Abstract
Most familial early-onset Alzheimer’s disease cases are caused by mutations in the presenilin 1 (PS1) gene. Subcellular localization of the endogenous PS1 is essential for understanding its function, interactions with proteins, and role in Alzheimer’s disease. Although numerous studies revealed predominant localization of PS1 to endoplasmic reticulum and Golgi, there are conflicting reports on the localization of PS1 to the cell surface. We found that endogenous PS1 is highly expressed in T lymphocytes (Jurkat cells). Using a variety of methods, we present evidence that endogenous PS1 is localized to the cell surface in addition to intracellular membrane compartments. Moreover, PS1 appeared in high levels on the surface of lamellipodia upon adhesion of the cells to a collagen matrix. The redistribution of PS1 in adhered cells was strikingly similar to that of the well characterized adhesion protein CD44. Cell surface PS1 formed complexes in vivo with actin-binding protein filamin (ABP-280), which is known to form bridges between cell surface receptors and cytoskeleton and mediate cell adhesion and cell motility. Taken together, our results suggest a role of PS1 in cell adhesion and/or cell–matrix interaction.
The presenilin (PS1 and PS2) genes have been identified as major causal genes for early-onset familial Alzheimer’s disease (FAD) (1–3). However, the biological functions of presenilins are unknown.
The presenilins are integral membrane proteins with a proposed structure of seven to eight hydrophobic transmembrane domains and a hydrophilic loop located between transmembrane domains 6 and 7 (1). More than 60% of amino acid residues in the sequence of PS1 and PS2 are conserved (1, 3). FAD mutations are found throughout the entire molecule of PS1 (4–6).
Northern blot analysis and in situ hybridization studies of PS1 and PS2 mRNAs demonstrate a widespread, uniform expression of RNAs both in the brain and peripheral tissues of humans and rodents (1, 7–9). A high level of expression of endogenous presenilins was detected only in neurons (8, 9).
Light and electron microscopy studies revealed predominant localization of PS1 and PS2 to endoplasmic reticulum–Golgi compartments and to coated transport vesicles in neurons and in various cell types transfected by PS1 or PS2 cDNAs (9–12). In addition, immunocytochemical studies of transfected cells have identified PS-1 in the nuclear membrane, interphase kinetochores, and centrosomes (13). Conflicting results were reported for localization of PS1 to the plasma membrane (10, 14–16).
PS1 reveals approximately 50% homology with Caenorhabditis elegans protein sel-12 (17), which facilitates signaling mediated by the Notch/lin-12 family receptors. Notch receptors are cell surface proteins that regulate cell–cell interactions and cell fate choices during T cell and neural development (18, 19). The expression of Notch 1 mRNA is decreased significantly in the presomitic mesoderm of PS1 null mice characterized by massive neuronal loss in specific subregions of the mutant brain (20, 21). Vito et al. (22) demonstrated that the PS2 gene contributes to T cell receptor (TCR)-induced apoptosis.
Several groups reported interactions of presenilins with amyloid β protein precursor, catenin, and filamin—proteins that are involved in cell adhesion and cell–cell contacts (23–26).
In this paper we show that endogenous PS1 is highly expressed and is concentrated at the surface of lamellipodia in Jurkat cells adhered to a collagen matrix. Cell surface PS1 forms complexes with the actin-binding protein filamin (ABP-280), which mediates cell adhesion and cell motility. These results suggest a role of PS1 in cell adhesion and cell–matrix interaction.
MATERIALS AND METHODS
Cell Cultures and Brain Extracts.
Jurkat cells, clone FHCRC E6–1, a human leukemia T cell line, and HEp-2 human epithelial cells were obtained from the American Type Culture Collection. Jurkat cells, grown in RPMI 1640 medium containing 10% FBS, were plated on chambered coverslips covered with a saturated solution of rat tail collagen, type 1. Brain extracts from wild-type PS1(+/+) and homozygous PS1 knockout (−/−) mice were kindly provided by S. Sisodia (The Johns Hopkins University, Baltimore).
Antibodies.
The following rabbit polyclonal antibodies and one mAb were generated against synthetic peptides corresponding to amino-terminal regions of human PS1: PS1-N was generated against residues 27–44; 231f was generated against residues 2–20 (provided by B. Yankner, Harvard Medical School); R222 was generated against residues 2–12 (provided by N. Robakis, Mount Sinai School of Medicine); Ab14 was generated against residues 3–15 (provided by S. Gandy, Nathan Kline Institute); and mAb MKAD 3.4 was generated against residues 45–48 (provided by T. Honda, Yokohama Research Center, Japan). Rabbit antiserum PS1-L (antibody to loop region of PS1) was generated against residues 331–350 of human PS1 (12). Antibodies PS-N, 231f, and PS1-L were purified by using affinity chromatography on columns with immobilized peptides. Preabsorbed antibodies were obtained by using immobilized recombinant S-Tag-PS1 (30). Anti-αβ-TCR mAb clone T10139.1A-31 (PharMingen), anti-Golgi 58-kDa protein mAb clone 58K-9 and anti-CD44 mAb A3D8 (Sigma), anti-filamin mAb PM6/317 (Research Diagnostics), and anti-transthyretin rabbit polyclonal antibody (Boehringer Mannheim) were purchased.
Interaction of CD44 and TCR with PS1.
