Abstract
Reciprocal epithelial–mesenchymal interactions shape site-specific development of skin. Here we show that site-specific HOX expression in fibroblasts is cell-autonomous and epigenetically maintained. The distal-specific gene HOXA13 is continually required to maintain the distal-specific transcriptional program in adult fibroblasts, including expression of WNT5A, a morphogen required for distal development. The ability of distal fibroblasts to induce epidermal keratin 9, a distal-specific gene, is abrogated by depletion of HOXA13, but rescued by addition of WNT5A. Thus, maintenance of appropriate HOX transcriptional program in adult fibroblasts may serve as a source of positional memory to differentially pattern the epithelia during homeostasis and regeneration.
[Keywords: HOXA13, WNT5A, chromatin immunoprecipitation, gene regulation, epithelial–mesenchymal interaction]
Epithelial tissues such as skin demonstrate remarkable anatomic diversity in their structure and function. For instance, while long terminal hairs decorate the scalp, palmo-plantar skin lacks hairs but possesses thickened epidermal barriers for mechanical stress. These anatomic differences lead to many body site-specific manifestations of diseases and guide their appropriate treatments (Bolognia et al. 2003). Because the epidermis is continually turned over, the apparent stability of site-specific features raises the fundamental question of how positional identity is acquired and maintained in the skin. Classic heterotypic recombination experiments in chick suggested that a primary dermal signal may dictate the positional identity of epithelial differentiation (Dhouailly 1984). We and others have used a genomic approach to examine the anatomic diversity of gene expression of cell types in the dermis and identified fibroblasts as a cell type that demonstrates consistent and large-scale differences of the expression of cell–cell signaling proteins in a site-specific manner (Chang et al. 2002; Chi et al. 2003; Rinn et al. 2006). Systematic comparison of the global transcriptional programs of fibroblasts from anatomic sites that finely map the body suggest that fibroblasts are differentiated based on their position along three anatomic divisions: anterior–posterior, proximal–distal, and dermal–nondermal (Rinn et al. 2006). Moreover, primary adult fibroblasts retained many features of the embryonic pattern of expression of HOX genes (Chang et al. 2002; Rinn et al. 2006), a family of homeodomain transcription factors that act to specify positional identity during development. These data support the idea, first proposed by Chuong (1993), that a “HOX code” may dictate the positional identity of skin and thus influence site-specific epidermal differentiation. However, fundamental elements of this hypothesis, such as the stability of HOX expression, gene regulatory function, and inductive activities governed by HOX genes in adult fibroblasts remain poorly understood. Here we show that expression of HOXA13 is required and the transcriptional targets are sufficient to maintain epidermal inductive properties of distal fibroblasts.
Results and Discussion
To test the hypothesis that ongoing HOX expression in dermal fibroblasts contributes to skin positional identity, we first examined the stability of HOX gene expression. Microarray analysis of serially passaged primary fibroblasts derived from foot or thigh showed that expression levels of all HOX genes are stable for at least 35 passages in vitro (Fig. 1A). We next tested whether the site-specific gene expression patterns were influenced by secreted factors in the media by culturing foot fibroblasts in conditioned media from thigh fibroblasts and vice versa (proximal vs. distal), followed by microarray analysis. Although the addition of conditioned media from any source predictably altered fibroblast gene expression, unsupervised hierarchical clustering of the gene expression patterns show a clear separation of thigh fibroblasts from foot fibroblasts, irrespective of the source of conditioned media (Fig. 1B). Their repertoire of HOX genes also remained distinctive and site-specific. Reciprocal, heterotypic conditioned media treatment of foreskin and lung fibroblasts also showed that their site-specific gene expression programs were essentially unaltered (Supplemental Fig. 1). These experiments suggest that secreted factors from adult fibroblasts are unable to reprogram site-specific gene expression of other fibroblasts, at least in the assay conditions used.
Figure 1.
