contribute to synthesis or breakdown of signaling molecules. On the other hand, the list also includes many genes participating in transcription regulation, among them high-mobility group AT-hook 2, aryl hydrocarbon receptor, retinoid X receptor, γ, ID3, nuclear receptor-interacting protein 1, and RUNX1. Both of these functional classes are typical for cancer genes. We noted only one gene clearly related to apoptosis (and lost in PTC), the well-known BCL2. Interestingly enough, some immunohistochemical studies report its up-regulation in PTC (Aksoy et al. 2005).
Although the selected genes were obtained by analysis of PTC, many of them may be found also in other types of thyroid tumors (M Oczko-Wojciechowska, J Starzyński, M Jarząb, Z Wygoda, A Czarniecka, G Gala, M Kalemba, E Gubala & B Jarząb, unpublished data). This is convincingly illustrated by the overlapping results of our analysis and one of the studies which dealt with follicular thyroid tumors only (Barden et al. 2003).
Accuracy of discriminating PTC from benign/normal thyroid tissue
Our study is the first to define the classification accuracy for thyroid cancer by 95% CIs and one of the few dealing with the problem of diagnostic accuracy of microarray-derived classifiers (Kerr & Churchill 2001). Although the estimation of CIs by Monte Carlo analysis has not gained a general acceptance still, it is necessary to stress the very good accuracy of PTC diagnosis in our study with the lower range of the CI at 95%, obtained using a sufficiently large study group, mimicking the real clinical setting. From a clinical point of view, for a PTC classifier, an even higher accuracy is required, as the risk of diagnosing PTC in a thyroid nodule is only about 5% (Hegedus 2004).
Our results stress the importance of multi-gene approaches for the molecular diagnosis of cancer. We observed that lower limits of accuracy CIs were decreased in case of classification by gene sets with less than ten genes. The initial conclusion from these data is that any combination of more than five to ten genes increases the reliability of distinguishing between malignant and benign tissue samples. This result is similar to that obtained by Hua et al. (2005), who demonstrated on simulated and real breast cancer data that for different classifiers the number of features lower than five was usually much less effective than larger classifiers. Recent paper reports a six-gene molecular classifier, efficient for molecular diagnosis of thyroid cancer (Kebebew et al. 2006).
Bootstrap-based multi-gene classification of PTC microarray data
Selection of genes is an important goal of microarray studies contributing to broader understanding of the cancer transcriptome as well as yielding novel molecular cancer markers. Such studies have been successfully performed in PTC and large numbers of discriminating physiologically relevant genes were proposed (Huang et al. 2001, Wasenius et al. 2003, Aldred et al. 2004, Chevillard et al. 2004, Finley et al. 2004a,b, Wreesmann et al. 2004, Baris et al. 2005, Detours et al. 2005, Giordano et al. 2005). However, in the majority of these studies, the selection of important genes was based on either fold-change or significance criteria obtained using classical statistical tests. These approaches either favor genes with large amplitudes, sometimes coming from a minor proportion of samples, or genes with low within-group variance, thus rather stably expressed in all analyzed tumor samples. Bearing in mind, complexity of molecular changes in tumors, the widespread skepticism about a single ‘cancer marker’ as well as possible differences in histological subtypes or other features of PTC, we decided to use SVM, a routine machine-learning approach to construct classifiers based on multiple features of the analyzed objects. This method allows integrating the information carried by many genes in the gene sets. Thus, effective molecular multi-gene classifiers may be built that rely on inter-gene interactions rather than on combining single ‘best markers’. SVMs have been confirmed as an effective method of multi-gene set selection and this is supported by our comparison to other class prediction methods. Our procedure helps us to optimize the list of markers which are to be implemented to real-time quantitative PCR-supported fine needle biopsy (Lubitz & Fahey 2006).
From the diagnostic point of view, the major drawback of the SVM-based methods are the fluctuations of gene content between classifiers of different size or based on slightly different training sets. To overcome this problem, we extended the original algorithm with bootstrap iterations, as recommended (Braga-Neto & Dougherty 2004). A bootstrap iteration depends on creating a temporary learning set (bootstrap sample) by performing selection from the original set with return of samples. Then, the classification rule is derived based on a bootstrap sample and applied to the rest of the original set. Multiple selections of slightly different training sets represent the variability, which may be observed between different thyroid cancer collections, laboratories, etc. Indeed, our current data generated using the bootstrap technique show much better agreement with the results of other thyroid cancer studies (Oczko-Wojciechowska et al. submitted) than data created by leave-one-out cross-validation of the whole dataset (Jarzab et al. 2005).
Originally, in a bootstrap iteration one counts only the number of misclassifications. Since in all bootstrap iterations every step of data processing (gene selection and classifier training) has to be repeated (Simon et al. 2003), some additional knowledge can be gained. The procedure used by us enables ranking of genes which are most often present in the classifiers obtained from the different subsets of the training set (BBFR). Furthermore, it also estimates the accuracy with appropriate CIs. Moreover, it allows ranking the samples according to the frequency of misclassifications (BBOD). The use of BBFR resulted in delineation of genes, which were either novel or not recognized before for their contribution to the PTC gene expression profile, even if they were included in the large gene lists given in previous genomic studies. BBOD allowed us to reveal ‘difficult’ samples in the analyzed group. The two thyroid samples with the poorest accuracy of diagnosis were derived from patients with familial thyroid tumors, which suggest that their gene expression profiles may differ from sporadic ones. For the remaining samples, in 175 out of 180 cases (>97%) the percentage of correct diagnoses was >90%.
Recently, Zhang et al. (2006) have published a SVM-based recursive method of gene selection. This method, called R-SVM, differs from the standard RFE algorithm, used here, in modified criteria applied in elimination steps. Moreover, the final gene subset is created on the basis of any resample method used at the validation stage, which is similar to our approach presented here. Nevertheless, our bootstrap-based method allows detecting outlier samples and provides the estimation of CIs for the classification accuracy, which is much more informative than the accuracy estimator alone.
