Abstract
The circadian clock enables the anticipation of daily recurring environmental changes by presetting an organism’s physiology and behavior. Driven and synchronized by a central pacemaker in the brain, circadian output genes fine-tune a wide variety of physiological parameters in peripheral organs. However, only a subset of circadianly transcribed genes seems to be directly regulated by core clock proteins. Assuming that yet unidentified transcription factors may exist in the circadian transcriptional network, we set out to develop a novel technique, differential display of DNA-binding proteins (DDDP), which we used to screen mouse liver nuclear extracts. In addition to several established circadian transcription factors, we found DNA binding of heat-shock factor 1 (HSF1) to be highly rhythmic. HSF1 drives the expression of heat-shock proteins at the onset of the dark phase, when the animals start to be behaviorally active. Furthermore, Hsf1-deficient mice have a longer free-running period than wild-type littermates, suggesting a combined role for HSF1 in the mammalian timekeeping and cytoprotection systems. Our results also suggest that the new screening method DDDP is not limited to the identification of circadian transcription factors but can be applied to discover novel transcriptional regulators in various biological systems.
[Keywords: Circadian rhythms, HSF1, in vitro screen]
Circadian oscillation of biological processes has been described in organisms ranging from photosynthetic bacteria to vertebrates and reflects the existence of underlying intrinsic biological clocks with near 24-h oscillation periods (Hardin 2006). Circadian control of physiology and behavior in mammals is driven by a master pacemaker located in the suprachiasmatic nuclei (SCN) of the hypothalamus. Rhythmicity in the SCN is entrained by external Zeitgeber cues, such as daily changes in light intensity, and further transmitted to peripheral organs (Saper et al. 2005). Studies using transgenic rodents and fibroblast cell lines indicate that peripheral tissues of mammals also have an autonomous capacity for circadian gene expression (Balsalobre et al. 1998; Yamazaki et al. 2000). The underlying circadian oscillators function in a cell-autonomous and self-sustained manner, similar to those operative in SCN neurons (Nagoshi et al. 2004; Welsh et al. 2004; Liu et al. 2007).
Circadian rhythms appear to be generated by feedback loops in clock gene expression (Hardin et al. 1990). In mammals, the CLOCK and BMAL1 transcription factors activate the expression of Per and Cry genes. Once the PER and CRY proteins reach a critical concentration and/or activity, they attenuate the CLOCK/BMAL1-mediated activation of their own genes in a negative feedback loop (Reppert and Weaver 2002). In addition, several post-translational events, such as the control of protein phosphorylation, degradation, and nuclear entry, contribute critically to the generation of daily oscillations in clock gene products (Lee et al. 2001; Gallego and Virshup 2007).
The proteins that constitute the core clock oscillator regulate directly or indirectly the transcription of output genes, the expression products of which constitute the circadian transcriptome and proteome in every tissue (Kornmann et al. 2001, 2007; Akhtar et al. 2002; Duffield et al. 2002; Panda et al. 2002; Storch et al. 2002; McCarthy et al. 2007; Miller et al. 2007). In the liver, for example, factors involved in processing and detoxification of nutrients have been found to be rhythmically expressed (Gachon et al. 2006). Interestingly, circadian expression of the majority of liver genes is tissue-specific, and only a small fraction of these genes seems to be a direct target of the transcription factors that drive the core oscillator (Panda et al. 2002).
Insight into a transcriptional network such as the circadian system requires the identification of all factors that are involved in its regulation. Gene expression is primarily controlled by transcription factors, sequence-specific DNA-binding proteins that bind to regulatory regions of genes and interact with the basic transcription machinery to facilitate or repress transcription. The expression and activity of transcription factors can, in principle, be regulated at the level of transcription, mRNA stability, translation, protein stability, or by post-translational mechanisms. Transcription factors whose expression is regulated at the level of mRNA accumulation can be identified by functional genomics strategies such as microarray hybridization, serial analysis of gene expression, or massive parallel signature sequencing (Velculescu et al. 1995; Brenner et al. 2000; Panda et al. 2003). However, circadian transcription does not always result in circadian mRNA accumulation, due to a potentially long half-life of the mRNA. Moreover, all regulatory mechanisms that occur on the protein level—e.g., phosphorylation, acetylation, glycosylation, farnesylation, ubiquitinylation, proteolytic cleavage, ligand binding, multimerization, subcellular localization, etc.—escape this kind of analysis. Here we set out to circumvent some of these shortcomings and developed a new technique, differential display of DNA-binding proteins (DDDP), for the identification of circadian transcription factors based on their in vitro DNA-binding activity. Using this novel procedure, we were able to identify several well-established clock proteins as well as transcription factors that until now have not been implicated in circadian transcription in peripheral tissues. One of them, heat-shock factor 1 (HSF1), displays highly circadian DNA-binding activity in the dark phase of the day. Maximal DNA binding coincides with the uptake of food and maximal core body temperature in the animals. Our results suggest that every day the mammalian body goes through a proteotoxic stress event that elicits the expression of cell-protective proteins. Moreover, a significantly longer free-running period of Hsf1-deficient mice compared with heterozygous or wild-type littermates directly implicates HSF1 in the mammalian core clock mechanism.