Stability of site-specific gene expression in fibroblasts. (A) Stability of HOX gene expression after serial passage. The expression level of HOX genes plotted for passage 5 (X-axis) and passage 35 (Y-axis) for thigh (left) and foot fibroblasts (right). (B) Stability of site-specific gene expression patterns with heterotypic conditioned media. Unsupervised hierarchical clustering of thigh fibroblasts (P) without coculture (−), cultured with their self-conditioned media (P, P) or with conditioned media from foot fibroblasts (P, D) and vice versa for foot fibroblasts. Expression level of each gene above (red) or below (green) the global median (black) is denoted by the color scale (fourfold to 0.25-fold on linear scale or +2 to −2 on log2 scale). (C) UPRT-mediated RNA capture. Only UPRT+ cells can incorporate 4TU into thio-uracil (sU) in RNA, enabling retrieval of cell-type-specific RNA by thio-directed RNA biotinylation and purification. (D) UPRT-mediated RNA biotinylation. The 18S RNA band is shown by Northern blot probed with streptavidin-horseradish peroxidase (top) or ethidium bromide (bottom). (E) Verification of cell-type-specific RNA capture from 1:1 mixture of UPRT+ and UPRT− RNA. In the absence of the biotinylation reaction, little RNA was retrieved from either population. With biotin-mediated RNA pull down, RNA from UPRT+ cells was specifically enriched by eightfold to 10-fold compared with that of the mixed population. (F) Little change in the expression of genes diagnostic of proximal–distal limb origin in human foot fibroblasts after coculture with thigh fibroblasts. (G) Synergistic effects of trichostatin A (TSA) and Aza-C (Aza) on HOXA13 mRNA expression.
To address whether site-specific gene expression of fibroblasts may be reprogrammed by coculture and physical contact between heterotypic fibroblasts, we adapted a genetically encoded method of RNA labeling and capture (Fig. 1C–E; Cleary et al. 2005). Briefly, introduction of the Toxoplasma gondii UPRT gene allows all RNAs in the UPRT+ cells to be specifically biotinylated and captured from a pool of UPRT+ and UPRT− cells; we could therefore study the cell-type-specific consequences of coculture on gene expression without artifacts introduced by cell suspension or cell sorting. Using UPRT-mediated RNA capture and microarray analysis, we found that the global gene expression profiles of UPRT+ foot fibroblasts are not appreciably affected by coculture with thigh fibroblasts when compared with UPRT− foot fibroblasts (Fig. 1F). This result suggests that the majority of the site-specific gene expression in fibroblasts is controlled by a cell-autonomous mechanism. Importantly, site-specific expression of HOX genes in fibroblasts can be altered by treatment with trichostatin, a histone deacetylase inhibitor, and is potentiated by Aza-C, a DNA methylation inhibitor (Fig. 1G). These results are consistent with data indicating that site-specific HOX expression is epigenetically maintained by chromatin modifications in fibroblasts (Bernstein et al. 2005; Rinn et al. 2007).