PTC and normal/benign difference versus inter-platform difference
To assure a sufficient number of tissue samples, it was necessary to combine data obtained using different generations of GeneChips, which cannot be compared by a direct approach (Eszlinger et al. 2006). The use of multi-gene classifiers allows, however, overcoming this difficulty. We showed earlier that the classifier selected using the U133 platform (Jarzab et al. 2005) performs well on U95-obtained data and has high classification accuracy (Eszlinger et al. 2006). In the present paper, we demonstrate that it is possible, after correctly matching genes from two different generation microarrays, to derive an efficient multi-gene classifier. When we included both benign and malignant samples from both platforms, the vast majority of these samples were properly classified. Using Affymetrix GeneChips, Barden et al. (2003) and Finley et al. (2004a,b) had previously reported 20 of 43 genes now confirmed by us as diagnostically relevant for PTC. This is a level of agreement rarely noted for inter-group comparisons of microarray results.
Our analysis has been performed on microarray data pre-processed by the standard MAS5 algorithm. Although many authors demonstrate the superiority of other pre-processing methods (e.g. RMA or GC-RMA; Irizarry et al. 2003), for inter-platform comparisons, the MAS5 method still seems to be a reasonable approach. In the MAS5 algorithm, each array is processed independently and the bootstrap procedure does not have to involve this step. Use of RMA pre-processing, which has to operate on the whole dataset, would pose the question of whether this step should also be bootstrapped. Presently, this is not feasible due to huge computational demand of pre-processing for large sample sets.
Redundancy of multi-gene cancer classifiers
This is inherently linked to the huge differences in gene expression profiles of several tumors, originating from the same tissue. This was indicated for the first time by Ein-Dor et al. (2005) in breast cancer. These authors re-analyzed the data of van't Veer et al. (2002) and showed that multiple similar classifiers may be obtained; they have comparable classification potency as van’t Veer’s original 70-gene classifier but a different gene content. Ein-Dor et al. stressed also that even slight differences in the training set composition influenced the selected genes. Our analysis demonstrates that similar redundancy is present in PTC. This fact is frequently overlooked by authors interpreting the results of gene expression profile studies that involved only a few genes or which were obtained in small groups of patients. In this paper, we propose a method of ranking genes according to their importance in multi-gene classifiers and with appropriate CIs indicating the robustness of the result.
To conclude, the primary goal of this study was to validate a novel SVM-based approach to differentiation of PTC from benign thyroid lesions. This goal was achieved with a very satisfactory degree of accuracy, over 95%. Simultaneously, we were able to rank the genes most essential for the molecular diagnosis of PTC. Although the presented list of genes can be enlarged, we believe the first 40 genes are especially suitable for further prospective studies in fine needle biopsy material and may serve to construct multi-gene classifiers with potential application in clinical setting. The comparison with other published microarray studies yields sufficient validation for the vast majority of them.
Acknowledgements
We gratefully acknowledge Aleksander Sochanik, PhD, for the thorough language revision of the manuscript. This work was partially supported by Polish Ministry of Education and Science under grant 3T11A 019 29 (K F) and 2P05A 022 30 (B J). This work was partially supported by the Deutsche Krebshilfe grant 106542 (R P and K K) and the Interdisciplinary Center for Clinical Research at the Faculty of Medicine of the University of Leipzig (projects B20, Z03). This work was partially supported within GENRISK-T project, contract number 036495 (A S, B J). Authors declare no potential conflict of interest.
References
- Aksoy M, Giles Y, Kapran Y, Terzioglu T, Tezelman S. Expression of bcl-2 in papillary thyroid cancers and its prognostic value. Acta Chirurgica Belgica. 2005;105:644–648. doi: 10.1080/00015458.2005.11679794. [DOI] [PubMed] [Google Scholar]
- Aldred MA, Ginn-Pease ME, Morrison CD, Popkie AP, Gimm O, Hoang-Vu C, Krause U, Dralle H, Jhiang SM, Plass C, et al. Caveolin-1 and caveolin-2, together with three bone morphogenetic protein-related genes, may encode novel tumor suppressors down-regulated in sporadic follicular thyroid carcinogenesis. Cancer Research. 2003;63:2864–2871. [PubMed] [Google Scholar]
- Aldred MA, Huang Y, Liyanarachchi S, Pellegata NS, Gimm O, Jhiang S, Davuluri RV, de la Chapelle A, Eng C. Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. Journal of Clinical Oncology. 2004;22:3531–3539. doi: 10.1200/JCO.2004.08.127. [DOI] [PubMed] [Google Scholar]
- Ambroziak M, Pachucki J, Stachlewska-Nasfeter E, Nauman J, Nauman A. Disturbed expression of type 1 and type 2 iodothyronine deiodinase as well as titf1/nkx2-1 and pax-8 transcription factor genes in papillary thyroid cancer. Thyroid. 2005;15:1137–1146. doi: 10.1089/thy.2005.15.1137. [DOI] [PubMed] [Google Scholar]