Results
Generation and validation of a random DNA library
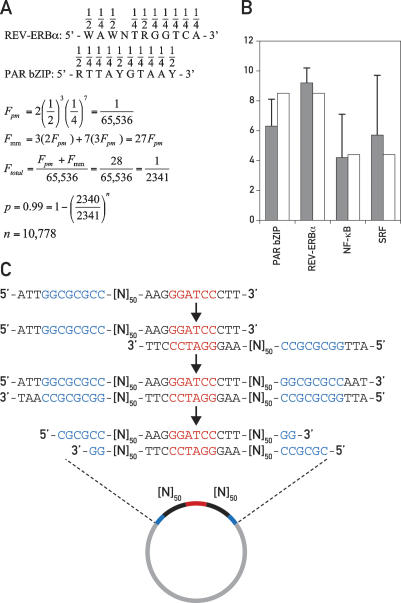
In general, transcription factors bind to their specific cognate sequences with dissociation equilibrium constants (KDs) in the low nanomolar to subnanomolar range (10−8 M to 10−10 M). For example, the PAR bZIP proteins DBP, HLF, and TEF bind to their perfect consensus DNA sequence RTTAYGTAAY with a KD of ∼1.5 × 10−10 M (Falvey et al. 1996). However, a sequence with a single mismatch at any of the 10 positions still binds these proteins with a KD of 10−8 M to 10−9 M (Fonjallaz et al. 1996). Other transcription factors, such as the orphan nuclear receptor REV-ERBα, bind sequences with a similar complexity (WAWNTRGGTCA) and with similar affinities (Forman et al. 1994; L. Marcacci and U. Schibler, unpubl.). Recognition sequences matching the PAR bZIP or REV-ERBα consensus sequences perfectly or with one mismatch exist statistically in random DNA once in every 2341 base pairs (bp) (Fig. 1A). If the sequences of both complementary strands are taken into consideration, at least one such binding site should be found with a probability of >99% in ∼11,000 bp of random DNA (Fig. 1A). The number of well-conserved bases in the binding site of a eukaryotic transcription factor is typically five to 10 (Wingender et al. 1996; Berg et al. 2004). The binding sequences for PAR bZIP proteins and REV-ERBα are, therefore, rather complex, and other, simpler binding sites like E-boxes (CAC GTG) are expected to be found with an even higher probability in random DNA.
Figure 1.
Construction of a random DNA library. (A) Consensus binding sites for REV-ERBα and PAR bZIP proteins showing the probability for each nucleotide position in random DNA. The following calculation determines the number of base pairs (n) that have to be tested in random DNA to obtain with a probability (p) of >99% a binding site of a complexity equal to a REV-ERBα or PAR bZIP-binding site if a single mismatch is allowed. The total frequency (Ftotal) of such binding sites is composed of the frequency of a perfect match (Fpm) plus the frequency of all sequences with one mismatch (Fmm). (B) Calculated (white bars) and obtained (gray bars) frequencies of exemplary transcription factor-binding sites in in silico generated 20,000-bp random DNA libraries; standard deviations are indicated by error bars. (C) Construction of a random DNA plasmid library. Oligonucleotides with 50 random nucleotide positions ([N]50), a 5′-flanking region containing an AscI restriction site (blue), and a palindromic 3′-flanking region containing a BamHI restriction site (red) are self-annealed, rendered double-stranded, and cloned into the AscI site of a bacterial plasmid vector (see Materials and Methods).
