Abstract
Mitosis is triggered in vertebrate cells by the cyclin B1–Cdc2 complex. The activation of this complex at the end of G2 phase is accompanied by its translocation from the cytoplasm to the nucleus. We used digitonin-permeabilized human cells to analyze the mechanism by which cyclin B1–Cdc2 is imported into the nucleus. Cyclin B1–Cdc2 import was not blocked by inhibitors of the importin α-dependent import pathway or by dominant negative versions of the GTPase Ran or importin β. However, the rate of cyclin B1 import was decreased by immunodepletion of importin β from cytosol. Purified importin β promoted cyclin B1 import in the absence of cytosol or Ran and in the presence of the dominant negative Ran mutant. We conclude that cyclin B1 import is mediated by an unusual importin β-dependent mechanism that does not require Ran.
Keywords: cell cycle, cyclin-dependent kinases
The initiation of mitosis in vertebrate cells is triggered by the cyclin-dependent protein kinase Cdc2 (Cdk1; for reviews, see refs. 1–5). The activation of Cdc2 begins with the binding of cyclin B1, whose levels gradually increase during S and G2 phases. The cyclin B1–Cdc2 complex is held in an inactive state before mitosis by inhibitory phosphorylation of Cdc2 at Thr-14 and Tyr-15. At the end of G2 phase, these residues are abruptly dephosphorylated by the phosphatase Cdc25, and the active cyclin B1–Cdc2 complex is then competent to initiate the events of mitosis.
The timing of mitosis is also influenced by changes in cyclin B1–Cdc2 localization. Cyclin B1 is located in the cytoplasm during S and G2 phases and then rapidly accumulates in the nucleus at the onset of prophase (6–10). The translocation of cyclin B1 to the nucleus appears to be required for the initiation of mitosis. In Xenopus, the mitosis-promoting activity of cyclin B1 is blocked by mutations that prevent its nuclear localization (11), whereas mutations that lead to constitutive nuclear localization of cyclin B1 are sufficient to induce premature mitotic events under some conditions (11–13).
The region of cyclin B1 responsible for its cytoplasmic localization during interphase has been mapped to a region encompassing amino acids 88–154; deletion of this cytoplasmic retention signal (CRS) causes cyclin B1 to localize to the nucleus throughout the cell cycle (14). The CRS may act in part by anchoring cyclin B1 to cytoplasmic structures such as microtubules (15). Recent evidence also suggests that the CRS region contains a leucine-rich nuclear export signal that mediates active nuclear export of cyclin B1 by a mechanism that is sensitive to the drug leptomycin B and involves the export receptor CRM1/exportin 1 (13, 16, 17). The accumulation of cyclin B1 in the nucleus at prophase may result from inactivation of export due to the phosphorylation of multiple serine residues flanking the leucine-rich nuclear export signal (11, 17).
Transport of proteins into the nucleus is mediated by soluble receptor proteins that interact with nuclear cargo and carry it through the pore (for reviews, see refs. 18–21). Proteins carrying the classical nuclear localization signal (NLS) bind to a heterodimeric receptor composed of importin α (p62, karyopherin-α, or Srp1α) and importin β (p97 or karyopherin β). This complex is translocated through the nuclear pore and disassembled in the nucleus by mechanisms that requires the small GTPase Ran (TC4) and an accessory protein NTF2 (p10).
Recent studies have led to the identification of alternative import mechanisms that involve importin β-related proteins; for example, the heterogeneous nuclear ribonucleoprotein A1 contains an M9 import signal that interacts with the importin β-related receptor transportin (22). Several additional importin β-related proteins have recently been identified and have been implicated not only in nuclear import but also in export (20, 23, 24). A dominant negative mutant form of importin β can block several import and export pathways (25, 26), suggesting that these pathways involve shared components.
Nucleocytoplasmic transport appears to be governed by differences in the localization of key regulators of the Ran GTPase. The Ran guanine nucleotide exchange factor RCC1 is found in the nucleus, whereas the GTPase-activating protein RanGAP1 is in the cytoplasm; as a result, the concentration of RanGTP is predicted to be higher in the nucleus. This asymmetric distribution of RanGTP may provide the basis for the directionality of nucleocytoplasmic transport (for reviews, see refs. 18, 20, and 27–29). Nuclear export requires the association of RanGTP with exportin–cargo complexes, which are transported out of the nucleus and then dissociate when GTP hydrolysis in the cytoplasm converts Ran to the GDP form. Similarly, the directionality of nuclear import may be driven by nuclear RanGTP, which dissociates importin–cargo complexes after their translocation through the nuclear pore.