- Aratake Y, Nomura H, Kotani T, Marutsuka K, Kobayashi K, Kuma K, Miyauchi A, Okayama A, Tamura K. Coexistent anaplastic and differentiated thyroid carcinoma: an Immunohistochemical Study. American Journal of Clinical Pathology. 2006;125:399–406. [PubMed] [Google Scholar]
- Arnaldi LA, Borra RC, Maciel RM, Cerutti JM. Gene expression profiles reveal that DCN, DIO1, and DIO2 are underexpressed in benign and malignant thyroid tumors. Thyroid. 2005;15:210–221. doi: 10.1089/thy.2005.15.210. [DOI] [PubMed] [Google Scholar]
- Arturi F, Russo D, Giuffrida D, Ippolito A, Perrotti N, Vigneri R, Filetti S. Early diagnosis by genetic analysis of differentiated thyroid cancer metastases in small lymph nodes. Journal of Clinical Endocrinology and Metabolism. 1997;82:1638–1641. doi: 10.1210/jcem.82.5.4062. [DOI] [PubMed] [Google Scholar]
- Baloch ZW, Livolsi VA. Follicular-patterned lesions of the thyroid: the bane of the pathologist. American Journal of Clinical Pathology. 2002;117:143–150. doi: 10.1309/8VL9-ECXY-NVMX-2RQF. [DOI] [PubMed] [Google Scholar]
- Barden CB, Shister KW, Zhu B, Guiter G, Greenblatt DY, Zeiger MA, Fahey TJ., III Classification of follicular thyroid tumors by molecular signature: results of gene profiling. Clinical Cancer Research. 2003;9:1792–1800. [PubMed] [Google Scholar]
- Baris O, Savagner F, Nasser V, Loriod B, Granjeaud S, Guyetant S, Franc B, Rodien P, Rohmer V, Bertucci F, et al. Transcriptional profiling reveals coordinated up-regulation of oxidative metabolism genes in thyroid oncocytic tumors. Journal of Clinical Endocrinology and Metabolism. 2004;89:994–1005. doi: 10.1210/jc.2003-031238. [DOI] [PubMed] [Google Scholar]
- Baris O, Mirebeau-Prunier D, Savagner F, Rodien P, Ballester B, Loriod B, Granjeaud S, Guyetant S, Franc B, Houlgatte R, et al. Gene profiling reveals specific oncogenic mechanisms and signaling pathways in oncocytic and papillary thyroid carcinoma. Oncogene. 2005;24:4155–4161. doi: 10.1038/sj.onc.1208578. [DOI] [PubMed] [Google Scholar]
- Basolo F, Fiore L, Fusco A, Giannini R, Albini A, Merlo GR, Fontanini G, Conaldi PG, Toniolo A. Potentiation of the malignant phenotype of the undifferentiated ARO thyroid cell line by insertion of the bcl-2 gene. International Journal of Cancer. 1999;81:956–962. doi: 10.1002/(sici)1097-0215(19990611)81:6<956::aid-ijc19>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Belfiore A, Gangemi P, Costantino A, Russo G, Santonocito GM, Ippolito O, Di Renzo MF, Comoglio P, Fiumara A, Vigneri R. Negative/low expression of the Met/hepatocyte growth factor receptor identifies papillary thyroid carcinomas with high risk of distant metastases. Journal of Clinical Endocrinology and Metabolism. 1997;82:2322–2328. doi: 10.1210/jcem.82.7.4104. [DOI] [PubMed] [Google Scholar]
- Berlingieri MT, Pierantoni GM, Giancotti V, Santoro M, Fusco A. Thyroid cell transformation requires the expression of the HMGA1 proteins. Oncogene. 2002;21:2971–2980. doi: 10.1038/sj.onc.1205368. [DOI] [PubMed] [Google Scholar]
- Boser B, Guyon I & Vapnik V 1992 A training algorithm for optimal margin classifiers. Fifth Annual Workshop on Computational Learning Theory, Pittsburgh.
- Braga-Neto U, Dougherty E. Is cross-validation valid for small sample microarray classification? Bioinformatics. 2004;20:374–380. doi: 10.1093/bioinformatics/btg419. [DOI] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Machine Learning. 1996;24:123–140. [Google Scholar]
- Cerutti JM, Delcelo R, Amadei MJ, Nakabashi C, Maciel RM, Peterson B, Shoemaker J, Riggins GJ. A preoperative diagnostic test that distinguishes benign from malignant thyroid carcinoma based on gene expression. Journal of Clinical Investigation. 2004;113:1234–1242. doi: 10.1172/JCI19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KT, Lin JD, Chao TC, Hsueh C, Chang CA, Weng HF, Chan EC. Identifying differentially expressed genes associated with metastasis of follicular thyroid cancer by cDNA expression array. Thyroid. 2001;11:41–46. doi: 10.1089/10507250150500658. [DOI] [PubMed] [Google Scholar]
- Cherian MG, Jayasurya A, Bay BH. Metallothioneins in human tumors and potential roles in carcinogenesis. Mutation Research. 2003;533:201–209. doi: 10.1016/j.mrfmmm.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Chevillard S, Ugolin N, Vielh P, Ory K, Levalois C, Elliott D, Clayman GL, El-Naggar AK. Gene expression profiling of differentiated thyroid neoplasms: diagnostic and clinical implications. Clinical Cancer Research. 2004;10:6586–6597. doi: 10.1158/1078-0432.CCR-04-0053. [DOI] [PubMed] [Google Scholar]
- Czarnocka B, Pastuszko D, Janota-Bzowski M, Weetman AP, Watson PF, Kemp EH, McIntosh RS, Asghar MS, Jarzab B, Gubala E, et al. Is there loss or qualitative changes in the expression of thyroid peroxidase protein in thyroid epithelial cancer? British Journal of Cancer. 2001;85:875–880. doi: 10.1054/bjoc.2001.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl E, Winterhager E, Reuss B, Traub O, Butterweck A, Willecke K. Expression of the gap junction proteins connexin31 and connexin43 correlates with communication compartments in extraembryonic tissues and in the gastrulating mouse embryo, respectively. Journal of Cell Science. 1996;109:191–197. doi: 10.1242/jcs.109.1.191. [DOI] [PubMed] [Google Scholar]