This calculation prompted us to use random DNA fragments as probes for in vitro protein–DNA-binding experiments. Testing a sufficiently large number of random DNA probes should result in an unbiased display of most cellular DNA-binding activities. When using nuclear protein extracts harvested around the clock, this experimental setup could then be used to screen for circadian transcription factors. However, first we attempted to verify the validity of our calculations using bioinformatics tools. We used the Mersenne twister algorithm for the generation of pseudo-random numbers (Makoto and Takuji 1998) to create six random DNA libraries of 20,000 bp each. Statistically, one random DNA library of 20,000 bp contains 8.5 perfect or imperfect binding sites each for PAR bZIP proteins (RTTAYG TAAY) and for REV-ERBα (WAWNTRGGTCA) and 4.4 binding sites each for nuclear factor κB (NF-κB, GG GAMTTYCC) and serum response factor (SRF, CCA WATAWGG). When we inspected the in silico generated libraries for binding sites for these transcription factors, the obtained frequencies came close to the calculated ones (Fig. 1B).
Next, we constructed a random DNA library of 150 plasmids, each containing a 100-bp random DNA insert in the form of 2 × 50 bp interrupted by a central BamHI site (Fig. 1C). Oligonucleotides were synthesized that started with an 11-bp sequence including an AscI restriction site followed by 50 positions containing N, where N is any of the four nucleotides. The oligonucleotides ended with a 12-bp palindromic sequence including a BamHI site that was used to self-anneal the oligonucleotides. After self-annealing, the ssDNA overhangs containing the 50 N-positions and the 5′-sequence harboring the AscI site were filled in by Klenow polymerase with deoxynucleotides, and the double-stranded molecule was cloned into the AscI site of a plasmid. DNA was prepared from 150 bacterial colonies, and the plasmids were serially numbered so that they could be reidentified and their inserts be sequenced if necessary.
Our next objective was to demonstrate the existence of functional binding sites for arbitrarily chosen mammalian transcription factors in the DNA library by EMSA competition experiments. Ten equimolar mixtures of 15 plasmids each were used in EMSA experiments as nonradioactive competitor DNA in combination with radioactive probes for the transcription factors TEF, SRF, or NF-κB (Ellis et al. 2002; Gachon et al. 2004; Eisner et al. 2006). One plasmid pool competed efficiently for binding to recombinant TEF protein (Supplemental Fig. 1A), and several pools competed for binding to SRF and NF-κB in nuclear extracts from mouse liver (Supplemental Fig. 1C; data not shown). The competing pools were split up, and the plasmids were used individually as competitor DNA in another EMSA experiment. Two of the competing pools were chosen arbitrarily for the continuation of the experiment with NF-κB and SRF. In each pool one single plasmid could be identified that competed specifically with the radioactive probe (Supplemental Fig. 1B,D). The random DNA inserts of these plasmids were sequenced and inspected for matching transcription factor-binding sites. The plasmid that competed for TEF binding contained the PAR bZIP-binding site ATTACGTAAC that perfectly matches the PAR bZIP consensus sequence RTTAYGTAAY. Both plasmids that competed for binding to SRF contained SRF-binding sites with two mismatches each (CCTAATATAG and ACTAATAAGG, mismatches are underlined). Nevertheless, when these fragments were themselves radioactively labeled and used as EMSA probes, they formed strong complexes with SRF (Supplemental Fig. 1E). The two fragments that competed for NF-κB binding contained three and four mismatches, respectively (GG GATTTTGG and GGGTTTTTGT, mismatches are underlined). Again, both formed complexes with NF-κB when they were themselves used as radioactive probes (data not shown). We felt encouraged by these results to expand the library to 40,000 bp and to use it to probe mouse liver nuclear extracts for circadian transcription factors.
DDDP
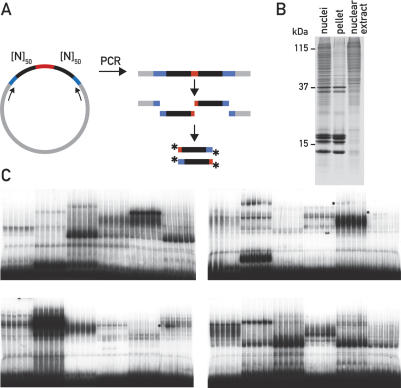
The 400 inserts of the final random library were separately amplified by PCR, and the resulting DNA fragments were cleaved in both flanking regions with AscI and BamHI, respectively. The resulting 5′-overhanging ends were radioactively labeled, so that each clone gave rise to an equimolar mixture of two EMSA probes containing 50 random base pairs each (Fig. 2A). Liver nuclei were prepared from mice sacrificed at Zeitgeber times ZT1, ZT5, ZT9, ZT13, ZT17, and ZT21 (ZT0 and ZT12 are the times when the lights are switched on and off, respectively), and nuclear nonhistone proteins were extracted nearly quantitatively in a cocktail containing 0.3 M NaCl, 1 M urea, and 1% Nonidet P-40 (Fig. 2B). As demonstrated previously, this procedure is significantly more efficient in recovering nonhistone proteins than methods based on high salt extraction (Lavery and Schibler 1993).