The mechanism by which the cyclin B1–Cdc2 complex is imported into the nucleus is not understood. Neither cyclin B1 nor Cdc2 contain consensus NLS or M9 import sequences. In the present work, we used a cell-free nuclear import assay to study the mechanism of cyclin B1 import in mammalian cells. Our results suggest that cyclin B1 is imported by an unusual mechanism that requires importin β but does not require importin α or Ran.
MATERIALS AND METHODS
Construction of Recombinant Baculoviruses.
Baculoviruses encoding Cdc2 and Cdc2K− have been described (30); Cdc2K− is a mutant form of Cdc2 in which Lys-33 and Lys-34 (in the ATP-binding site) are changed to Met and Ile, respectively. Baculoviruses encoding cyclin B1 were created by using the following plasmids according to the Bac-to-Bac protocol (GIBCO/BRL). HMCycB1(pFastBac) was constructed by placing six His residues followed by four copies of a Myc epitope tag sequence (EQKLISEEDLN) at the N terminus of the human cyclin B1 cDNA by using the NcoI site at the start codon. Cyclin B1 derivatives were created by oligonucleotide-mediated mutagenesis with uracil-containing template DNA (31) derived from HMCycB1(pFastBac). To create HMCycB1E, Ser-126, -128, -133, and -147 were replaced with Glu residues. HMCycB1A contains Ala substitutions at the same residues. HMCycB1(ΔCRS) was constructed by creating AatII sites at amino acids 88 and 154 (CCTATGCTGGTG changed to CCTGACGTCGTG and AATGATGTGGAT changed to AATGACGTCGAT). The resulting DNA was digested with AatII and ligated to fuse amino acid 87 in-frame to amino acid 155. Cyclin B1 lacking the Myc tag was created by digestion of HMCycB1(pFastBac) with NcoI, followed by ligation.
Protein Expression.
NLS-tagged BSA, recombinant IBB domain, Ran Q69L, importin β, and importin β-(71–876) were prepared as described (25, 32). Cyclin B1 and Cdc2 were expressed in Sf9 cells and lysates were prepared as described (30). Cdc2 was purified as described (30), except that the Superose 12 column was omitted. Cyclin B1 lysates were prepared and supplemented to 300 mM NaCl after Dounce homogenization. The 100,000 × g supernatant was loaded onto a HiTrap chelating column (Pharmacia) loaded with cobalt and equilibrated in buffer A (25 mM Hepes⋅NaOH, pH 7.4/300 mM NaCl/10% glycerol/0.1 mM PMSF). The column was washed in buffer B (25 mM Hepes⋅NaOH, pH 6/300 mM NaCl/10% glycerol/0.1 mM PMSF), and cyclin B1 was eluted with a linear gradient of imidazole (0 to 200 mM) in buffer A. Fractions containing cyclin B1 were pooled and dialyzed against buffer A (containing 1 mM DTT) to remove imidazole. All purified cyclin B1 proteins were fully functional as activators of Cdc2 in vitro.
Nuclear Import Assay.
Nuclear import reactions were performed as described (33). Briefly, cells were grown on poly(l-lysine)-coated glass coverslips for 24 to 48 h. Cells were washed twice in transport buffer [20 mM Hepes⋅KOH, pH 7.3/110 mM KOAc/2 mM Mg(OAc)2/1 mM EGTA/2 mM DTT/aprotinin (1 μg/ml)/pepstatin A (1 μg/ml)] and then treated with digitonin (40 μg/ml) for 6 min on ice. After permeabilization, the cells were washed twice with transport buffer and inverted on top of 15 μl of import reaction in transport buffer containing 300 nM import substrate, cytosol (3 mg/ml), and an ATP-regenerating system [1 mM ATP/5 mM creatine phosphate/creatine kinase (10 units/ml)] and incubated at 30°C for 30 min. Cells were washed twice in transport buffer, fixed in 3.7% paraformaldehyde for 20 min at room temperature, and permeabilized with 0.1% Triton X-100 in PBS. Cells were then blocked in PBS/3% BSA for 10 min at room temperature and incubated with primary antibody for 1 h at room temperature [1:500 dilution of anti-Myc 9E10 monoclonal antibody (Babco, Richmond, CA) or 1:10 dilution of anti-cyclin B1 monoclonal antibody in PBS/3% BSA]. Cells were then washed in PBS, incubated in secondary antibody [1:100 dilution of FITC-conjugated anti-mouse (Boehringer Mannheim) in PBS/3% BSA] for 30 min at room temperature, washed with PBS, stained with Hoechst 33258 at 50 ng/ml, mounted in 90% glycerol containing o-phenylenediamine, and examined with a Nikon fluorescence microscope.