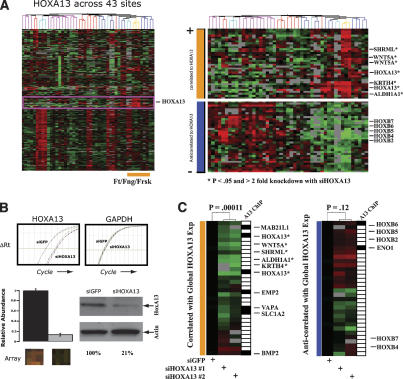
We next addressed the requirement of ongoing HOX activity for site-specific gene expression in fibroblasts. By comparing global gene expression profiles of primary fibroblasts from 43 anatomic sites, we previously defined a “distal gene expression neighborhood” that comprises 52 probes representing 44 unique genes specifically induced in cells from distal anatomic sites such as fingers, feet, and foreskin (Fig. 2A). HOXA13 is member of this distal neighborhood; interestingly, HOXA13 mutations in human patients produce malformations in these exact same anatomic sites and is termed hand-foot-genital syndrome (Mortlock and Innis 1997). We hypothesized that HOXA13 positively regulates transcriptional activation of this distal module, as these 44 genes are highly correlated in their expression across all 43 fibroblasts sites. We also suspected that HOXA13 may negatively regulate the transcription of genes that are anti-correlated to the express level of HOXA13 across our fibroblast compendium (Fig. 2A, bottom right). Interestingly, the most highly anti-correlated genes to HOXA13 include HOXB2, HOXB4, HOXB5, HOXB6, and HOXB7, suggesting a negative interaction between two HOX loci located on separate chromosomes, similar to what has been seen for the HOXC and HOXD loci (Rinn et al. 2007). siRNA-mediated depletion of HOXA13 in foreskin fibroblasts followed by microarray analysis showed that almost all genes in the distal neighborhood are substantially reduced in expression upon acute loss of HOXA13 (P < 0.00011, hypergeometric distribution) (Fig. 2B,C; Supplemental Table 1). The 32 genes anti-correlated with HOXA13 did not exhibit coordinate regulation upon depletion of HOXA13; however, HOXB2, HOXB5, and HOXB6 showed a trend toward increased expression. These data suggest that ongoing HOXA13 expression is required for the activation of a distal-specific pattern of gene expression in fibroblasts.
Figure 2.
HOXA13 regulates a distal gene expression program. (A, top right) Identification of genes most similar in expression pattern to HOXA13 across fibroblasts from 43 anatomic sites, which comprise a distal-specific neighborhood. (Bottom right) We also identified a set of genes that show the most opposite pattern of expression to HOXA13. (B) siRNA-mediated depletion of HOXA13 as measured by qRT–PCR (top), microarray (bottom left), and immunoblot (bottom right). (C) Effect of HOXA13 depletion on distal gene expression. Expression level of each gene above (red) or below (green) the average mock siRNA treatment (black) is denoted by the color scale (fourfold to 0.25-fold on linear scale or +2 to −2 on log2 scale). Genes differing significantly (twofold and P < 0.05) are denoted by asterisk. The significance of the entire distal neighborhood or anti-neighborhood being coordinately repressed or induced is 0.00011 versus. 0.12 (hypergeometric distribution), respectively. Direct binding of HOXA13 to a given promoter as determined by ChIP–chip is denoted by a black box.
To determine which of these transcriptional changes are directly regulated by HOXA13, we performed chromatin immunopreciptation (ChIP) of HOXA13, followed by hybridization to microarrays tiling over 30,000 human promoters (“ChIP–Chip” analysis) (Fig. 2C; Supplemental Table 2). The promoters of seven of the 44 genes in the distal neighborhood were occupied by HOXA13, which is significantly more enriched than expectation by chance alone (P < 1 × 10−3, hypergeometric distribution). HOXA13-occupied promoters in fibroblasts include HOXA13 itself; HOXB6 the most anti-correlated gene to HOXA13; BMP2, a HOXA13 target gene important in distal limb development (Knosp et al. 2004); and MAB21L1, a cell fate determining protein. Together, these results identify both direct and indirect targets of HOXA13 in the distal-specific gene expression program of adult human fibroblasts (Supplemental Tables 2, 3).