Full-length PS1 cDNA was cloned into the EcoRI site of the pET29a expression vector containing S-Tag peptide and a thrombin cleavage site upstream of cloning insert (Novagen) (27). S-Tag-PS-1 fusion protein was expressed in Escherichia coli strain BL21 (DE3) and affinity-purified from inclusion bodies by using S-protein agarose. HEp-2 or Jurkat cells (107) were lysed in buffer A: 10 mM Tris⋅HCl, pH 7.4/1% Triton X-100/1% NP-40/0.1% SDS/1% sodium deoxycholate/150 mM NaCl containing a protease inhibitor mixture (5 μg/ml leupeptin/5 μg/ml aprotinin/2 μg/ml pepstatin A/0.25 mM PMSF, Sigma). Cell lysates were mixed with fusion protein S-Tag-PS1 bound to the S-agarose and incubated for 2 hr at room temperature. PS1-bound proteins were eluted by thrombin treatment of washed resin (31). For immunoprecipitation, 106 Jurkat cells were lysed in buffer A containing the protease inhibitor mixture and mixed with mAb to CD44 (mouse IgG) or mAb to TCR (mouse IgM). Cell lysates were incubated for 1 hr at 4°C with primary antibodies and 30 min with goat anti-mouse IgG or anti-mouse IgM-agarose (Sigma). The resin was washed three times with modified RIPA buffer (10 mM Tris⋅HCl, pH 7.4/1% Triton X-100/0.1% SDS/1% sodium deoxycholate/150 mM NaCl) containing a protease inhibitor cocktail. Immunoprecipitates were eluted from the resin by boiling in 2× Laemmli sample buffer (40% glycerol/6% SDS/4% 2-mercaptoethanol) for 10 min and electrophoresed in 10% Tris-glycine gels, and separated proteins were transferred to a poly(vinylidene difluoride) membrane (Bio-Rad). Western blotting, using enhanced chemiluminescence detection, was performed as per the manufacturer’s instructions (Amersham). Molecular sizes were determined by using a Kaleidoscope-prestained molecular standard (Bio-Rad).
Flow Cytometry Analysis.
Jurkat cells were incubated first with 0.5% BSA in PBS and then with primary antibodies for 2 hr at room temperature and 1 hr with corresponding FITC-conjugated secondary antibodies. Data were collected on a Becton Dickinson FACScan cytofluorimeter. Jurkat cells were gated for forward and side-angle scatters, and 9,000 fluorescent particles of gated population were analyzed. The percentage of positive cells was estimated by measuring the variation in the mean values of the logarithm of intensity of fluorescence.
Biotinylation and Cross-Linking.
Jurkat cells were grown to a density of 5 × 106 cells/ml and then washed three times with ice-cold PBS (pH 8.0). Cells (108) were resuspended in 1 ml PBS containing 2 mg of EZ-Link Sulfo-NHS-LC-LC-Biotin (Pierce) and incubated for 45 min with shaking at room temperature. After washing with ice-cold PBS, cells were lysed in 1 ml of RIPA buffer containing protease inhibitor cocktail. Biotinylated proteins were isolated by using 2-ml columns with either Immuno Pure immobilized monomeric avidin and elution with 2 mM d-biotin, or streptavidin-agarose and elution by boiling in Laemmli sample buffer with 0.1 M 2-mercaptoethanol. Permeabilization of Jurkat cells with lysolecithin (Sigma) and chemical cross-linking with the reversible, homobifunctional cross-linker DTSSP (Pierce) were performed as recommended by Altin and Pagler (28). Before SDS/PAGE, all samples containing DTSSP were boiled for 10 min in sample buffer containing 100 mM 2-mercaptoethanol.
Laser Confocal Microscopy.
Jurkat cells adhered to a collagen matrix in eight-chambered coverslips were fixed for 30 min in 2% paraformaldehyde/0.1% glutaraldehyde buffered with PBS and permeabilized with 0.5% Triton X-100 for 15 min at room temperature. The cells then were blocked with 5% albumin and incubated with primary antibody. PS1 was visualized by using affinity-purified polyclonal antibodies PS1-N at 1:100 dilution or PS1-L at 1:200 dilution. For dual immunolabeling, we used anti-CD44 mAb, anti-TCR mAb, or antifilamin mAb, followed by FITC- or tetramethylrhodamine B isothiocyanate-conjugated secondary antibodies. Immunofluorescence images were visualized with either a ×40 1.4 numerical aperture (NA) or a ×60 1.3 NA oil-coupled objective on a Noran Confocal Odyssey system through a Nikon inverted-diaphragm microscope.
Scanning Electron Microscopy.
Jurkat cells adhered to a collagen matrix in eight-chambered coverslips were fixed in 2% paraformaldehyde/0.1% glutaraldehyde, blocked with 5% BSA, and incubated with primary antibodies at 4°C for 16 hr. Gold–silver immunostaining was performed by following the Goldmark Protocol (Goldmark Biologicals, Phillipsburg, NJ). Cells were fixed with 2% paraformaldehyde/1.5% glutaraldehyde in 0.1 M cacodylate, pH 7.4, for 1 hr, washed twice in 0.1 M cacodylate, pH 7.4, postfixed with freshly prepared 1% osmiumtetroxide (Polysciences) in 0.1 M cacodylate, pH 7.4, for 1 hr at 22°C, and dehydrated with ethanol. A 1:1 mixture of hexamethyldisilazane and ethanol then was added for 30 min, followed by 100% hexamethyldisilazane for 2 hr at room temperature. After complete evaporation of hexamethyldisilazane, specimens were visualized in a scanning electron microscope model JSM-5300 (JEOL) at 10 kV with tilt angles of 40–52°.