To prepare interphase cytosol, asynchronous HeLa S3 cells were cultured in DMEM plus 10% FCS, harvested with a rubber policeman, and washed twice in PBS. The cells were washed once in cell wash buffer [10 mM Hepes⋅KOH, pH 7.3/110 mM KOAc/2 mM Mg(OAc)2/2 mM DTT] and resuspended in an equal volume of lysis buffer [5 mM Hepes⋅KOH, pH 7.3/10 mM KOAc/2 mM Mg(OAc)2/2 mM DTT/1 mM PMSF/aprotinin (1 μg/ml)/leupeptin (1 μg/ml)/pepstatin A (1 μg/ml)]. Cells were incubated on ice for 10 min and lysed by Dounce homogenization. The homogenate was clarified by centrifugation (100,000 × g, 1 h, 4°C), dialyzed extensively against transport buffer, and stored at −80°C.
To prepare mitotic HeLa cells, cells were arrested in S phase by treatment with 2 mM thymidine for 17 h and then released by washing with PBS. Cells were grown in fresh medium for 9 h and then blocked with a second thymidine treatment for 14 h. Cells were released from the thymidine block by washing in PBS followed by addition of fresh medium. After a 6-h release, the cells were arrested in mitosis by the addition of nocodazole (200 ng/ml) for approximately 16 h. Cell lysates were prepared as described above.
To deplete importin β from cytosol, mitotic cytosol was incubated with monoclonal anti-importin β (mAb3E9, Affinity BioReagents, Neshanic Station, NJ) for 1.5 h at 4°C. Protein A-Sepharose (Sigma) was added to the cytosol for an additional 2 h at 4°C and removed by centrifugation.
RESULTS
Nuclear Accumulation of Cyclin B1 Requires Cdc2-Dependent Phosphorylation.
We used permeabilized cells to dissect the mechanisms controlling cyclin B1 localization (33). HeLa cells growing on coverslips were treated with the detergent digitonin, which perforates the plasma membrane while leaving the nuclear envelope intact and competent for active transport. Digitonin-permeabilized cells were incubated for 30 min with purified human cyclin B1 tagged with an N-terminal Myc epitope. Cells were then fixed and analyzed by secondary immunofluorescence with antibodies against the Myc epitope.
In preliminary studies, we found that purified cyclin B1 or purified cyclin B1–Cdc2 complexes rapidly accumulated in the nuclei of permeabilized cells incubated with ATP and crude interphase cytosol (data not shown). Mitotic cytosol did not increase the amount of cyclin B1 in the nucleus. We therefore suspected that the addition of cyclin B1 or cyclin B1–Cdc2 was generating high levels of Cdc2 activity in interphase cytosol, resulting in a mitosis-like state that triggered nuclear accumulation of cyclin B1. To test this possibility, we analyzed the nuclear accumulation of complexes of cyclin B1 and Cdc2K−, a version of Cdc2 whose activity is abolished by mutations in the ATP binding site. The cyclin B1–Cdc2K− complex did not accumulate in the nucleus in the presence of interphase cytosol (Fig. 1A) but did accumulate in the presence of mitotic cytosol (Fig. 1B). Addition of an untagged active cyclin B1–Cdc2 complex to interphase cytosol allowed nuclear accumulation of the tagged cyclin B1–Cdc2K− complex (data not shown). We conclude that Cdc2 activity promotes nuclear cyclin B1 accumulation in this system.
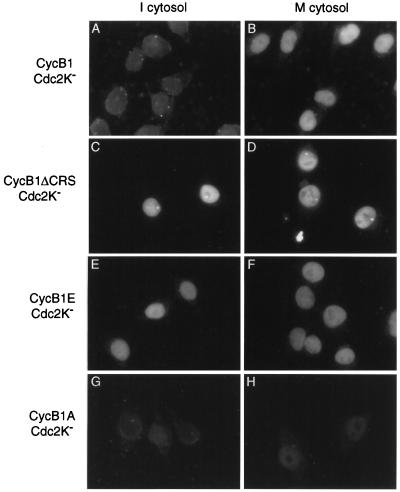
Figure 1.
Mitosis-dependent nuclear accumulation of cyclin B1–Cdc2 requires phosphorylation sites in the CRS. Digitonin-permeabilized HeLa cells were incubated for 30 min with preformed complexes of Myc-epitope-tagged cyclin B1 and kinase-deficient Cdc2K− (300 nM), plus either interphase cytosol (A, C, E, and G) or mitotic cytosol (B, D, F, and H). Complexes contained either wild-type cyclin B1 (A and B), a deletion mutant lacking the CRS domain (B1ΔCRS) (C and D), a mutant in which four Ser residues in the CRS are changed to Glu (B1E) (E and F), or a mutant in which these residues are changed to Ala (B1A) (G and H). Cells were fixed and analyzed by secondary immunofluorescence with antibodies against the Myc epitope tag.