- Detours V, Wattel S, Venet D, Hutsebaut N, Bogdanova T, Tronko MD, Dumont JE, Franc B, Thomas G, Maenhaut C. Absence of a specific radiation signature in post-Chernobyl thyroid cancers. British Journal of Cancer. 2005;92:1545–1552. doi: 10.1038/sj.bjc.6602521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B. Bootstrap methods:another look at the jackknife. Annals of Statistics. 1979;7:1–26. [Google Scholar]
- Efron B. Estimating the error rate of prediction rule: improvement on cross-validation. Journal of the American Statistical Association. 1983;78:316–331. [Google Scholar]
- Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- Eszlinger M, Krohn K, Paschke R. Complementary DNA expression array analysis suggests a lower expression of signal transduction proteins and receptors in cold and hot thyroid nodules. Journal of Clinical Endocrinology and Metabolism. 2001;86:4834–4842. doi: 10.1210/jcem.86.10.7933. [DOI] [PubMed] [Google Scholar]
- Eszlinger M, Krohn K, Frenzel R, Kropf S, Tonjes A, Paschke R. Gene expression analysis reveals evidence for inactivation of the TGF-beta signaling cascade in autonomously functioning thyroid nodules. Oncogene. 2004;23:795–804. doi: 10.1038/sj.onc.1207186. [DOI] [PubMed] [Google Scholar]
- Eszlinger M, Wiench M, Jarzab B, Krohn K, Beck M, Lauter J, Gubala E, Fujarewicz K, Swierniak A, Paschke R. Meta- and reanalysis of gene expression profiles of hot and cold thyroid nodules and papillary thyroid carcinoma for gene groups. Journal of Clinical Endocrinology and Metabolism. 2006;91:1934–1942. doi: 10.1210/jc.2005-1620. [DOI] [PubMed] [Google Scholar]
- Fedele M, Pierantoni GM, Berlingieri MT, Battista S, Baldassarre G, Munshi N, Dentice M, Thanos D, Santoro M, Viglietto G, et al. Overexpression of proteins HMGA1 induces cell cycle deregulation and apoptosis in normal rat thyroid cells. Cancer Research. 2001;61:4583–4590. [PubMed] [Google Scholar]
- Finley DJ, Arora N, Zhu B, Gallagher L, Fahey TJ., III Molecular profiling distinguishes papillary carcinoma from benign thyroid nodules. Journal of Clinical Endocrinology and Metabolism. 2004a;89:3214–3223. doi: 10.1210/jc.2003-031811. [DOI] [PubMed] [Google Scholar]
- Finley DJ, Zhu B, Barden CB, Fahey TJ., III Discrimination of benign and malignant thyroid nodules by molecular profiling. Annals of Surgery. 2004b;240:425–436. doi: 10.1097/01.sla.0000137128.64978.bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fourn V, Ferrand M, Franc JL. Differential expression of thyroperoxidase mRNA splice variants in human thyroid tumors. Biochimica et Biophysic Acta. 2004;1689:134–141. doi: 10.1016/j.bbadis.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Franc B, De La Salmoniere P, Lange F, Hong C, Louvel A, De Roquancourt A, Wild F, Hejblum G, Chevret S, Chastang C. Interobserver and intraobserver reproducibility in the histopathology of follicular thyroid carcinoma. Human Pathology. 2003;34:1092–1100. doi: 10.1016/s0046-8177(03)00403-9. [DOI] [PubMed] [Google Scholar]
- Freund Y & Schapire R 1996 Experiments with a new boosting algorithm. Proceedings of the 13th International Conference on Machine Learning Bari 325–332.
- Frohlich E, Machicao F, Wahl R. Action of thiazolidinediones on differentiation, proliferation and apoptosis of normal and transformed thyrocytes in culture. Endocrine-Related Cancer. 2005;12:291–303. doi: 10.1677/erc.1.00973. [DOI] [PubMed] [Google Scholar]
- Furuya F, Shimura H, Miyazaki A, Taki K, Ohta K, Haraguchi K, Onaya T, Endo T, Kobayashi T. Adenovirus-mediated transfer of thyroid transcription factor-1 induces radioiodide organification and retention in thyroid cancer cells. Endocrinology. 2004;145:5397–5405. doi: 10.1210/en.2004-0631. [DOI] [PubMed] [Google Scholar]
- Ghinea N, Baratti-Elbaz C, De Jesus-Lucas A, Milgrom E. TSH receptor interaction with the extracellular matrix: role on constitutive activity and sensitivity to hormonal stimulation. Molecular Endocrinology. 2002;16:912–923. doi: 10.1210/mend.16.5.0820. [DOI] [PubMed] [Google Scholar]
- Gingras R, Richard C, El-Alfy M, Morales CR, Potier M, Pshezhetsky AV. Purification, cDNA cloning, and expression of a new human blood plasma glutamate carboxypeptidase homologous to N-acetyl-aspartyl-alpha-glutamate carboxypeptidase/prostate-specific membrane antigen. Journal of Biological Chemistry. 1999;274:11742–11750. doi: 10.1074/jbc.274.17.11742. [DOI] [PubMed] [Google Scholar]
- Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, Zhu Z, Ciampi R, Roh M, Shedden K, et al. Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene. 2005;24:6646–6656. doi: 10.1038/sj.onc.1208822. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Green LM, Bianski BM, Murray DK, Rightnar SS, Nelson GA. Characterization of accelerated iron-induced damage in gap junction-competent and -incompetent thyroid follicular cells. Radiation Research. 2005;163:172–182. doi: 10.1667/rr3297. [DOI] [PubMed] [Google Scholar]
- Griffith OL, Melck A, Jones SJ, Wiseman SM. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. Journal of Clinical Oncology. 2006;24:5043–5051. doi: 10.1200/JCO.2006.06.7330. [DOI] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;64:389–422. [Google Scholar]