RESULTS
By screening random peptide displayed libraries, we recently identified several groups of peptides that interact with PS1 (27). We compared the sequences of these peptides with National Center for Biotechnology Information databases by using pblast alignment algorithms. The first group of peptides was characterized by a common consensus sequence (shown in bold): TPVLIAFVSSGSWPV, GVSSGGARPVGR, FVSSMDLZZIIRDSS, RPLRHLSGSSGE, VFHNLVLLSSGSDSS, FTSASSGSRFRSHLF, and GRQFVGVSLGSFGVL. The consensus sequence of these peptides shares homology with the sequence VSSGS, which is found in the extracellular domain of T cell surface glycoprotein CD44 (29). The consensus sequence of the next group of peptides (RPGVTGGSPSVDTSP, SEISAWSGGHPS, GNERSFAPWWFGGHA, and HRSTGGRASVPAS) was similar to the WTGGSP sequence found in the VDJ junction region of the T cell receptor β chain (30). Another peptide, TLIPRSFCPTHDRDC, was the most frequent peptide in our search and displayed the strongest binding to PS1 (28). This peptide showed a high homology with the sequence CATHPRD found in the T cell receptor β chain (31).
The observation of an in vitro interaction between PS1 and recombinant peptides whose sequences share homology with the CD44 and TCR suggested that PS1 may interact with CD44 and TCR in T cells. To verify this suggestion, we analyzed Jurkat cells (human leukemia T lymphocytes) that express both CD44 and TCR.
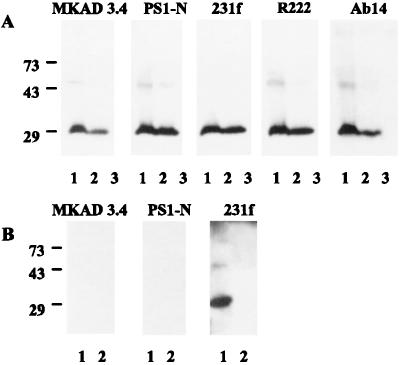
To determine the specificity of anti-PS1 antibodies, we performed Western blot analysis of endogenous PS1 of Jurkat cells and PS1 in HEp-2 cells transiently transfected with full-size PS1 cDNA (Fig. 1A). N-terminal antibodies that were used in this paper (231f, PS1-N, and MKAD 3.4) were compared with other well characterized N-terminal antibodies, AB14 (8) and R222. Cell extracts were incubated for 6 hr at room temperature in a loading buffer containing 0.1% SDS before electrophoresis. All tested antibodies revealed a very similar pattern of PS1 fragments: a strong staining of a 29-kDa N-terminal fragment and weak or no staining of 48- to 52-kDa full-length PS1 (Fig. 1A, lanes 1 and 2). No immunoreactive PS1 fragments were detected in naive HEp-2 cells (Fig. 1A, lane 3).
Figure 1.
Characterization of N-terminal polyclonal PS1 antibodies by Western blot analysis. (A) Western blot analysis of endogenous and overexpressed human PS1. Lanes: 1, cell extract of Jurkat cells; 2, cell extract of HEp-2 cells transfected with full-size PS1 cDNA (Vector pcDNA3; Invitrogen); 3, cell extract of untransfected HEp-2 cells. (B) Western blot analysis of endogenous PS1 of mouse brain. Lanes: 1, brain extract of wild-type PS1(+/+) mice antibodies; 2, brain extract of PS1 knockout (−/−) mice.
To verify further the specificity of N-terminal antibodies, we used brain extracts of wild-type PS1(+/+) mice and PS1 knockout (−/−) mice (Fig. 1B). Antibodies PS1-N and MKAD 3.4 were generated against synthetic peptides, corresponding to residues 27–44 and 45–48 of human PS1, which have no homology with mouse PS1 sequence. Therefore, PS1-N and MKAD 3.4 should not recognize mouse PS1 in brain extracts of PS1(+/+) mice. In contrast, antibody 231f, which is specific for residues 2–20, cross-reacts with both human and mouse PS1. This antibody recognized N-terminal PS1 fragment and full-length PS1 in brain extracts of wild-type PS1(+/+) mice and did not reveal PS1 immunoreactivity in brain extracts of PS1 knockout (−/−) mice (Fig. 1B).
For the analysis of interaction of PS1 with CD44 and TCR and colocalization studies, we used Jurkat and HEp-2, which highly express CD44 (32).
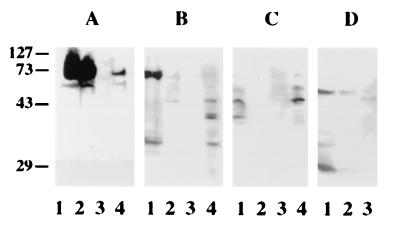
To determine whether PS1 interacts with CD44 and TCR, whole-cell extracts were incubated with recombinant PS1 immobilized through the N-terminal S tag to S protein agarose (27). Because “S-tag” was fused with PS1 through the thrombin-cleavable linker, complexes of cell proteins with PS1 were eluted selectively by thrombin cleavage. Western blot analysis of eluted proteins clearly showed that both CD44 and TCR bound recombinant PS1 (Fig. 2 A–C). Although CD44 was partially degraded in Jurkat cell extracts, the pattern of immunoreactivity was very similar in cell extracts and eluates (Fig. 2B). No CD44 or TCR immunoreactivity was detected in control experiments with S protein agarose columns that contained no PS1 (data not shown). In the second approach, CD44 or TCR was immunoprecipitated from extracts of Jurkat cells by using corresponding mAbs. Immunoprecipitates were analyzed by Western blotting with N-terminal PS1-N antibody. As shown in Fig. 2D, PS1 immunoreactivity was detected in CD44 and TCR immunoprecipitates from Jurkat cells. It should be noted that 28-kDa N-terminal fragment was detected in CD44 but not in TCR precipitates.