We also tested whether the phosphorylation of cyclin B1 was required for nuclear accumulation in our system. The CRS of human cyclin B1 contains four Ser residues in positions that correspond to the Ser residues known to be phosphorylated in the CRS of Xenopus cyclin B1 (11, 34). We prepared mutant versions of human cyclin B1 in which these residues were changed either to Glu (cyclin B1E) or Ala (cyclin B1A). In addition, we prepared a cyclin B1 mutant lacking the entire CRS region (amino acids 88–154) (cyclin B1ΔCRS). Each form of cyclin B1 was purified from lysates of baculovirus-infected insect cells and prebound to purified Cdc2K−. The cyclin B1ΔCRS–Cdc2K− complex localized to the nucleus in the presence of interphase or mitotic cytosol (Fig. 1 C and D), confirming that the CRS promotes cytoplasmic localization in vitro as it does in vivo. Similarly, complexes of Cdc2K− and the cyclin B1E mutant accumulated in the nucleus in the presence of interphase cytosol (Fig. 1E). Nuclear accumulation of cyclin B1A–Cdc2K− was minimal in the presence of mitotic cytosol (Fig. 1H). These results are consistent with recent evidence from Xenopus oocytes that phosphorylation of serine residues in the CRS promotes nuclear accumulation by inhibiting the leucine-rich nuclear export signal in this region (17). Thus, the net nuclear accumulation of cyclin B1 in our system, like that in vivo, appears to be determined by the interplay of nuclear import, export, and Cdc2-dependent phosphorylation of the CRS.
Cyclin B1–Cdc2 Does Not Enter the Nucleus by the NLS Receptor Pathway.
Having established the basic features of our cyclin import system, we next proceeded to the key question in our work: What is the mechanism by which cyclin B1–Cdc2 complexes are transported into the nucleus? For these experiments, we analyzed both the wild-type cyclin B1–Cdc2 complex and the cyclin B1E–Cdc2K− complex, which allowed us to focus on the import process in the absence of export and the requirement for CRS phosphorylation.
To investigate whether the nuclear import of cyclin B1 is mediated by the classical NLS-dependent pathway, we tested cyclin B1 import in the presence of a peptide carrying an NLS sequence, which prevents binding of NLS-containing proteins to their receptor, importin α. We also tested the effects of adding the IBB domain of importin α, which binds the C terminus of importin β and prevents its interaction with importin α (32, 35). Both inhibitors effectively blocked nuclear import of fluorescently labeled BSA conjugated to multiple copies of an NLS peptide (Fig. 2 G–I). However, import of cyclin B1E–Cdc2K− or wild-type cyclin B1–Cdc2 was not inhibited by the NLS peptide or the IBB domain (Fig. 2 A–F), indicating that cyclin B1 does not enter the nucleus by the NLS/importin α-mediated transport pathway.
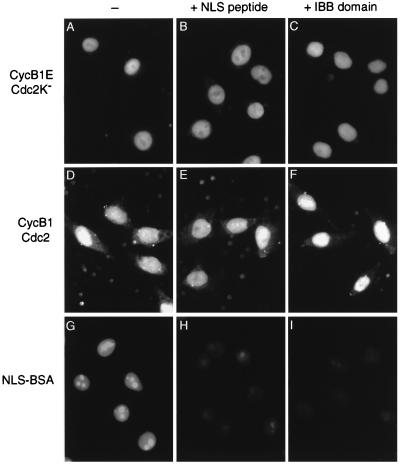
Figure 2.
Nuclear import of cyclin B1–Cdc2 does not occur by the NLS-dependent pathway. Nuclear import reactions were performed with interphase cytosol and cyclin B1E–Cdc2K− (A–C), wild-type cyclin B1–Cdc2 (D–F), or FITC-labeled NLS-BSA (G–I), in the absence of inhibitors (A, D, and G) or in the presence of 100 μM NLS peptide (B, E, and H) or 2 μM IBB domain (C, F, and I). Cells were analyzed by direct immunofluorescence (G–I) or were fixed and analyzed by secondary immunofluorescence with antibodies against the Myc epitope tag (A–F).
Cyclin B1 Nuclear Import Is Not Blocked by Ran Q69L or a Dominant Negative Version of Importin β.