To begin to understand how HOX-dependent gene expression in fibroblasts may impact epithelial–mesenchymal interactions, we focused our analysis on WNT5A, a member of the distal neighborhood whose expression requires ongoing HOXA13 function, but is not a direct transcriptional target of HOXA13, at least for the genomic region of WNT5A promoter that was interrogated. WNT5A is a member of the WNT family of extracellular cell–cell signaling proteins, and it has been considered an atypical WNT because it can either activate or inhibit a canonical WNT signaling cascade, depending on the presence of its coreceptor ROR2 kinase (Mikels and Nusse 2006). Importantly, murine Wnt5a is highly expressed in distal limbs, external genitalia, and distal snout during development, and deletion of Wnt5a in mice leads to distal truncation of all these structures (Yamaguchi et al. 1999). Despite the clear evidence implicating Wnt5a as a mediator distal outgrowth, the mechanism that specifies distal-specific expression of Wnt5a has not been identified. Our microarray analysis thus provides the first experimental evidence that HoxA13 may activate Wnt5a expression, leading to distal patterning. To confirm this hypothesis, first we performed whole-mount in situ hybridization of HoxA13 and Wnt5a during murine development and showed that these two genes are indeed spatially and temporally coexpressed in the distal limb bud (Fig. 3A). Second, we examined the effect of HoxA13 loss of function on Wnt5a expression by using HoxA13GFP/GFP mice, where a GFP sequence was inserted into a HoxA13 coding sequence, thereby disrupting HoxA13 function (Stadler et al. 2001). Wnt5a mRNA was detected in the cartilage condensate, scattered cells in the dermis, and in the epidermis of wild-type distal embryonic limb buds; this pattern was similar to the HoxA13 expression domain, as visualized by GFP expression in HoxA13GFP/GFP limb buds (Fig. 3B). Importantly, Wnt5a expression was substantially reduced in HoxA13GFP/GFP limb buds, indicating that HoxA13 is required for Wnt5a expression (Fig. 3B). In contrast, microarray expression of HOXA13-depleted fibroblasts showed a 2.5-fold induction of WNT receptor FZD8, and in situ hybridization confirmed the overexpression of murine FZD8 mRNA in HoxA13GFP/GFP limb buds (Fig. 3B). This finding suggests that loss of Wnt5a expression in HoxA13 mutant limbs is unlikely due to cell death or general tissue dysfunction, but is more likely a specific gene regulatory function of HoxA13.
Figure 3.
HoxA13 regulates Wnt5a expression in vivo. (A) Whole-mount in situ hybridization of E10.5 mouse limb buds for HoxA13 or Wnt5a mRNA expression. (B) Expression of GFP, Wnt5a, or Fzd8 in wild-type or HoxA13GFP/GFP (null) E16.5 limb bud sections.
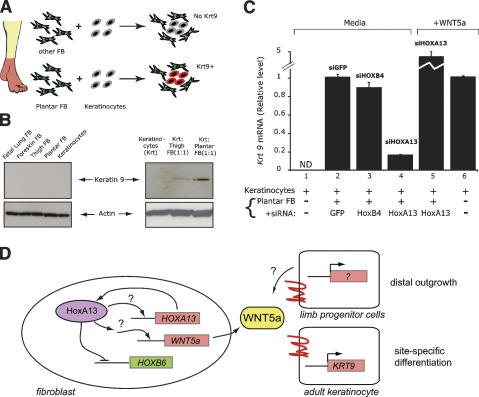
Finally, we examined whether HOXA13-induced expression of WNT5A in adult human fibroblasts continue to provide a distal-specific function. In postnatal human skin from an anatomically distal site, HOXA13 protein is expressed in the dermis but not in the epidermis (Rinn et al. 2006). Previous studies have shown that coculture of primary dermal fibroblast from palms or soles can reprogram keratinocytes from other sites to express acral-specific differentiation genes, such as keratin 9 (K9) (Fig. 4A; Yamaguchi et al. 1999). Mutations in K9 cause epidermolytic palmoplantar keratoderma (Reis et al. 1994), underscoring the importance of proper regulation of K9. Therefore, we tested whether such a distal-specific inductive function involved the HOXA13–WNT5A pathway. Consistent with previous reports (Yamaguchi et al. 1999), we found that plantar fibroblasts could induce K9 expression in cocultured keratinocytes, albeit a modest induction compared with K9 level in vivo. This effect required the coculture of fibroblasts and keratinocytes, and is more efficient using fibroblasts of palmo-plantar origin (Fig. 4B). This inductive effect appears to be regulated at the transcriptional level as induction of K9 mRNA paralleled protein accumulation (Fig. 4C). Depletion of HOXA13 in plantar fibroblasts before coculture, but not treatment with control siRNAs against GFP or HOXB4, substantially reduced the ability of these cells to induce epidermal K9 transcription, and this defect can be fully rescued by addition of recombinant purified WNT5A protein to the coculture (Fig. 4C). In fact, direct addition of WNT5A protein to keratinocytes in the absence of cocultured fibroblasts could also induce K9 transcription (Fig. 4C), and this effect occurred without perturbing the general differentiation program of keratinocytes (Supplemental Fig. 2). Microarray analysis of the transcriptional response of keratinocytes to WNT5A confirmed that WNT5A predominantly regulates genes involved in cell differentiation (Supplemental Table 4). These results suggest that HOXA13 control of WNT5A in fibroblasts is one of the mechanisms that can regulate site-specific epidermal differentiation.