Figure 2.
Interaction of CD44 and TCR with PS1 in vitro. (A) HEp2 cells. Western blot analysis of cell proteins bound to immobilized recombinant PS1 with mAb to CD44 (HEp-2 cells). Lanes: 1, cell extract; 2, unbound fraction; 3, last wash; 4, elution with thrombin. (B) Jurkat cells. Western blot analysis of cell proteins bound to immobilized recombinant PS1 with mAb to CD44. Lanes: 1, cell extract; 2, unbound fraction; 3, last wash; 4, elution with thrombin. (C) Jurkat cells. Western blot analysis of cell proteins bound to immobilized recombinant PS1 with mAb to TCR. Lanes: 1, cell extract; 2, unbound fraction; 3, last wash; 4, elution with thrombin. (D) Detection of PS1 in CD44 and TCR immunoprecipitates from Jurkat cells. Western blot analysis with PS1-N antibody. Lanes: 1, cell extract; 2, immunoprecipitation with anti-CD44 antibody; 3, immunoprecipitation with anti-TCR antibody.
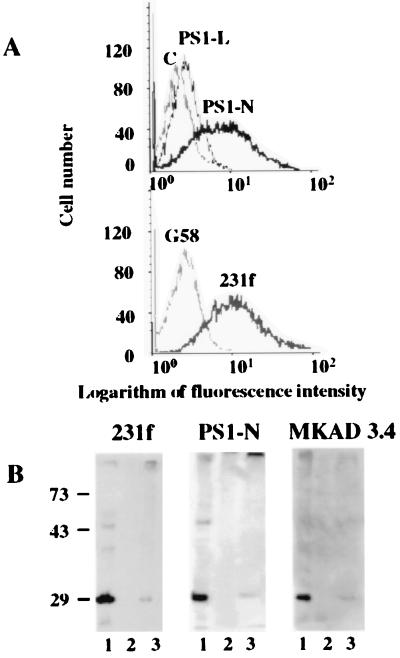
Our observation that PS1 forms complexes with CD44 and TCR in vitro (Fig. 2) suggests that PS1 may interact with these receptors intracellularly, on the cell surface, or both. To determine whether PS1 is present on the cell surface of Jurkat cells, we used flow-cytometric analysis. We found that anti-PS1 N-terminal antibodies bind to the surface of Jurkat cells when we compared the distributions of the immunofluorescence signals for PS1-N and 231f antibodies with the signals obtained with a mAb to the intracellular G58 Golgi protein or with preabsorbed PS1-N antibody (Fig. 3A). Loop antibody PS1-L revealed very weak binding to Jurkat cells (Fig. 3A). The percentage of positive cells stained with PS1-N or 231f antibody reached 40–50% of the total Jurkat cell population.
Figure 3.
Cell-surface expression of PS1 in Jurkat cells. (A) Flow cytometry analysis. Cells were incubated with anti-rabbit N-terminal PS1-N or loop PS1-L antibody to PS1 followed by staining with fluorescent anti-rabbit IgG immunoglobulins. The binding was monitored by flow cytometry. Control experiments were performed by using preabsorbed PS1-N antibody (C) or mouse mAb to the intracellular 58-kDa Golgi protein (G58). The graphs show one of three independent representative experiments. (B) Western blot analysis of biotinylated cell-surface proteins. Jurkat cells were treated with biotin and control Jurkat cells were not. Control and biotinylated cells were lysed and chromatographed on streptavidin-agarose, and eluates were analyzed by Western blotting with anti-PS1 N-terminal polyclonal antibodies 231f and PS1-N and mAb MKAD 3.4. Lanes: 1, whole Jurkat cell lysate; 2, control Jurkat cell proteins eluted from streptavidin-agarose; 3, biotinylated Jurkat cell proteins eluted from streptavidin-agarose.
The association of the N terminus of PS1 with the plasma membrane was examined further by biotin labeling of live Jurkat cells (Fig. 3B). Biotinylated and control Jurkat cells subsequently were lysed, and lysates were incubated with streptavidin-agarose. Biotinylated proteins were eluted from streptavidin-agarose by boiling resin in gel-loading buffer containing 2% SDS and 0.2 M DTT. Western blotting of eluted biotinylated proteins with N-terminal antibodies 231f, PS1-N, and mAb MKAD 3.4 revealed the N-terminal 28-kDa fragment of PS1 in the biotin-labeled sample (Fig. 3B, lane 3). No PS1 immunoreactivity was detected in the unlabeled control samples (Fig. 3B, lane 2).