Import of most nuclear cargo is blocked by the addition of Ran Q69L, a version of Ran that is unable to hydrolyze GTP and is, therefore, predominantly in the GTP-bound state. This protein is thought to inhibit import by causing premature dissociation of import complexes. To test the role of Ran in cyclin B1 import, we analyzed the import of cyclin B1 by permeabilized cells in the presence of Ran Q69L. The import of NLS-labeled BSA was blocked, but there was no effect on the nuclear import of cyclin B1E–Cdc2K− or wild-type cyclin B1–Cdc2 (Fig. 3 C, F, and I). Similarly, import of cyclin B1–Cdc2 was not blocked by guanosine 5′-[γ-thio]triphosphate, a nonhydrolyzable analog of GTP (Fig. 3 B, E, and H). We conclude that the import of cyclin B1–Cdc2, unlike that of most previously described nuclear proteins, is not inhibited by RanGTP.
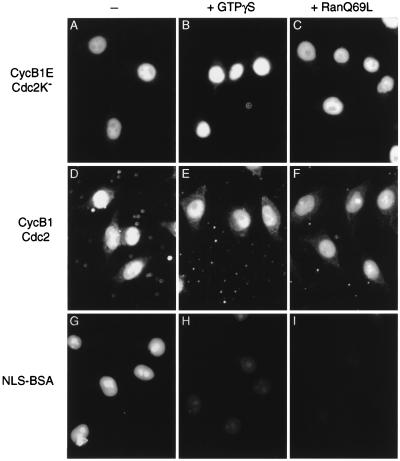
Figure 3.
Nuclear import of cyclin B1–Cdc2 is not blocked by a GTPase-deficient Ran mutant. Nuclear import reactions were performed with interphase cytosol and cyclin B1E–Cdc2K− (A–C), wild-type cyclin B1–Cdc2 (D–F), or FITC-NLS-BSA (G–I), in the absence of inhibitors (A, D, and G) or in the presence of 0.2 mM guanosine 5′-[γ-thio]triphosphate (B, E, and H) or 2 μM Ran Q69L (C, F, and I). Cells were analyzed by direct immunofluorescence (G–I) or were fixed and analyzed by secondary immunofluorescence with antibodies against the Myc epitope tag (A–F).
The concentration of Ran Q69L used in these experiments (2 μM) has been shown to inhibit import of several proteins in other laboratories (36–38). We also failed to detect inhibition of cyclin import in the presence of 8 μM Ran Q69L (data not shown).
We next tested the effects of adding an N-terminally truncated version of importin β [importin β-(71–876)] that lacks the ability to bind Ran and has been shown to block several nuclear import and export pathways (25, 26). The dominant negative importin β, at a concentration of 2 μM, effectively inhibited import of NLS-BSA, resulting in nuclear rim staining as observed previously (Fig. 4F). However, dominant negative importin β did not inhibit import of cyclin B1E–Cdc2K− or wild-type cyclin B1–Cdc2 (Fig. 4 B and D). Higher concentrations of the inhibitor (8 μM) also failed to block cyclin B1 import (data not shown).
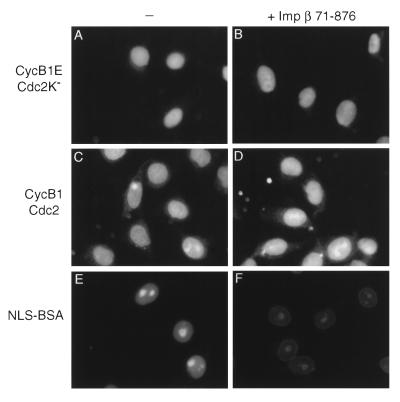
Figure 4.
Nuclear import of cyclin B1–Cdc2 is not blocked by dominant negative importin β. Nuclear import reactions were performed with interphase cytosol and cyclin B1E–Cdc2K− (A and B), wild-type cyclin B1–Cdc2 (C and D), or FITC-NLS-BSA (E and F), in the absence of inhibitor (A, C, and E) or in the presence of 2 μM importin β-(71–876) (B, D, and F). Because importin β-(71–876) is Myc-tagged, cyclin B1 in this experiment was detected with monoclonal anti-cyclin B1 antibodies. Endogenous cyclin B1 is not present at significant levels in the permeabilized cells and cytosol used in these experiments (data not shown).
Nuclear Import of Cyclin B1 Is an Energy-Dependent Process That Requires Importin β.