Figure 4.
HOXA13 is required for fibroblast induction of site-specific keratin. Epidermal induction of plantar-specific K9. (A) Schematic of experiment. K9, a palmo-plantar differentiation marker, is induced in keratinocytes by coculture with plantar fibroblasts, but not more proximal fibroblasts. (B) Induction of K9 protein by coculture with plantar fibroblasts. (C) Relative induction of K9 mRNA in coculture assay upon depletion of HOXA13 in fibroblasts or addition of WNT5A. (ND) Not detectable. (D) Model of HOXA13 transcriptional network. In the developing limb, cells that make WNT5a may also respond to WNT5a themselves. The ectodermal cells in limb bud also express WNT5A in a HOXA13-dependent manner (Fig. 3), suggesting that the distal transcriptional program may occur in both mesenchymal and ectodermal cells of the distal limb bud.
Here we provide several lines of evidence supporting a fibroblast HOX code that contributes to the positional identity of skin. Consistent with heterotopic epithelial–mesenchymal recombination experiments, where the mesenchyme can reprogram heterotopic epithelia, we showed that the positional-specific HOX code in fibroblasts is stably maintained in the face of heterotypic signals. Ongoing HOXA13 activity is required to maintain a distal-specific gene expression program in adult fibroblasts. As illustrated by WNT5A, the same HOXA13-dependent gene is used to pattern distal outgrowth early in development and can be used again to specify distal epidermal differentiation in adulthood (Fig. 4D). Our analysis also gave some of the first insights into the human HOXA13 transcriptional networks in fibroblasts, revealing the potential autoregulation of HOXA13, trans-repression of HOXB6, and continued regulation of embryonic target genes such as BMP2, which is required for footpad epidermal differentiation (Plikus et al. 2004).
We found that the HOXA13 protein occupied the HOXB6 promoter, but not the WNT5A promoter. Interestingly, HoxB5 and HoxB6 demonstrate an antagonistic pattern of expression as Wnt5a expression in lung development (Sakiyama et al. 2000). Therefore, we speculate that HOXA13 activation of WNT5A may occur directly via more distant enhancer sequences or indirectly via HOXB6. Wnt5a expression is substantially reduced, but not completely absent in HoxA13 knockout limb. Wnt5a is also expressed more proximally than the HoxA13 expression domain, suggesting that other transcription factors also induce Wnt5a expression. These additional layers of regulation may explain the more severe distal truncation phenotype in Wnt5a knockouts.
Previous studies have shown that DKK1, an antagonist of canonical Wnt signaling, is also induced in palmoplantar fibroblasts (Yamaguchi et al. 2004). The combination of DKK1, WNT5A, and repression of FZD8 in palmoplantar skin could block canonical WNT-dependent appendage formation and promote site-specific epidermal differentiation, such as induction of K9. The specialized fibroblasts that support hair follicle differentiation and cycling, termed dermal papilla cells, control the site-specific identity of the hairs and also express HOX genes (Jahoda 1992; Rendl et al. 2005), suggesting a possible role of HOX genes in patterning epidermal appendage development.