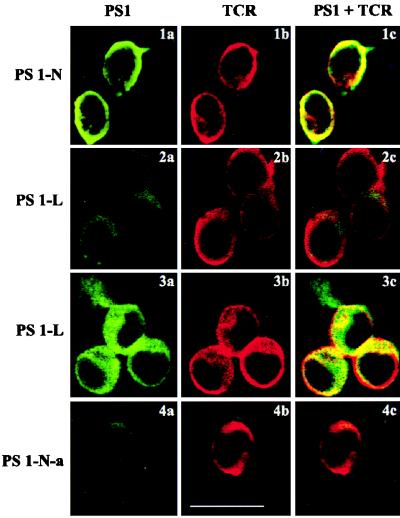
To localize PS1 in Jurkat cells, we used laser confocal microscopy. We observed partial colocalization of PS-N with TCR immunoreactivity on the plasma membrane of nonpermeabilized Jurkat cells (Fig. 4, 1a–1c). Preabsorbed PS1-N antibody (Fig. 4, 4a) and loop antibody PS1-L (Fig. 4, 2a) demonstrated weak surface staining of Jurkat cells. However, the staining of permeabilized cells with the PS1-L antibody was quite different. As seen in Fig. 4, 3a–3c, intense immunoreactivity was apparent inside the cells. Immunofluorescent staining was abundant in the cytoplasm and cell projections. The nuclei were not stained. Partial colocalization with TCR was observed on the plasma membrane, but only in the regions of cell–cell contacts (Fig. 4, 3c). TCR immunoreactivity in cell projections was not detected (Fig. 4, 3b). Partial colocalization of PS1-N and CD44 also was detected on the plasma membrane of nonpermeabilized and permeabilized Jurkat cells (data not shown).
Figure 4.
Immunolocalization of PS1 in Jurkat cells. PS1 (green) and TCR (red) were localized in Jurkat cells by confocal immunofluorescent microscopy. (1a–1c, 2a–2c, and 4a–4c) Nonpermeabilized cells. (3a–3c) Cells permeabilized by 0.5% Triton X-100. Cells were coincubated with mAb to TCR and PS-N (1a–1c), PS-L (2a–2c and 3a–3c), and preabsorbed antibody PS1-N-a (4a–4c). (Right) Colocalization of PS1 with TCR is shown (1c, 2c, 3c, and 4c). (Bar = 10 μm.)
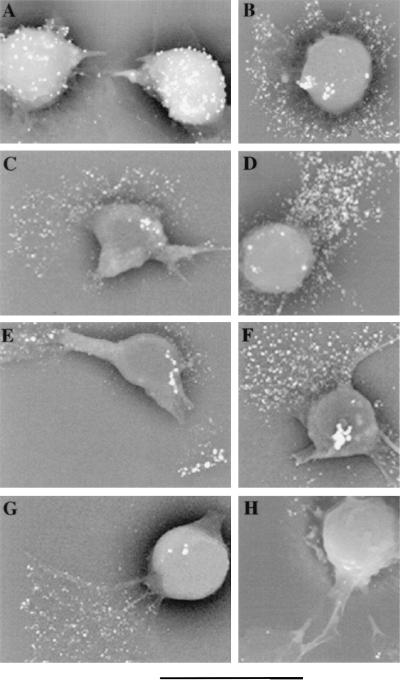
Overnight incubation of Jurkat cells on a collagen matrix induced cell polarization with the formation of cytoplasmic projections (uropods and lamellipodia). To assess directly the distribution of PS1 on the surface of adherent Jurkat cells, scanning immunoelectron microscopy was performed. Adhesion of Jurkat cells on a collagen matrix did not result in redistribution of TCR to lamellipodia (Fig. 5A). TCR immunoreactivity was detected on the surface of the cell body, whereas the surface of lamellipodia contained only a few immunogold particles. However, adhered cells demonstrated a significant concentration of CD44 immunoreactivity at the leading edge of lamellipodia and filopodia (Fig. 5B).
Figure 5.
Scanning immunoelectron micrographs of Jurkat cells adherent to a collagen matrix. After overnight adhesion to collagen, cells were incubated with mAb to TCR (A), CD44 (B), and PS1-N (C– G) and with affinity-purified rabbit polyclonal antibody to transthyretin (H). Bound antibodies were visualized with 5-nm gold-labeled antibody to mouse IgM (TCR), mouse IgG (CD44), and rabbit IgG to PS1-N and transthyretin followed by silver enhancing. (Bar = 11.258 μm; ×3,500.)
Immunogold staining of Jurkat cells with the N-terminal PS-N antibody revealed a redistribution of PS1 similar to the one observed for CD44. Adhered Jurkat cells contained only a few gold particles on the surface of the cell body, whereas a significant level of PS1 immunoreactivity was found on the surface of lamellipodia (Fig. 5 C–G). A similar redistribution of PS1 immunoreactivity in adhered Jurkat cells was observed with polyclonal N-terminal 231f antibody and N-terminal mAb MKAD 3.4 (data not shown). Control antibody to transthyretin showed very little immunostaining of the cell surface (Fig. 5H).
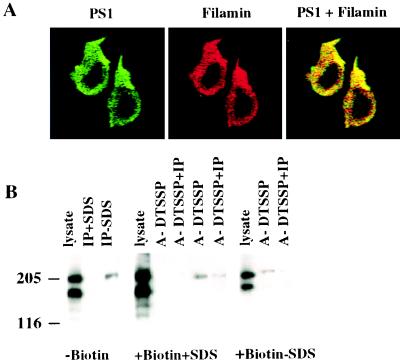
A recent report demonstrated that in vitro PS1 interacts with the actin-binding protein filamin (26). However, the biological relevance of such interaction remained unclear. In lymphocytes, filamin is localized to the lamellipodia and filopodia of spreading cells and links cytoskeleton to the plasma membrane by binding cell-surface receptors (33, 34). Therefore, our observation that PS1 redistributes to the surface of lamellipodia of adhered lymphocytes may suggest that filamin is involved in this process as a binding partner. To determine whether PS1 and filamin are colocalized in adhered, permeabilized Jurkat cells, we performed double-label immunofluorescence experiments by using laser confocal microscopy (Fig. 6A). PS1 and filamin exhibit partial colocalization intracellularly as seen by the presence of red, green, and yellow patterns. Fig. 6A clearly shows the highest staining intensity for both proteins at lamellipodia.