In further studies of the cyclin B1 import mechanism, we found that import of cyclin B1E–Cdc2K− complexes required the presence of crude cytosol and was prevented by depletion of ATP, incubation on ice, or addition of the lectin wheat germ agglutinin (which binds to glycosylated residues present on many nucleoporins; data not shown). We also found that a 16-fold excess of untagged cyclin B1 (4 μM) completely inhibited the nuclear accumulation of Myc-epitope-tagged cyclin B1–Cdc2 complexes (data not shown). These results indicate that cyclin B1 import occurs by an active, saturable, and cytosol-dependent process.
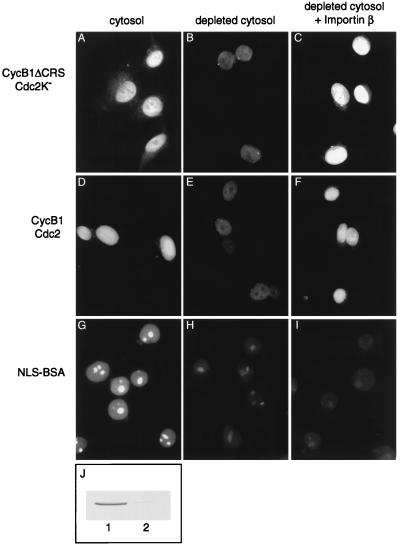
We partially purified the major cytosolic activity responsible for cyclin B1 import by ion-exchange chromatography on S-Sepharose, Q-Sepharose, and hydroxyapatite. While additional purification steps were being developed, Moore et al. (39) reported that cyclin B1 binds directly to importin β in vitro and that importin β is able to promote cyclin B1 import by permeabilized human cells (in a reaction containing Ran; the requirement for Ran was not tested). We therefore explored the possibility that the cytosol component required for cyclin B1–Cdc2 import in our system is importin β. Consistent with this possibility, we found that importin β comigrated with cyclin B1 import activity in our partially purified fractions (data not shown). In addition, immunodepletion of the majority of importin β from crude cytosol led to a significant decrease in the nuclear accumulation of cyclin B1ΔCRS–Cdc2K− and cyclin B1–Cdc2 complexes (Fig. 5 B and E). Cyclin B1 import was restored by adding back purified importin β protein (Fig. 5 C and F). Although these experiments do not rule out a contribution by other transporters, they indicate that importin β is required for much of the cyclin B1–Cdc2 transport we observe under these conditions.
Figure 5.
Nuclear import of cyclin B1–Cdc2 is reduced by depletion of importin β. Nuclear import reactions were performed with cyclin B1ΔCRS–Cdc2K− (A–C), wild-type cyclin B1–Cdc2 (D–F), or FITC-NLS-BSA (G–I) in the presence of mitotic cytosol (A, D, and G), mitotic cytosol that had been depleted of importin β (B, E, and H), or depleted cytosol supplemented with 0.5 μM importin β (C, F, and I). Cells were analyzed by direct immunofluorescence (G–I) or secondary immunofluorescence with monoclonal anti-cyclin B1 antibodies (A–F). (J) Western blot of importin β in the undepleted (lane 1) or depleted cytosol (lane 2). As shown (40, 41), addition of importin β to depleted cytosol did not restore import of NLS-BSA, presumably because other components (such as importin α) are also depleted with importin β.
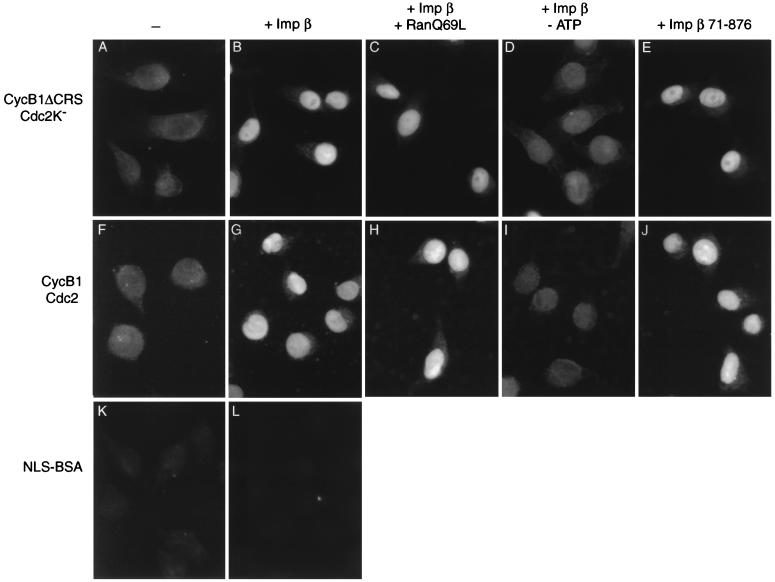
Importin β alone was also able to promote cyclin B1–Cdc2 import by digitonin-permeabilized cells in the absence of cytosol and other exogenous factors (such as Ran and importin α; Fig. 6 B and G) and in the presence of Ran Q69L (Fig. 6 C and H). ATP depletion reduced import by importin β (Fig. 6 D and I). Interestingly, the truncated importin β-(71–876) protein, which lacks the ability to bind Ran (25, 26), was able to promote cyclin B1 import to the same extent as wild-type importin β [Fig. 6 E and J; presumably, importin β-(71–876) did not enhance cyclin B1 import in our previous experiments with crude cytosol (Fig. 4) because importin β activity was not limiting]. These results further support our hypothesis that cyclin B1 import by importin β is independent of Ran.