Figure 2.
DDDP. (A) Generation of EMSA probes from individual clones of the random DNA library. Random DNA inserts were amplified by PCR, digested with AscI and BamHI, and radioactively labeled by the incorporation of α32PdCTP (asterisks). (B) NUN extraction of mouse liver nuclei. The Coomassie-stained SDS–polyacrylamide gel shows mouse liver nuclei after lysis in 1% SDS, the redissolved pellet after the NUN extraction containing mainly DNA and histones, and the nuclear extract containing the soluble nonhistone protein fraction that was subsequently used for EMSA experiments. The molecular weight of three marker proteins is indicated. (C) Four typical gels from the first round of DDDP screening. Each autoradiograph displays EMSAs with nuclear extracts harvested around the clock with six random probes; circadian probes are marked by asterisks.
Each probe was subsequently used in EMSA reactions together with six nuclear extracts prepared around the clock. In total, 2400 EMSA reactions were performed and displayed by native polyacrylamide gel electrophoresis. Four typical autoradiographies of dried gels are shown in Figure 2C. All gel displays were visually inspected for DNA-binding activities that varied during the course of 1 d. As expected, multiple DNA–protein complexes were observed in virtually every lane. We tested all 400 random DNA inserts with a first set of mouse liver nuclear extracts and found 53 of them to display apparent circadian DNA-binding activity. These probes were then retested with a different set of circadian nuclear extracts to eliminate potentially false positives. The plasmids corresponding to the 43 probes that displayed circadian binding activity with two different sets of extracts were sequenced and analyzed for transcription factor-binding sites with Match (http://www.gene-regulation.com/pub/programs.html#match), a program that uses a library of mononucleotide weight matrices from TRANSFAC 6.0 (Wingender et al. 1996). The sequences of all circadian probes are listed in Supplemental Table 1.
We noticed in the course of the experiments that the presence of phosphatase inhibitors in each preparation step is essential in order to obtain highly active nuclear extracts yielding reproducible results. Probes 201–400 were therefore tested with nuclear extracts that had been prepared in the presence of phosphatase inhibitors, and these experiments resulted in roughly six times as many circadian protein–DNA complexes as probes 1–200 that had been examined with nuclear extracts prepared in the absence of phosphatase inhibitors.
Identification of established circadian transcription factors by DDDP
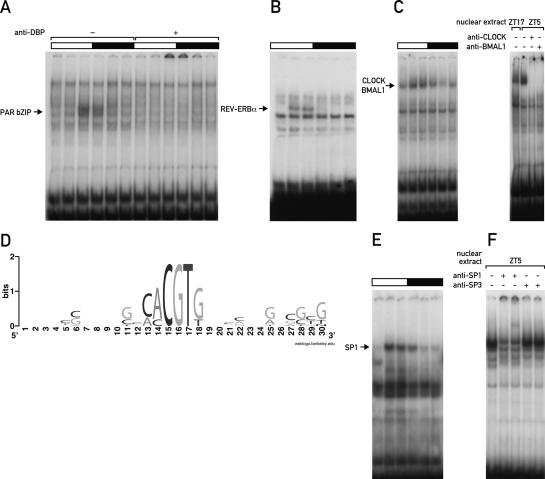
Two of the probes identified in the screen showed a high-amplitude circadian protein–DNA complex with a maximal binding activity at ZT9 (Fig. 3A). Sequence analysis revealed the perfect PAR bZIP-binding site ATTACG TAAC in one probe and a PAR bZIP-binding site with two mismatches (CTTATGCAAC, mismatches are underlined) in the other. In liver, DBP is the most abundant PAR bZIP protein, and most of the other two PAR bZIP members TEF and HLF form heterodimers with DBP. An anti-DBP antibody selectively supershifted the circadian DNA-binding activity, confirming PAR bZIP proteins as the corresponding circadian transcription factors (Fig. 3A). Another probe contained the imperfect REV-ERBα-binding site GATGTGGGACA (mismatches are underlined) and generated a circadian protein–DNA complex whereby maximal binding occurred at ZT5–ZT9 (Fig. 3B). REV-ERBα has been shown before to form a DNA–protein complex of comparable size whose circadian phase was nearly identical (Preitner et al. 2002; Ueda et al. 2002).