Figure 6.
Interaction of cell-surface PS1 with filamin in Jurkat cells. (A) Colocalization of PS1 and filamin in Jurkat cells. Jurkat cells were plated on chambered coverslips covered with saturated solution of rat tail collagen, type 1. Cells were fixed and permeabilized with 0.5% Triton X-100. PS1 (green) and filamin (red) were visualized in Jurkat cells by multiple optical sectioning (0.2 μ) by using confocal immunofluorescent microscopy. (×1,600.) Antibodies used were anti-N-terminal rabbit antibody (PS1-N) and antifilamin mAb PM 6/317. (B) Binding of cell-surface PS1 to filamin in Jurkat cells. Jurkat cells were treated with biotin (+Biotin) or were not treated (−Biotin). Cells were permeabilized by lysolecithin and treated with the reversible cross-linker (+DTSSP), whereas control cells were not treated with cross-linker (−DTSSP). The cells subsequently were lysed with or without 0.4% SDS (+SDS or −SDS), cell lysates were subjected to chromatography on immobilized monomeric avidin (A), and bound proteins were eluted with 2 mM d-biotin and immunoprecipitated with anti-PS1 N-terminal antibody 231f (IP). Immunoprecipitates were boiled for 10 min in Laemmli sample buffer containing 100 mM 2-mercaptoethanol, separated on a 6% SDS/PAGE gel, and Western blotted with antifilamin mAb PM 6/317. The molecular mass markers are shown on the left.
Because confocal microscopy provided only a descriptive view of the possible functional interaction of PS1 with filamin, we directly examined interaction of cell-surface biotinylated PS1 with intracellular filamin. First, we detected filamin immunoreactivity in PS1 immunoprecipitates from nonbiotinylated Jurkat cells lysed with RIPA buffer lacking SDS (Fig. 6B, “−Biotin”). However, there were very weak or no filamin bands in PS1 immunoprecipitates from Jurkat cells lysed with RIPA buffer containing 0.4% SDS (Fig. 6B, “−Biotin”). These results suggested that native filamin-PS1 complexes are sensitive to SDS treatment. It should be noted that PS1 coimmunoprecipitated with nondegraded filamin. To determine whether PS1 and filamin interact intracellularly, we performed cross-linking experiments. We first biotinylated Jurkat cells, then permeabilized biotinylated cells by lysolecithin, and, finally, treated cells with the reversible, homobifunctional cross-linker, dithiobis(sulfosuccinimidylpropionate) (DTSSP). The cells subsequently were lysed. Biotinylated proteins were isolated on columns with immobilized monomeric avidin and immunoprecipitated with N-terminal PS1 antibody 231f. Fig. 6B (“+Biotin+SDS”) shows that filamin forms SDS-resistant complexes with biotinylated surface proteins only after treatment with chemical cross-linker DTSSP (Fig. 6B, lane A+DTSSP). Although most complexes probably represent biotinylated β2-integrin, which is linked to filamin (34), we also detected filamin in PS1 immunoprecipitates from extracts of biotinylated cells treated with DTSSP (Fig. 6B, lane A+DTSSP+IP). There was no filamin immunoreactivity in the control samples that were not treated with DTSSP (Fig. 6B, lanes A-DTSSP and A-DTSSP+IP). Complexes of filamin with biotinylated PS1 also were detected in cell lysates that were not treated by SDS and DTSSP (Fig. 6B, “+Biotin-SDS,” lane A-DTSSP+IP).
DISCUSSION
To date, more than 40 mutations in the PS1 gene have been reported (1, 4–6). However, the normal functions of presenilins as well as the nature of the dysfunction caused by these mutations are unknown. We show that PS1, which is highly expressed in cultured Jurkat cells, redistributes to lamellipodia upon adhesion of these cells to a collagen matrix such as other proteins that mediate cell adhesion and cell movement (33, 35). Thus, redistribution of PS1 to the surface of lamellipodia suggests that cell-surface PS1 may be involved in cell adhesion of Jurkat cells. Additional evidence for this suggestion was obtained by analysis of the interaction of PS1 with filamin. Zhang et al. (26) reported that the hydrophilic loop of PS1 interacts with the actin-binding protein filamin (ABP-280) in vitro. However, the biological relevance of such interaction was not clear. Filamin is a member of a family of actin-binding proteins that link actin filaments with adhesion molecules on the cell surface (33). In leukocytes, filamin binds the cytoplasmic tail of β2-integrin and, through this interaction, mediates the extension of lamellipodia at the leading edge (34). We found that both PS1 and filamin are concentrated at lamellipodia of adhered Jurkat cells, and cell-surface PS1 interacts with filamin. Thus, our results suggest that PS1, which is expressed at the surface of Jurkat cells, is anchored to the cytoskeleton by filamin. It was shown that PS1 may interact with other actin-binding proteins, catenins (25).