Figure 6.
Nuclear import of cyclin B1–Cdc2 by importin β is Ran-independent and energy-dependent. Nuclear import reactions were performed with cyclin B1ΔCRS–Cdc2K− (A–E), wild-type cyclin B1–Cdc2 (F–J), or FITC-NLS-BSA (K and L) in the absence (A, F, and K) or presence of 2.7 μM importin β (B–D, G–I, and L) or importin β-(71–876) (E and J). (C and H) Ran Q69L (2 μM) was included in the reaction. (D and I) Hexokinase (0.4 unit/μl) and glucose (5 mM) were included to deplete residual ATP. Cells were analyzed by direct immunofluorescence (K and L) or secondary immunofluorescence with monoclonal anti-cyclin B1 antibodies (A–J).
DISCUSSION
We conclude that cyclin B1 is transported into the nucleus of permeabilized human cells by an unusual mechanism that requires importin β but does not require importin α or Ran. Immunodepletion of importin β from crude cytosol reduced the rate of cyclin B1 import, importin β alone was sufficient to stimulate cyclin B1 import by permeabilized cells, and import was not affected by the importin α IBB domain, which blocks the interaction between importins α and β. In addition, Moore et al. (39) recently found that cyclin B1 interacts directly with the N-terminal half of importin β, at a site distinct from the C-terminal region that interacts with importin α. It therefore appears that the nuclear import of cyclin B1 is mediated by a direct importin α-independent interaction with importin β. Thus, in this respect, cyclin B1 import is similar to that of certain ribosomal and viral proteins that are thought to be imported by importin β in the absence of importin α (38, 42, 43). However, the cyclin B1 import mechanism is clearly distinct from these other mechanisms in one important respect: it is not inhibited by GTP-bound Ran. We found that the GTPase-deficient Ran Q69L mutant did not inhibit the import of cyclin B1 by permeabilized cells, either in the presence of crude cytosol or in the presence of importin β alone. We also found that a version of importin β that cannot bind Ran is able to promote cyclin B1 import as effectively as wild-type importin β. We therefore believe that cyclin B1 import by importin β is not only independent of importin α but also independent of Ran.
Kose et al. (44) recently demonstrated that importin β, when it is not associated with Ran or importin α, can shuttle through the nuclear pore by a mechanism that is not blocked by RanGTP. It seems reasonable to propose that this Ran-independent translocation of importin β provides the basis of the cyclin B1 import mechanism. Perhaps the binding of cyclin B1 to the N-terminal half of importin β allows cyclin B1 to enter the nucleus by a Ran-independent process.
In most nuclear import pathways, nuclear RanGTP serves to induce the dissociation of importin–cargo complexes after their transit through the pore. Our observations suggest that RanGTP is not able to cause the dissociation of the cyclin B1–importin β complex, and it appears likely that the dissociation of this complex requires some other component. This dissociation step may be energy-dependent, explaining our observation that cyclin B1 import by purified importin β is reduced in the absence of ATP.
The localization of cyclin B1 during the cell cycle represents the net effect of opposing import and export processes. Because the ΔCRS version of cyclin B1 localizes constitutively to the nucleus, it appears likely that some basal level of cyclin B1 import occurs throughout the cell cycle (although it remains possible that the rate of import increases at the onset of mitosis). Active export of cyclin B1 during interphase ensures that imported cyclin B1 is returned to the cytoplasm; decreased export during prophase may then contribute to the net shift in cyclin localization from cytoplasm to nucleus. Recent studies support the hypothesis that phosphorylation of four Ser residues in the CRS region of Xenopus cyclin B1 inhibits the nuclear export signal in this region (17). Consistent with this possibility, we have found that replacement of the equivalent Ser residues in human cyclin B1 with Ala residues reduces the nuclear accumulation of cyclin B1 in mitotic cytosol; mutation of these residues to Glu leads to constitutive nuclear localization.