Figure 3.
Identification of established circadian transcription factors by DDDP. (A) Circadian EMSA of probe 98 containing a perfect PAR bZIP-binding site without or with anti-DBP antiserum. Light and dark phases are indicated by bars above the autoradiographs. (B) Circadian EMSA of probe 82 containing an imperfect REV-ERBα-binding site. (C) Circadian EMSA of probe 270 containing a noncanonical E-box (left panel) and supershifts with anti-CLOCK and anti-BMAL1 antibodies (right panel). (D) WebLogo consensus sequence of the CLOCK/BMAL1-binding sites that were obtained in the DDDP screen. The letter size indicates the relative conservation of the respective nucleotide position. (E) Circadian EMSA of probe 220 containing a highly GC-rich binding sequence. (F) Supershift experiment with anti-SP1 and anti-SP3 antibodies.
CLOCK/BMAL1 heterodimers exhibit a characteristic EMSA pattern with a maximal binding activity at around ZT5–ZT9 and a shift toward a more slowly migrating form at around ZT13–ZT17 (Ripperger and Schibler 2006). This change in RF-value is likely to be due to post-translational modifications, since both CLOCK and BMAL1 are circadianly phosphorylated (Lee et al. 2001). We observed a CLOCK/BMAL1-like EMSA pattern with 11 probes and confirmed with one probe the presence of both proteins in the DNA–protein complexes by supershift (Fig. 3C). Sequence analysis of the corresponding random DNA inserts revealed that only four of the 11 probes contained a perfect E-box sequence (CACGTG), and the remaining seven probes contained E-boxes with a single mismatch resulting in either the binding motif CACGTT or CACGGG. Both mismatched sequences represent so-called noncanonical E-box motifs that have been found to bind E-box-binding transcription factors in certain promoters (Harris et al. 2000; Coulson et al. 2003). Interestingly, the noncanonical E-box motif CAC GTT formed the strongest CLOCK/BMAL1 complex in the screen, and this motif has been reported by Yoo et al. (2005) to be bound by CLOCK/BMAL1 in vivo and to drive the expression of the Per2 gene in transgenic mice.
The identification of several well-known circadian transcription factors by DDDP demonstrates that the new method is well suited for the identification of differentially regulated DNA-binding proteins. As predicted, transcription factors that bind to simple recognition sites may be found multiple times in the random DNA library, potentially providing additional information about specific sequence requirements for binding. This was the case for the CLOCK/BMAL1 heterodimer, which was found to tolerate specifically the two sequence variants CACGTT and CACGGG. Moreover, sequence alignment revealed a clear sequence bias in the flanking regions. A consensus sequence for all E-box motifs found in the screen was created with WebLogo (Crooks et al. 2004) and is shown in Figure 3D.
A GC-rich element-binding protein displays differential DNA-binding activity throughout the day
Computational analysis revealed that the remaining probes fell into two groups. One group contained highly guanine-rich stretches on one DNA strand; for example, GGGGGTGG in probe 220 (Fig. 3E) and probe 285 or GGGGCGGGGG in probe 274, resembling GC- and GT-boxes, the preferred binding motifs of SP-like transcription factors (Suske 1999). Indeed, two different antibodies against SP1 resulted in a specific supershift of a circadian band from probe 220, whereas no supershift with anti-SP3 antibodies was observed (Fig. 3F). Although these findings pointed toward a role of SP1 in circadian transcription, we did not detect circadian binding of SP1 in vivo. Chromatin immunoprecipitation (ChIP) experiments on the promoter of the purported SP1 target gene Dhfr revealed SP1 binding specifically at its target sites; however, binding levels seemed to be constant throughout the day (data not shown). Likewise, pre-mRNA levels of the SP1 target genes Dhfr, Cdkn1a, and Bbc3 were analyzed and did not show any notable circadian regulation (data not shown). It is therefore not clear in the moment how the differential DNA binding that SP1 exhibits in vitro might translate to a circadian regulatory activity in vivo. Moreover, other transcription factors could be additionally involved in circadian binding of GC-rich probes, especially since not the entire fraction of the circadian DNA–protein complexes was supershifted by SP1 antibodies (Fig. 3F). Further experiments are necessary to address the contribution of potential binding partners or post-translational modifications to the function of SP1 at target gene promoters.