Because the proper position-specific expression of HOX genes would appear to have critical importance in dictating skin positional identity, mechanisms that ensure the fidelity of HOX expression in skin should be investigated in future studies.
In summary, human adult fibroblasts maintain features of the embryonic HOX pattern over decades in vivo and over numerous cell divisions in vitro, which suggests that they are a valuable system to investigate the mechanisms of transcriptional memory. The maintenance of region-specific HOX gene expression in adult fibroblasts may serve as a source of positional memory for skin during homeostasis and regeneration. These data are consonant with emerging evidence in other model organisms that HOX genes not only orchestrate segmental patterning in development, but continue to act as micromanagers of differentiation in subsequent developmental stages (Akam 1998). Whether ectopic expression of HOX genes in adult skin may suffice to reprogram positional identity will be examined in future studies. Finally, additional cell types likely also contribute to skin positional identity, and their roles also await clarification in future studies.
Materials and methods
Heterotypic cell interactions
Primary fibroblasts (Rinn et al. 2006) and UPRT-mediated RNA capture (Cleary et al. 2005) were as described. Media conditioned for 4 d were applied to fresh cells for 4 d. Cells were treated with 5-AzaC for 5 d, replacing the drug every 48 h, or with trichostatin A for 48 h.
HOXA13 transcriptional network
After siRNA-mediated depletion of HOXA13, RNA was isolated and hybridized to Stanford cDNA microarrays (Rinn et al. 2006). ChIP using HOXA13 antibody (Knosp et al. 2004) was hybridized to human HG18 promoter tiling arrays (Nimblegen, Inc.). Candidate ChIP peaks of different enrichment values were validated by ChIP followed by qPCR (Supplemental Fig. 2). Candidate HOXA13 targets were compared with genes altered by HOXA13 RNAi or HOXA13 overexpression (Williams et al. 2005), yielding a HOXA13 transcriptional network supported by independent lines of evidence. In situ hybridization of wild-type and HOXA13GFP/GFP mice (Stadler et al. 2001) were as described (Albrecht et al. 1997). Array data are available at GEO (GSE9922) and Stanford Microarray Database.
K9 induction
KRT9 qPCR was performed 24 h after coculture of keratinocytes and fibroblasts in KGM media or addition of 300 ng of recombinant WNT5A.
Full methods are available in the Supplemental Material.
Acknowledgments
We thank A. Oro, P. Khavari, and members of Program in Epithelial Biology for helpful comments. This work was supported by NIH (AR050007 to H.Y.C), National Science Foundation Predoctoral Award (J.K.W.), and Scleroderma Research Foundation (H.Y.C.). J.L.R. is a Fellow of the Damon Runyon Cancer Research Foundation. H.Y.C. is the Kenneth G. and Elaine A. Langone Scholar of the Damon Runyon Cancer Research Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1610508
References
- Akam M. Hox genes: From master genes to micromanagers. Curr. Biol. 1998;8:R676–R678. doi: 10.1016/S0960-9822(98)70433-6. [DOI] [PubMed] [Google Scholar]
- Albrecht U.E.G., Eichele G., Helms J.A., Lu H.-C. Visualization of gene expression patterns by in situ hybridization. In: Daston G.P., editor. Molecular and cellular methods in developmental toxicology. CRC Press; Boca Raton, FL: 1997. pp. 23–48. [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bolognia J., Jorizzo J., Rapini R. Dermatology. Mosby; New York: 2003. [Google Scholar]