The protein kinases that phosphorylate the CRS of cyclin B1 remain to be identified. One of these kinases may be cyclin B1–Cdc2 itself. Two of the four Ser residues phosphorylated in the CRS are located within cyclin-dependent kinase consensus phosphorylation sequence motifs and are phosphorylated by cyclin B1–Cdc2 in vitro (45). Other sites in the Xenopus CRS, however, are not phosphorylated by Cdc2 and may, therefore, be targeted by another protein kinase. In our system, where the addition of active Cdc2 triggered cyclin B1 import by interphase cytosol, it appears that other protein kinases acting on the CRS must either be present in interphase cells or activated by Cdc2. The existence of multiple CRS kinases would provide a mechanism by which multiple regulatory pathways can influence the timing of mitotic entry.
Acknowledgments
We thank Maxence Nachury and Pete Takizawa for helpful discussions, S. Adam for the importin β-(71–876) expression vector, Paige Nittler for assistance with DNA constructs, Ali Fattaey for monoclonal anti-cyclin B1 antibody, and Julia Charles and Sue Jaspersen for comments on the manuscript. This work was supported by funding from the Sandler Family Supporting Foundation, the National Institute of General Medical Sciences, the Deutsche Forschungsgemeinschaft (K.W.), and a National Research Service Award Postdoctoral Fellowship (C.G.T.).
ABBREVIATIONS
- CRS
cytoplasmic retention signal
- NLS
nuclear localization signal
References
- 1.Nurse P. Nature (London) 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 2.King R W, Jackson P K, Kirschner M W. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 3.Dunphy W G. Trends Cell Biol. 1994;4:202–207. doi: 10.1016/0962-8924(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 4.Lew D J, Kornbluth S. Curr Opin Cell Biol. 1996;8:795–804. doi: 10.1016/s0955-0674(96)80080-9. [DOI] [PubMed] [Google Scholar]
- 5.Morgan D O. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 6.Pines J, Hunter T. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ookata K, Hisanaga S, Okano T, Tachibana K, Kishimoto T. EMBO J. 1992;11:1763–1772. doi: 10.1002/j.1460-2075.1992.tb05228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailly E, Pines J, Hunter T, Bornens M. J Cell Sci. 1992;101:529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- 9.Gallant P, Nigg E A. J Cell Biol. 1992;117:213–224. doi: 10.1083/jcb.117.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallant P, Fry A M, Nigg E A. J. Cell Sci. 1995. Suppl. 19, 21–28. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Meyer A N, Donoghue D J. Proc Natl Acad Sci USA. 1997;94:502–507. doi: 10.1073/pnas.94.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin P, Hardy S, Morgan D O. J Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pines J, Hunter T. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ookata K, Hisanaga S, Bulinski J C, Murofushi H, Aizawa H, Itoh T J, Hotani H, Okumura E, Tachibana K, Kishimoto T. J Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagting A, Karlsson C, Clute P, Jackman M, Pines J. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Bardes E S G, Moore J D, Brennan J, Powers M A, Kornbluth S. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 19.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 20.Weis K. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 21.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 22.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 23.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Görlich D, Dabrowski M, Bischoff E R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chi N C, Adam E J H, Adam S A. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- 26.Kutay U, Izaurralde E, Bischoff E R, Mattaj I W, Görlich D. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Görlich D. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 28.Izaurralde E, Kutay U, Von Kobbe C, Mattaj I W, Görlich D. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melchior F, Gerace L. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 30.Desai D, Gu Y, Morgan D O. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weis K, Ryder U, Lamond A I. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 33.Adam S A, Sterne-Marr R, Gerace L. Methods Enzymol. 1992;219:97–110. doi: 10.1016/0076-6879(92)19013-v. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Meyer A N, Donoghue D J. Mol Biol Cell. 1995;6:1111–1124. doi: 10.1091/mbc.6.9.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Görlich D, Henklein P, Laskey R A, Hartmann E. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 36.Palacios I, Weis K, Klebe C, Mattaj I W, Dingwall C. J Cell Biol. 1996;133:485–494. doi: 10.1083/jcb.133.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakielny S, Dreyfuss G. Curr Biol. 1998;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- 38.Jäkel S, Görlich D. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore J D, Yang J, Truant R, Kornbluth S. J Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi N C, Adam E J H, Adam S A. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palacios I, Hetzer M, Adam S A, Mattaj I W. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Truant R, Cullen B R. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmeri D, Malim M H. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izumi T, Maller J L. Mol Cell Biol. 1991;11:3860–3867. doi: 10.1128/mcb.11.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]