Localization of PS1 to the plasma membrane of Jurkat cells raises an interesting question about the membrane topology of PS1. Although the N terminus and the hydrophilic loop region of PS1 were reported to be oriented toward the cytoplasm in the intracellular endoplasmic reticulum–Golgi pool (36, 37), our results show that the topology of PS1 may be different at the plasma membrane. Furthermore, we found the highest level of N-terminal PS1 immunoreactivity at the surface of lamellipodia that lack endoplasmic reticulum. Therefore, our results suggest that in Jurkat cells, PS1 may exhibit multiple topological membrane orientations. More than one topological membrane orientation has been reported previously for other proteins: prion protein, microsomal epoxide hydrolase, cytochrome P450s, P-glycoprotein, and ductin (38, 41).
The expression of PS1 at the surface of lamellipodia and filopodia of Jurkat cells is of considerable interest because similar structures are implicated in neuronal growth and directional neuronal movement (33, 42). PS1 was detected recently in neuritic processes and filopodia-like structures of growth cones in a primary culture of rat hippocampal neurons (43, 44). However, in this work, surface localization of PS1 was not studied. Our results indicate that cell-surface expression of PS1 in neurons and its possible role in neuronal growth and synaptic plasticity is the next logical step of investigation.
The possibility that PS1 may be involved in cell adhesion is especially compelling because cell-adhesion molecules may play a role in memory formation. Recently, Grotewiel et al. (45) identified a new α-integrin, Volado, which is required for short-term memory processes in Drosophila. Conceivably, failure of normal adhesion function of PS1 would result in the memory deficit observed in AD.
Acknowledgments
We thank Mr. Gregory Rudamen and Mr. David Colflesh for their excellent technical assistance with scanning electron microscopy and laser confocal microscopy. This work was supported, in part, by Alzheimer’s Association Grant IIRG-94-118 (to A.L.S.), National Institutes of Health Grant R01 AG13706 (to D.G.), and a Long Island Alzheimer Foundation grant (to D.G.).
ABBREVIATIONS
- FAD
familial Alzheimer’s disease
- PS1 and PS2
presenilin 1 and 2
- TCR
T cell receptor
- PS1-N
N-terminal antibody to PS1
- PS1-L
antibody to loop region of PS1
- DTSSP
dithiobis(sulfosuccinimidylpropionate)
References
- 1.Sherington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–759. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 2.Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Oshima J, Pettingel W H, Yu C-E, Jondro P D, Schmidt S D, Wang K, et al. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 3.Rogaev E I, Sherington R Y, Rogaeva E A, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, et al. Nature (London) 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 4.Campion D, Flaman J-M, Brice A, Hannequin D, Dubois B, Martin C, Moreau V, Charbonnier F, Didierjean O, Tardieu S, et al. Hum Mol Gen. 1995;4:2373–2377. doi: 10.1093/hmg/4.12.2373. [DOI] [PubMed] [Google Scholar]
- 5.Van Broeckhoven C. Nat Genet. 1995;11:230–232. doi: 10.1038/ng1195-230. [DOI] [PubMed] [Google Scholar]
- 6.Hutton M, Hardy J. Hum Mol Genet. 1997;6:1639–1646. doi: 10.1093/hmg/6.10.1639. [DOI] [PubMed] [Google Scholar]
- 7.Cribbs D, Chen L, Bende S M, LaFerla F M. Am J Pathol. 1996;148:1797–1806. [PMC free article] [PubMed] [Google Scholar]
- 8.Lee M K, Slunt H H, Martin L J, Thinakaran G, Kim G, Gandy S E, Seeger M, Koo E, Price D L, Sisodia S S. J Neurosci. 1996;16:7513–7525. doi: 10.1523/JNEUROSCI.16-23-07513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacs D M, Fausett H J, Page K J, Kim T W, Moir R D, Merriam D E, Hollister R D, Hallmark O G, Mancini R, Felsenstein K M, et al. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 10.Cook D G, Sung J C, Golde T E, Felsenstein K M, Wojczyk B S, Tanzi R E, Trojanowski J Q, Lee V M, Doms R W. Proc Natl Acad Sci USA. 1996;93:9223–9228. doi: 10.1073/pnas.93.17.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lah J J, Heilman C J, Nash N R, Rees H D, Yi H, Counts S E, Levey A. J Neurosci. 1997;17:1971–1980. doi: 10.1523/JNEUROSCI.17-06-01971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber L L, Leissring M A, Yang A J, Glabe C G, Cribbs D H, LaFerla F M. Exp Neurol. 1997;143:37–44. doi: 10.1006/exnr.1996.6348. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Xu M, Zhou H, Ma J, Potter H. Cell. 1997;90:917–927. doi: 10.1016/s0092-8674(00)80356-6. [DOI] [PubMed] [Google Scholar]
- 14.Takashima A, Sato M, Mercken M, Tanaka S, Kondo S, Honda T, Sato K, Murayama M, Noguchi K, Nakazato Y, et al. Biochem Biophys Res Commun. 1996;227:423–426. doi: 10.1006/bbrc.1996.1523. [DOI] [PubMed] [Google Scholar]