HSF1 is a circadian transcription factor
Our screen identified three circadian probes that did not contain binding motifs for known circadian transcription factors and that did not fall into the group of probes containing GC-rich sequence stretches. The computational analysis revealed in these probes multiple potential transcription factor-binding sites, making it impossible to test even a limited number of candidates in supershift experiments. Therefore we developed a short procedure to narrow down the exact position of a binding site within a random probe from 100 bp to 10–30 bp to facilitate further sequence analysis (Supplemental Fig. 2). Briefly, the two 50-bp random DNA regions of an insert were separately amplified by PCR and used as competitor DNA in an EMSA experiment with the original radioactive probe consisting of both 50-bp fragments. Subsequently, six 20-bp oligonucleotides spanning the sequence of the competing fragment were designed in such a way that each oligonucleotide overlaps by 10 bp with its counterpart on the opposite strand (Supplemental Fig. 2A). The oligonucleotides were annealed with each other or made double-stranded and were then used in various combinations as competitor DNA (Supplemental Fig. 2B).
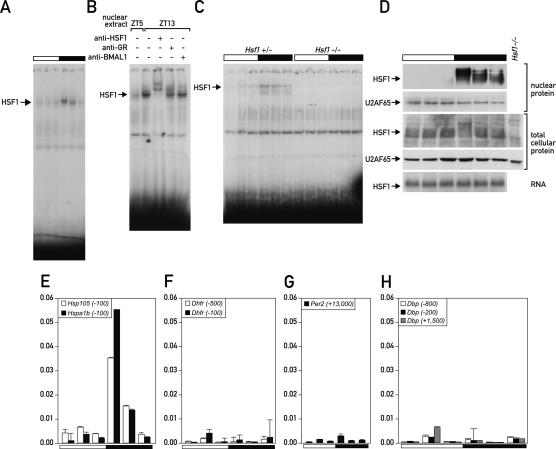
Probes 50, 317, and 325 (see Supplemental Table 1) formed a highly circadian DNA–protein complex with a maximal binding activity at ZT13 (Fig. 4A). Analysis of probe 50 with the procedure described above revealed a 20-bp sequence that was sufficient for efficient competition (Supplemental Fig. 2C). This sequence contained an almost perfect tripartite heat-shock element (HSE) (GTTCTAGAACTTGCC, mismatch is underlined), variations of which were also found in probes 317 and 325, similarly containing one mismatch each.
Figure 4.
HSF1 is a circadian transcription factor whose mRNA is constitutively expressed. (A) Circadian EMSA of probe 50 containing a tripartite HSE. (B) Supershift of probe 50 with antibodies against HSF1, GR, and BMAL1. (C) EMSA with nuclear extracts from livers of Hsf1+/− and Hsf1−/− mice. (D, top four panels) Western blot of nuclear and total cellular HSF1 protein with antibodies against HSF1 and against U2AF65 as a control for equal loading. The bottom panel shows a Northern blot of whole-cell Hsf1 mRNA. (E) ChIP of HSF1 on the promoters of its target genes Hsp105 and Hspa1b. (F–H) ChIP of HSF1 on the control genes Dhfr (F), Per2 (G), and Dbp (H). The numbers in parentheses after the gene names indicate the distance from the respective transcription start site.
HSEs are short inverted repeats of the sequence NGAAN that are found in multiple copies around the transcriptional start sites of heat-shock protein genes (Sakurai and Takemori 2007). The major inducible HSE-binding protein in mouse is HSF1, a member of the heat-shock factor family of transcriptional activators, which in mammals comprises additionally HSF2 and HSF4. In response to stress conditions that lead to intracellular protein denaturation, typically heat or oxidative stress, HSF1 becomes hyperphosphorylated, homotrimerizes, and is translocated into the nucleus, where it drives the transient expression of heat-shock proteins (Anckar and Sistonen 2007).