- Hamada A, Mankovskaya S, Saenko V, Rogounovitch T, Mine M, Namba H, Nakashima M, Demidchik Y, Demidchik E, Yamashita S. Diagnostic usefulness of PCR profiling of the differentially expressed marker genes in thyroid papillary carcinomas. Cancer Letters. 2005;224:289–301. doi: 10.1016/j.canlet.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Haugen BR, Larson LL, Pugazhenthi U, Hays WR, Klopper JP, Kramer CA, Sharma V. Retinoic acid and retinoid X receptors are differentially expressed in thyroid cancer and thyroid carcinoma cell lines and predict response to treatment with retinoids. Journal of Clinical Endocrinology and Metabolism. 2004;89:272–280. doi: 10.1210/jc.2003-030770. [DOI] [PubMed] [Google Scholar]
- Hegedus L. Clinical practice. The thyroid nodule. New England Journal of Medicine. 2004;351:1764–1771. doi: 10.1056/NEJMcp031436. [DOI] [PubMed] [Google Scholar]
- Hoos A, Stojadinovic A, Singh B, Dudas ME, Leung DH, Shaha AR, Shah JP, Brennan MF, Cordon-Cardo C, Ghossein R. Clinical significance of molecular expression profiles of Hurthle cell tumors of the thyroid gland analyzed via tissue microarrays. American Journal of Pathology. 2002;160:175–183. doi: 10.1016/S0002-9440(10)64361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Xiong Z, Lowey J, Suh E, Dougherty ER. Optimal number of features as a function of sample size for various classification rules. Bioinformatics. 2005;21:1509–1515. doi: 10.1093/bioinformatics/bti171. [DOI] [PubMed] [Google Scholar]
- Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, Kornacker K, LiVolsi V, Frankel W, Kloos RT, Eng C, et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. PNAS. 2001;98:15044–15049. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illario M, Amideo V, Casamassima A, Andreucci M, di MT, Miele C, Rossi G, Fenzi G, Vitale M. Integrin-dependent cell growth and survival are mediated by different signals in thyroid cells. Journal of Clinical Endocrinology and Metabolism. 2003;88:260–269. doi: 10.1210/jc.2002-020774. [DOI] [PubMed] [Google Scholar]
- Ippolito A, Vella V, La Rosa GL, Pellegriti G, Vigneri R, Belfiore A. Immunostaining for Met/HGF receptor may be useful to identify malignancies in thyroid lesions classified suspicious at fine-needle aspiration biopsy. Thyroid. 2001;11:783–787. doi: 10.1089/10507250152484646. [DOI] [PubMed] [Google Scholar]
- Irizarry R, Hobbs B, Colli F, Beazer-Barclay Y, Antonellis K, Scherf U, Speed T. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jacques C, Baris O, Prunier-Mirebeau D, Savagner F, Rodien P, Rohmer V, Franc B, Guyetant S, Malthiery Y, Reynier P. Two-step differential expression analysis reveals a new set of genes involved in thyroid oncocytic tumors. Journal of Clinical Endocrinology and Metabolism. 2005;90:2314–2320. doi: 10.1210/jc.2004-1337. [DOI] [PubMed] [Google Scholar]
- Jarzab B, Wiench M, Fujarewicz K, Simek K, Jarzab M, Oczko-Wojciechowska M, Wloch J, Czarniecka A, Chmielik E, Lange D, et al. Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Research. 2005;65:1587–1597. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- Kawai K, Resetkova E, Enomoto T, Fornasier V, Volpe R. Is human leukocyte antigen-DR and intercellular adhesion molecule-1 expression on human thyrocytes constitutive in papillary thyroid cancer? Comparative studies in human thyroid xenografts in severe combined immunodeficient and nude mice. Journal of Clinical Endocrinology and Metabolism. 1998;83:157–164. doi: 10.1210/jcem.83.1.4489. [DOI] [PubMed] [Google Scholar]
- Kebebew E, Peng M, Reiff E, McMillan A. Diagnostic and extent of disease multigene assay for malignant thyroid neoplasms. Cancer. 2006;106:2592–2597. doi: 10.1002/cncr.21922. [DOI] [PubMed] [Google Scholar]
- Kehlen A, Lendeckel U, Dralle H, Langner J, Hoang-Vu C. Biological significance of aminopeptidase N/CD13 in thyroid carcinomas. Cancer Research. 2003;63:8500–8506. [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Bootstrapping cluster analysis: assessing the reliability of conclusions from microarray experiments. PNAS. 2001;98:8961–8965. doi: 10.1073/pnas.161273698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholova I, Ludvikova M, Ryska A, Topolcan O, Pikner R, Pecen L, Cap J, Holubec L., Jr Diagnostic role of markers dipeptidyl peptidase IV and thyroid peroxidase in thyroid tumors. Anticancer Research. 2003a;23:871–875. [PubMed] [Google Scholar]
- Kholova I, Ryska A, Ludvikova M, Cap J, Pecen L. Dipeptidyl peptidase IV expression in thyroid cytology: retrospective histologically confirmed study. Cytopathology. 2003b;14:27–31. doi: 10.1046/j.1365-2303.2003.01138.x. [DOI] [PubMed] [Google Scholar]
- Kiess M, Scharm B, Aguzzi A, Hajnal A, Klemenz R, Schwarte-Waldhoff I, Schafer R. Expression of ril, a novel LIM domain gene, is down-regulated in Hras-transformed cells and restored in phenotypic revertants. Oncogene. 1995;10:61–68. [PubMed] [Google Scholar]