- 15.Devji N N, Singer S J. Proc Natl Acad Sci USA. 1996;93:12575–12580. doi: 10.1073/pnas.93.22.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devji N N, Singer S J. Proc Natl Acad Sci USA. 1997;94:9926–9931. doi: 10.1073/pnas.94.18.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitan D, Doyle T G, Brousseau D, Lee M K, Thinakaran G, Slunt H H, Sisodia S S, Greenwald I. Proc Natl Acad Sci USA. 1996;93:14940–14944. doi: 10.1073/pnas.93.25.14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Von Boehmer H. Curr Biol. 1997;7:R308–R310. doi: 10.1016/s0960-9822(06)00145-x. [DOI] [PubMed] [Google Scholar]
- 19.Weinmaster G. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- 20.Wong P C, Zheng H, Chen H, Becher M W, Sirinathsinghji D J S, Trumbauer M E, Chen H Y, Price D L, Van der Ploeg L H T, Sisodia S S. Nature (London) 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 21.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 22.Vito P, Lacana E, D’Adamio L. Science. 1996;271:521–525. doi: 10.1126/science.271.5248.521. [DOI] [PubMed] [Google Scholar]
- 23.Weidemann A, Paliga K, Durrwang U, Czech C, Evin G, Masters C L, Beyreuther K. Nat Med. 1997;3:328–332. doi: 10.1038/nm0397-328. [DOI] [PubMed] [Google Scholar]
- 24.Xia W, Zhang J, Perez R, Koo E, Selkoe D J. Proc Natl Acad Sci USA. 1997;94:8208–8213. doi: 10.1073/pnas.94.15.8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Liyanage U, Medina M, Ho C, Simmons A D, Lovett M, Kosik K S. NeuroReport. 1997;8:1489–1494. doi: 10.1097/00001756-199704140-00033. [DOI] [PubMed] [Google Scholar]
- 26.Zhang V, Han S-V, McKeel D, Goate A, Wu J Y. J Neurosci. 1998;18:914–922. doi: 10.1523/JNEUROSCI.18-03-00914.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarzman A, Tsiper M, Vitek A, St. George-Hyslop P, Goldgaber D. In: Progress in Alzheimer’s and Parkinson’s Disease. Fisher A, Yorshida M, Hanin I, editors. New York: Plenum; 1998. pp. 141–148. [Google Scholar]
- 28.Altin J G, Pagler E B. Anal Biochem. 1995;224:382–389. doi: 10.1006/abio.1995.1054. [DOI] [PubMed] [Google Scholar]
- 29.Aruffo A, Stamenkovic I, Melnick M, Underhill C B, Seed B. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 30.Desai-Mehta A, Mao C, Rajagopalan S, Robinson T, Datta S K. J Clin Invest. 1995;95:531–541. doi: 10.1172/JCI117695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howell M D, Diveley J P, Lundeen K A, Esty A, Winters S T, Carlo D J, Brostoff S W. Proc Natl Acad Sci USA. 1991;88:10921–10925. doi: 10.1073/pnas.88.23.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesley J, Hyman R, Kincade P W. Adv Immunol. 1993;54:271–335. doi: 10.1016/s0065-2776(08)60537-4. [DOI] [PubMed] [Google Scholar]
- 33.Matsudaira P. Semin Cell Biol. 1994;5:165–174. doi: 10.1006/scel.1994.1021. [DOI] [PubMed] [Google Scholar]
- 34.Sharma C P, Ezzel R M, Arnaout A. J Immunol. 1995;154:3461–3470. [PubMed] [Google Scholar]
- 35.del Pozo M A, Cabanas C, Montoya M C, Ager A, Sanchez-Mateos P, Sanchez-Madrid F. J Cell Biol. 1997;137:493–508. doi: 10.1083/jcb.137.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doan A, Thinakaran G, Borchelt D R, Slunt H H, Ratovitsky T, Podlisny M, Selkoe D J, Seeger M, Gandy S E, Price D L, et al. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Greenwald I. Neuron. 1996;17:1015–1021. doi: 10.1016/s0896-6273(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 38.De Fea K A, Nakahara D H, Calayag M C, Yost C S, Mirels L F, Prusiner S B, Lingappa V R. J Biol Chem. 1994;269:16810–16820. [PubMed] [Google Scholar]
- 39.Wu D, Cederbaum Hepatology. 1992;15:515–524. doi: 10.1002/hep.1840150326. [DOI] [PubMed] [Google Scholar]
- 40.Von Dippe P, Amoui M, Stellwagen R H, Levy D. J Biol Chem. 1996;271:18176–18180. doi: 10.1074/jbc.271.30.18176. [DOI] [PubMed] [Google Scholar]
- 41.Levy D. Essays Biochem. 1996;31:49–60. [PubMed] [Google Scholar]
- 42.Goodman C S. Annu Rev Neurosci. 1996;19:341–377. doi: 10.1146/annurev.ne.19.030196.002013. [DOI] [PubMed] [Google Scholar]
- 43.Elder G A, Tezapsidis N, Carter J, Shioi J, Bouras C, Li H-C, Johnston J M, Efthimiopoulos S, Friedrich V L, Jr, Robakis N K. J Neurosci Res. 1996;45:308–320. doi: 10.1002/(SICI)1097-4547(19960801)45:3<308::AID-JNR13>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 44.Busciglio J, Hartmann H, Lorenzo A, Wong C, Baumann K, Sommer B, Staufenbiel M, Yankner B A. J Neurosci. 1997;17:5101–5107. doi: 10.1523/JNEUROSCI.17-13-05101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grotewiel M S, Beck C D O, Wu K H, Zhu X-R, Davis R L. Nature (London) 1998;391:455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]