- Chang H.Y., Chi J.T., Dudoit S., Bondre C., de van Rijn M., Botstein D., Brown P.O. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J.T., Chang H.Y., Wang N.N., Chang D.S., Dunphy N., Brown P.O. Genomewide view of gene silencing by small interfering RNAs. Proc. Natl. Acad. Sci. 2003;100:6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong C.M. The making of a feather: Homeoproteins, retinoids and adhesion molecules. Bioessays. 1993;15:513–521. doi: 10.1002/bies.950150804. [DOI] [PubMed] [Google Scholar]
- Cleary M.D., Meiering C.D., Jan E., Guymon R., Boothroyd J.C. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- Dhouailly D. Specification of feather and scale patterns. In: Malacinski G.M., Bryant S.V., editors. Pattern formation, a primer in developmental biology. Macmillan; New York: 1984. pp. 581–601. [Google Scholar]
- Jahoda C.A. Induction of follicle formation and hair growth by vibrissa dermal papillae implanted into rat ear wounds: Vibrissa-type fibres are specified. Development. 1992;115:1103–1109. doi: 10.1242/dev.115.4.1103. [DOI] [PubMed] [Google Scholar]
- Knosp W.M., Scott V., Bachinger H.P., Stadler H.S. HOXA13 regulates the expression of bone morphogenetic proteins 2 and 7 to control distal limb morphogenesis. Development. 2004;131:4581–4592. doi: 10.1242/dev.01327. [DOI] [PubMed] [Google Scholar]
- Mikels A.J., Nusse R. Purified Wnt5a protein activates or inhibits β-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock D.P., Innis J.W. Mutation of HOXA13 in hand-foot-genital syndrome. Nat. Genet. 1997;15:179–180. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- Plikus M., Wang W.P., Liu J., Wang X., Jiang T.X., Chuong C.M. Morpho-regulation of ectodermal organs: Integument pathology and phenotypic variations in K14-Noggin engineered mice through modulation of bone morphogenic protein pathway. Am. J. Pathol. 2004;164:1099–1114. doi: 10.1016/S0002-9440(10)63197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis A., Hennies H.C., Langbein L., Digweed M., Mischke D., Drechsler M., Schrock E., Royer-Pokora B., Franke W.W., Sperling K., et al. Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK) Nat. Genet. 1994;6:174–179. doi: 10.1038/ng0294-174. [DOI] [PubMed] [Google Scholar]
- Rendl M., Lewis L., Fuchs E. Molecular dissection of mesenchymal–epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.137l/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Bondre C., Gladstone H.B., Brown P.O., Chang H.Y. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama J., Yokouchi Y., Kuroiwa A. Coordinated expression of Hoxb genes and signaling molecules during development of the chick respiratory tract. Dev. Biol. 2000;227:12–27. doi: 10.1006/dbio.2000.9880. [DOI] [PubMed] [Google Scholar]
- Stadler H.S., Higgins K.M., Capecchi M.R. Loss of Eph-receptor expression correlates with loss of cell adhesion and chondrogenic capacity in Hoxa13 mutant limbs. Development. 2001;128:4177–4188. doi: 10.1242/dev.128.21.4177. [DOI] [PubMed] [Google Scholar]
- Williams T.M., Williams M.E., Kuick R., Misek D., McDonagh K., Hanash S., Innis J.W. Candidate downstream regulated genes of HOX group 13 transcription factors with and without monomeric DNA binding capability. Dev. Biol. 2005;279:462–480. doi: 10.1016/j.ydbio.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Itami S., Tarutani M., Hosokawa K., Miura H., Yoshikawa K. Regulation of keratin 9 in nonpalmoplantar keratinocytes by palmoplantar fibroblasts through epithelial–mesenchymal interactions. J. Invest. Dermatol. 1999;112:483–488. doi: 10.1046/j.1523-1747.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Itami S., Watabe H., Yasumoto K., Abdel-Malek Z.A., Kubo T., Rouzaud F., Tanemura A., Yoshikawa K., Hearing V.J. Mesenchymal–epithelial interactions in the skin: Increased expression of dickkopf1 by palmoplantar fibroblasts inhibits melanocyte growth and differentiation. J. Cell Biol. 2004;165:275–285. doi: 10.1083/jcb.200311122. [DOI] [PMC free article] [PubMed] [Google Scholar]