- Kim HS, Roh CR, Chen B, Tycko B, Nelson DM, Sadovsky Y. Hypoxia regulates the expression of PHLDA2 in primary term human trophoblasts. Placenta. 2007;28:77–84. doi: 10.1016/j.placenta.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Kittel A, Csapo ZS, Csizmadia E, Jackson SW, Robson SC. Co-localization of P2Y1 receptor and NTPDase1/CD39 within caveolae in human placenta. European Journal of Histochemistry. 2004;48:253–259. [PubMed] [Google Scholar]
- Klopper JP, Hays WR, Sharma V, Baumbusch MA, Hershman JM, Haugen BR. Retinoid X receptor-gamma and peroxisome proliferator-activated receptor-gamma expression predicts thyroid carcinoma cell response to retinoid and thiazolidinedione treatment. Molecular Cancer Therapeutics. 2004;3:1011–1020. [PubMed] [Google Scholar]
- Kohrle J. Local activation and inactivation of thyroid hormones: the deiodinase family. Molecular and Cellular Endocrinology. 1999;151:103–119. doi: 10.1016/s0303-7207(99)00040-4. [DOI] [PubMed] [Google Scholar]
- Kromminga A, Hagel C, Arndt R, Schuppert F. Serological reactivity of recombinant 1D autoantigen and its expression in human thyroid and eye muscle tissue: a possible autoantigenic link in Graves' patients. Journal of Clinical Endocrinology and Metabolism. 1998;83:2817–2823. doi: 10.1210/jcem.83.8.5018. [DOI] [PubMed] [Google Scholar]
- Lazar V, Bidart JM, Caillou B, Mahe C, Lacroix L, Filetti S, Schlumberger M. Expression of the Na+/I− symporter gene in human thyroid tumors: a comparison study with other thyroid-specific genes. Journal of Clinical Endocrinology and Metabolism. 1999;84:3228–3234. doi: 10.1210/jcem.84.9.5996. [DOI] [PubMed] [Google Scholar]
- Lelievre V, Muller JM, Falcon J. Adenosine modulates cell proliferation in human colonic adenocarcinoma. I. Possible involvement of adenosine A1 receptor subtypes in HT29 cells. European Journal of Pharmacology. 1998;341:289–297. doi: 10.1016/s0014-2999(97)01462-3. [DOI] [PubMed] [Google Scholar]
- Letsas KP, Frangou-Lazaridis M, Skyrlas A, Tsatsoulis A, Malamou-Mitsi V. Transcription factor-mediated proliferation and apoptosis in benign and malignant thyroid lesions. Pathology International. 2005;55:694–702. doi: 10.1111/j.1440-1827.2005.01899.x. [DOI] [PubMed] [Google Scholar]
- Lin F, Ren XD, Doris G, Clark RA. Three-dimensional migration of human adult dermal fibroblasts from collagen lattices into fibrin/fibronectin gels requires syndecan-4 proteoglycan. Journal of Investigative Dermatology. 2005;124:906–913. doi: 10.1111/j.0022-202X.2005.23740.x. [DOI] [PubMed] [Google Scholar]
- Liu W, Asa SL, Ezzat S. 1alpha,25-Dihydroxyvitamin D3 targets PTEN-dependent fibronectin expression to restore thyroid cancer cell adhesiveness. Molecular Endocrinology. 2005;19:2349–2357. doi: 10.1210/me.2005-0117. [DOI] [PubMed] [Google Scholar]
- Lubitz CC, Fahey TJI. Gene expression profiling of thyroid tumors – clinical applicability. Nature Clinical Practice. Endocrinology & Metabolism. 2006;2:472–473. doi: 10.1038/ncpendmet0271. [DOI] [PubMed] [Google Scholar]
- Mazzanti C, Zeiger MA, Costouros NG, Umbricht C, Westra WH, Smith D, Somervell H, Bevilacqua G, Alexander HR, Libutti SK. Using gene expression profiling to differentiate benign versus malignant thyroid tumors. Cancer Research. 2004;64:2898–2903. doi: 10.1158/0008-5472.can-03-3811. [DOI] [PubMed] [Google Scholar]
- De Micco C, Vassko V, Henry JF. The value of thyroid peroxidase immunohistochemistry for preoperative fine-needle aspiration diagnosis of the follicular variant of papillary thyroid cancer. Surgery. 1999;126:1200–1204. doi: 10.1067/msy.2099.101428. [DOI] [PubMed] [Google Scholar]
- Mitselou A, Peschos D, Dallas P, Charalabopoulos K, Agnantis NJ, Vougiouklakis T. Immunohistochemical analysis of expression of bcl-2 protein in papillary carcinomas and papillary microcarcinomas of the thyroid gland. Experimental Oncology. 2004;26:282–286. [PubMed] [Google Scholar]
- Musholt TJ, Brehm C, Hanack J, von WR, Musholt PB. Identification of differentially expressed genes in papillary thyroid carcinomas with and without rearrangements of the tyrosine kinase receptors RET and/or NTRK1. Journal of Surgical Research. 2006;131:15–25. doi: 10.1016/j.jss.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Noro B, Licheri B, Sgarra R, Rustighi A, Tessari MA, Chau KY, Ono SJ, Giancotti V, Manfioletti G. Molecular dissection of the architectural transcription factor HMGA2. Biochemistry. 2003;42:4569–4577. doi: 10.1021/bi026605k. [DOI] [PubMed] [Google Scholar]
- Onda M, Akaishi J, Asaka S, Okamoto J, Miyamoto S, Mizutani K, Yoshida A, Ito K, Emi M. Decreased expression of haemoglobin beta (HBB) gene in anaplastic thyroid cancer and recovery of its expression inhibits cell growth. British Journal of Cancer. 2005;92:2216–2224. doi: 10.1038/sj.bjc.6602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozog J, Jarzab M, Pawlaczek A, Oczko-Wojciechowska M, Wloch J, Roskosz J, Gubala E. Expression of DPP4 gene in papillary thyroid carcinoma (In Polish) Endokrynologia Polska/Polish Journal of Endocrinology. 2006;57:12–17. [PubMed] [Google Scholar]
- Plum A, Hallas G, Willecke K. Expression of the mouse gap junction gene Gjb3 is regulated by distinct mechanisms in embryonic stem cells and keratinocytes. Genomics. 2002;79:24–30. doi: 10.1006/geno.2001.6671. [DOI] [PubMed] [Google Scholar]