Supershifting with an anti-HSF1 antibody suggested that HSF1 was the major protein factor bound to the HSE-containing probes (Fig. 4B). In particular, no supershift was observed with an antibody against glucocorticoid receptor, whose binding sites can bear resemblance to HSEs (Luisi et al. 1991) and whose activity has been implicated in circadian transcription in liver (Balsalobre et al. 2000). Importantly, in nuclear extracts from livers of Hsf1−/− mice (Xiao et al. 1999), the HSE-binding activity was completely abolished (Fig. 4C), providing compelling evidence that HSF1 is indeed the circadian binding activity. Previous studies suggested that HSF1 is regulated exclusively at the post-transcriptional level, by chaperone interaction, post-translational modification, and nuclear import (Voellmy 2006). In keeping with these reports, we found Hsf1 mRNA levels as well as total cellular levels of HSF1 protein to be constant; however, as depicted in Figure 4D, the nuclear accumulation of hyperphosphorylated HSF1 was found to be highly circadian. Moreover, a major fraction of total HSF1 migrated more slowly at the time when nuclear HSF1 reached peak levels, presumably representing the nuclear, hyperphosphorylated form of HSF1 (Fig. 4D, cf. panels 1 and 3). Finally, we show that HSF1 binds to the promoters of heat-shock genes in a circadian manner in vivo. To this end, a ChIP experiment was performed with chromatin from mouse liver that was prepared at six time points around the clock. The promoters of two HSF1 target genes, Hsp105 and Hspa1b (Hsp70), were analyzed along with several control regions. Figure 4E shows that HSF1 binds strongly to the Hsp105 and Hspa1b promoters at the onset of the dark phase, whereas no binding to any of the control regions was detectable at any time point (Fig. 4F–H). Moreover, Hsp105 and Hspa1b have been described before to be circadianly transcribed with a phase compatible with that of HSF1 activity (Kornmann et al. 2007).
Feeding time and body temperature fluctuations affect the phase of circadian HSF1 activity
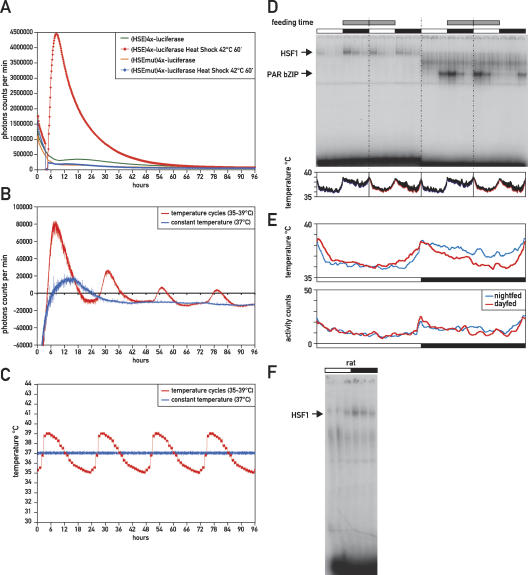
The circadian activation of HSF1, which bears all hallmarks of a regular heat-shock response, suggests that significant proteotoxic stress may be imposed day by day on the mammalian body. An obvious candidate for triggering HSF1 activation is the circadian fluctuations in body temperature, particularly since body temperature reaches peak values in the beginning of the dark phase (Damiola et al. 2000). We therefore asked if similar temperature fluctuations, which in mouse range approximately from 35°C to 39°C, are sufficient to drive rhythmic HSF1-dependent gene expression in tissue culture cells. To this end, a reporter plasmid was constructed that contained four copies of the HSE present in probe 50 (see Supplemental Table 1) in front of a minimal promoter driving a firefly luciferase gene. The responsiveness of this construct to temperature was verified by a 1-h heat shock at 42°C, which elicited a strong increase in luciferase expression compared with the expression levels obtained from a control plasmid with mutated HSEs (Fig. 5A; Supplemental Table 2). The cells were then subjected to circadian temperature fluctuations in the range between 35°C and 39°C (for details, see Materials and Methods), and luciferase expression as well as the actual temperature of the tissue culture medium was monitored in real time for 96 h (Fig. 5B,C). Luciferase expression from the mutated reporter construct was likewise measured and subtracted from the values obtained with the HSE-containing plasmid in order to account for HSE-independent effects on luciferase expression. The result shows that each increase in temperature was accompanied by a prominent peak of luciferase expression from the HSE-containing reporter plasmid, suggesting that physiologic temperature fluctuations can drive rhythmic HSF1 activity.
Figure 5.
Body temperature and feeding affect the phase of circadian HSF1 activity. (A) Real-time bioluminescence analysis of the HSE-luciferase reporter plasmids (HSE)4x-Luciferase and (HSEmut)4x-Luciferase in NIH3T3 mouse fibroblasts after a 60-min heat shock at 42°C or at constant temperature. (B) Real-time bioluminescence analysis of the HSE-luciferase reporter plasmid (HSE)4x-Luciferase in NIH3T3 cells that were entrained to temperature cycles (35°C–39°C) or kept at constant temperature (37°C). Luciferase expression of the non-temperature-responsive control plasmid (HSEmut)4x-Luciferase was subtracted, resulting in partially negative values for luciferase expression. (C) Real-time temperature monitoring of the tissue culture medium in the temperature entrainment experiment shown in B. (D) DNA-binding activity of HSF1 and PAR bZIP proteins in mice that were fed exclusively during the night or during the day. Feeding time is indicated by gray bars, while white and black bars indicate light and dark phases, respectively. Sampled real-time temperature measurements in living animals are shown below the autoradiograph. Colored lines indicate sampled body temperature of four night-fed (blue) or day-fed (red) animals during the last 7 d before they were sacrificed. Standard deviations are shown by black error bars on top of the colored lines. (E) The top panel shows an overlay of the sampled body temperature of night-fed (blue) and day-fed (red) animals. The bottom panel shows sampled activity counts for the same animals. (F) Circadian EMSA of HSF1 in rat.