- Prasad ML, Pellegata NS, Kloos RT, Barbacioru C, Huang Y, de la Chapelle A. CITED1 protein expression suggests papillary thyroid carcinoma in high throughput tissue microarray-based study. Thyroid. 2004;14:169–175. doi: 10.1089/105072504773297830. [DOI] [PubMed] [Google Scholar]
- Prasad ML, Pellegata NS, Huang Y, Nagaraja HN, de la CA, Kloos RT. Galectin-3, fibronectin-1, CITED-1, HBME1 and cytokeratin-19 immunohistochemistry is useful for the differential diagnosis of thyroid tumors. Modern Pathology. 2005;18:48–57. doi: 10.1038/modpathol.3800235. [DOI] [PubMed] [Google Scholar]
- Ramirez R, Hsu D, Patel A, Fenton C, Dinauer C, Tuttle RM, Francis GL. Over-expression of hepatocyte growth factor/scatter factor (HGF/SF) and the HGF/SF receptor (cMET) are associated with a high risk of metastasis and recurrence for children and young adults with papillary thyroid carcinoma. Clinical Endocrinology. 2000;53:635–644. doi: 10.1046/j.1365-2265.2000.01124.x. [DOI] [PubMed] [Google Scholar]
- Rocha AS, Soares P, Fonseca E, Cameselle-Teijeiro J, Oliveira MC, Sobrinho-Simoes M. E-cadherin loss rather than beta-catenin alterations is a common feature of poorly differentiated thyroid carcinomas. Histopathology. 2003;42:580–587. doi: 10.1046/j.1365-2559.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- Ruco LP, Stoppacciaro A, Ballarini F, Prat M, Scarpino S. Met protein and hepatocyte growth factor (HGF) in papillary carcinoma of the thyroid: evidence for a pathogenetic role in tumourigenesis. Journal of Pathology. 2001;194:4–8. doi: 10.1002/path.847. [DOI] [PubMed] [Google Scholar]
- Sarquis MS, Weber F, Shen L, Broelsch CE, Jhiang SM, Zedenius J, Frilling A, Eng C. High frequency of loss of heterozygosity in imprinted, compared with nonimprinted, genomic regions in follicular thyroid carcinomas and atypical adenomas. Journal of Clinical Endocrinology and Metabolism. 2006;91:262–269. doi: 10.1210/jc.2005-1880. [DOI] [PubMed] [Google Scholar]
- Scarpino S, Di NA, Rapazzotti-Onelli M, Pilozzi E, Ruco L. Papillary carcinoma of the thyroid: methylation is not involved in the regulation of MET expression. British Journal of Cancer. 2004;91:703–706. doi: 10.1038/sj.bjc.6601988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagdarsurengin U, Gimm O, Dralle H, Hoang-Vu C, Dammann R. CpG island methylation of tumor-related promotors occurs preferentially in undifferentiated carcinoma. Thyroid. 2006;16:633–642. doi: 10.1089/thy.2006.16.633. [DOI] [PubMed] [Google Scholar]
- Schelfhout LJ, Van Muijen GN, Fleuren GJ. Expression of keratin 19 distinguishes papillary thyroid carcinoma from follicular carcinomas and follicular thyroid adenoma. American Journal of Clinical Pathology. 1989;92:654–658. doi: 10.1093/ajcp/92.5.654. [DOI] [PubMed] [Google Scholar]
- Schmutzler C, Hoang-Vu C, Ruger B, Kohrle J. Human thyroid carcinoma cell lines show different retinoic acid receptor repertoires and retinoid responses. European Journal of Endocrinology. 2004;150:547–556. doi: 10.1530/eje.0.1500547. [DOI] [PubMed] [Google Scholar]
- Schnurr M, Toy T, Shin A, Hartmann G, Rothenfusser S, Soellner J, Davis ID, Cebon J, Maraskovsky E. Role of adenosine receptors in regulating chemotaxis and cytokine production of plasmacytoid dendritic cells. Blood. 2004;103:1391–1397. doi: 10.1182/blood-2003-06-1959. [DOI] [PubMed] [Google Scholar]
- Scouten WT, Patel A, Terrell R, Burch HB, Bernet VJ, Tuttle RM, Francis GL. Cytoplasmic localization of the paired box gene, Pax-8, is found in pediatric thyroid cancer and may be associated with a greater risk of recurrence. Thyroid. 2004;14:1037–1046. doi: 10.1089/thy.2004.14.1037. [DOI] [PubMed] [Google Scholar]
- Sedlak TW, Snyder SH. Messenger molecules and cell death: therapeutic implications. JAMA. 2006;295:81–89. doi: 10.1001/jama.295.1.81. [DOI] [PubMed] [Google Scholar]
- Shimura H, Suzuki H, Miyazaki A, Furuya F, Ohta K, Haraguchi K, Endo T, Onaya T. Transcriptional activation of the thyroglobulin promoter directing suicide gene expression by thyroid transcription factor-1 in thyroid cancer cells. Cancer Research. 2001;61:3640–3646. [PubMed] [Google Scholar]
- Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. Journal of National Cancer Institute. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- Slonim D, Tamayo P, Mesirov J, Golub T. Class prediction and discovery using gene expression data. Proceedings of the Fourth Annual International Conference on Computational Molecular Biology. 2000:263–272. [Google Scholar]
- Spessotto P, Cervi M, Mucignat MT, Mungiguerra G, Sartoretto I, Doliana R, Colombatti A. Beta 1 integrin-dependent cell adhesion to EMILIN-1 is mediated by the gC1q domain. Journal of Biological Chemistry. 2003;278:6160–6167. doi: 10.1074/jbc.M208322200. [DOI] [PubMed] [Google Scholar]
- Stassi G, Todaro M, Zerilli M, Ricci-Vitiani L, Di LD, Patti M, Florena A, di GF, Di GG, De MR. Thyroid cancer resistance to chemotherapeutic drugs via autocrine production of interleukin-4 and interleukin-10. Cancer Research. 2003;63:6784–6790. [PubMed] [Google Scholar]
- Takano T, Matsuzuka F, Miyauchi A, Yokozawa T, Liu G, Morita S, Kuma K, Amino N. Restricted expression of oncofetal fibronectin mRNA in thyroid papillary and anaplastic carcinoma: an in situ hybridization study. British Journal of Cancer. 1998;78:221–224. doi: 10.1038/bjc.1998.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Miyauchi A, Yokozawa T, Matsuzuka F, Maeda I, Kuma K, Amino N. Preoperative diagnosis of thyroid papillary and anaplastic carcinomas by real-time quantitative reverse transcription-polymerase chain reaction of oncofetal fibronectin messenger RNA. Cancer Research. 1999;59:4542–4545. [PubMed] [Google Scholar]
- Takano T, Hasegawa Y, Miyauchi A, Matsuzuka F, Yoshida H, Kuma K, Hayashi N, Nakamori S, Amino N. Quantitative analysis of osteonectin mRNA in thyroid carcinomas. Endocrine Journal. 2002;49:511–516. doi: 10.1507/endocrj.49.511. [DOI] [PubMed] [Google Scholar]
- Takano T, Miyauchi A, Yoshida H, Kuma K, Amino N. High-throughput differential screening of mRNAs by serial analysis of gene expression: decreased expression of trefoil factor 3 mRNA in thyroid follicular carcinomas. British Journal of Cancer. 2004;90:1600–1605. doi: 10.1038/sj.bjc.6601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharis SE, Margeli AP, Klijanienko JT, Kouraklis GP. Metallothionein expression in human neoplasia. Histopathology. 2004;45:103–118. doi: 10.1111/j.1365-2559.2004.01922.x. [DOI] [PubMed] [Google Scholar]
- Thomson RB, Igarashi P, Biemesderfer D, Kim R, bu-Alfa A, Soleimani M, Aronson PS. Isolation and cDNA cloning of Ksp-cadherin, a novel kidney-specific member of the cadherin multigene family. Journal of Biological Chemistry. 1995;270:17594–17601. doi: 10.1074/jbc.270.29.17594. [DOI] [PubMed] [Google Scholar]
- Thomson RB, Ward DC, Quaggin SE, Igarashi P, Muckler ZE, Aronson PS. cDNA cloning and chromosomal localization of the human and mouse isoforms of Ksp-cadherin. Genomics. 1998;51:445–451. doi: 10.1006/geno.1998.5402. [DOI] [PubMed] [Google Scholar]
- Tonoli H, Flachon V, Audebet C, Calle A, Jarry-Guichard T, Statuto M, Rousset B, Munari-Silem Y. Formation of three-dimensional thyroid follicle-like structures by polarized FRT cells made communication competent by transfection and stable expression of the connexin-32 gene. Endocrinology. 2000;141:1403–1413. doi: 10.1210/endo.141.4.7400. [DOI] [PubMed] [Google Scholar]
- Vapnik V. The Nature of Statistical Learning Theory. Springer-Verlag; New York: 1995. [Google Scholar]
- van't Veer L, Dai H, van de Vijver M, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wagner K, Arciaga R, Siperstein A, Milas M, Warshawsky I, Sethu S, Reddy K, Gupta MK. Thyrotropin receptor/thyroglobulin messenger ribonucleic acid in peripheral blood and fine-needle aspiration cytology: diagnostic synergy for detecting thyroid cancer. Journal of Clinical Endocrinology and Metabolism. 2005;90:1921–1924. doi: 10.1210/jc.2004-1793. [DOI] [PubMed] [Google Scholar]
- Wasenius VM, Hemmer S, Kettunen E, Knuutila S, Franssila K, Joensuu H. Hepatocyte growth factor receptor, matrix metalloproteinase-11, tissue inhibitor of metalloproteinase-1, and fibronectin are up-regulated in papillary thyroid carcinoma: a cDNA and tissue microarray study. Clinical Cancer Research. 2003;9:68–75. [PubMed] [Google Scholar]
- Watanabe TK, Katagiri T, Suzuki M, Shimizu F, Fujiwara T, Kanemoto N, Nakamura Y, Hirai Y, Maekawa H, Takahashi E. Cloning and characterization of two novel human cDNAs (NELL1 and NELL2) encoding proteins with six EGF-like repeats. Genomics. 1996;38:273–276. doi: 10.1006/geno.1996.0628. [DOI] [PubMed] [Google Scholar]
- Weber F, Shen L, Aldred MA, Morrison CD, Frilling A, Saji M, Schuppert F, Broelsch CE, Ringel MD, Eng C. Genetic classification of benign and malignant thyroid follicular neoplasia based on a three-gene combination. Journal of Clinical Endocrinology and Metabolism. 2005;90:2512–2521. doi: 10.1210/jc.2004-2028. [DOI] [PubMed] [Google Scholar]
- Widerak M, Ghoneim C, Dumontier MF, Quesne M, Corvol MT, Savouret JF. The aryl hydrocarbon receptor activates the retinoic acid receptoralpha through SMRT antagonism. Biochimie. 2006;88:387–397. doi: 10.1016/j.biochi.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Woodhouse EC, Amanatullah DF, Schetz JA, Liotta LA, Stracke ML, Clair T. Adenosine receptor mediates motility in human melanoma cells. Biochemical and Biophysical Research Communications. 1998;246:888–894. doi: 10.1006/bbrc.1998.8714. [DOI] [PubMed] [Google Scholar]
- Wreesmann VB, Sieczka EM, Socci ND, Hezel M, Belbin TJ, Childs G, Patel SG, Patel KN, Tallini G, Prystowsky M, et al. Genome-wide profiling of papillary thyroid cancer identifies MUC1 as an independent prognostic marker. Cancer Research. 2004;64:3780–3789. doi: 10.1158/0008-5472.CAN-03-1460. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lu X, Shi Q, Xu XQ, Leung HC, Harris LN, Iglehart JD, Miron A, Liu JS, Wong WH. Recursive SVM feature selection and sample classification for mass-spectrometry and microarray data. BMC Bioinformatics. 2006;7:197. doi: 10.1186/1471-2105-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Famulski KS, Parhar RS, Baitei E, Al-Mohanna FA, Farid NR, Shi Y. Microarray analysis of metastasis-associated gene expression profiling in a murine model of thyroid carcinoma pulmonary metastasis: identification of S100A4 (Mts1) gene overexpression as a poor prognostic marker for thyroid carcinoma. Journal of Clinical Endocrinology and Metabolism. 2004;89:6146–6154. doi: 10.1210/jc.2004-0418. [DOI] [PubMed] [Google Scholar]