Feeding has been shown to have a profound influence on the circadian clock and on body temperature in mammals (Damiola et al. 2000; Lowell and Spiegelman 2000). In order to examine a potential influence of feeding on the circadian phase of HSF1 activity, we performed a restricted feeding experiment in which one group of mice was fed exclusively during the night for 3 wk, and another group was fed exclusively during the day. In keeping with previously published results (Damiola et al. 2000), the phase of the circadian clock in liver adapted fully to the phase of the feeding regime, as exemplified here by a 12-h phase difference in the DNA-binding activity of PAR bZIP proteins between day-fed and night-fed animals (Fig. 5D). Moreover, shifting the onset of feeding to the beginning of the light phase induced a second peak of HSF1 activity at this time point (Fig. 5D). Interestingly, the peak at the beginning of the dark phase in night-fed animals is still present in day-fed animals, albeit to a somewhat lesser extent. In parallel, we measured body temperature in real time in a subgroup of both night-fed and day-fed animals that were used for the EMSA experiments. Mice that were fed exclusively during the day had a significantly lower body temperature during the night, whereas daytime temperature remained largely unaltered (Fig. 5D, bottom panel). However, the peaks of maximal body temperature persisted in day-fed animals, and only a sharper temperature increase toward the beginning of the light phase was observed (Fig. 5E). Moreover, the drop in nighttime temperature in day-fed animals did not seem to be a consequence of an overall lower nighttime activity level in these animals (Fig. 5E). Taken together, our experiments suggest that temperature and feeding strongly affect the phase of circadian HSF1 activity. Feeding might work at least partly through a modulation of body temperature, suggesting that temperature is a major entrainment factor for circadian HSF1 activity. Nevertheless, in rats, where fluctuations in body temperature have been reported to be significantly lower due to a higher body mass (Refinetti 1996), circadian DNA binding of HSF1 is as pronounced as in mice (Fig. 5F). This suggests that other stimuli besides heat, such as metabolic signals and/or cellular redox state, might contribute to circadian HSF1 activation (Yan et al. 2002; Ahn and Thiele 2003).
HSF1 influences the period length of the mammalian circadian clock
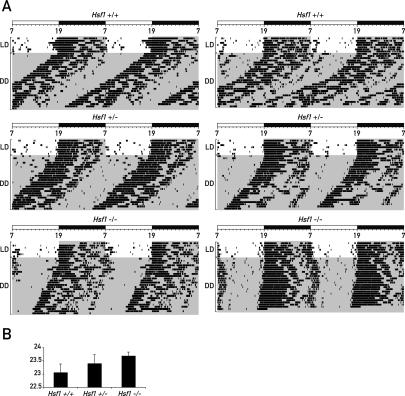
In order to examine whether HSF1 affects circadian behavior, we compared wheel-running activity of wild-type, heterozygous, and Hsf1-deficient mice in constant darkness (DD) (Fig. 6A). Under DD conditions, the average period length was markedly longer in Hsf1−/− mice than in heterozygous and wild-type littermates (Fig. 6B). Interestingly, the average period length of heterozygous Hsf1+/− mice lies exactly in between the period lengths of the wild-type and the knockout mice, indicating a dose-dependent effect of HSF1 on the core oscillator mechanism. Formally, we cannot exclude the possibility that another locus influencing period length cosegregates with the Hsf1 gene. Although a genome-wide complex trait analysis did, indeed, reveal a locus in close proximity to Hsf1 affecting running wheel activity (Shimomura et al. 2001), this locus appears to cause fragmentation rather than a lengthening in period duration of free-running locomotor activity. Furthermore, the corresponding allele turned out to be weakly penetrant and was only identified in a pairwise analysis together with another locus on a different chromosome.
Figure 6.
Circadian locomotor activity in mice lacking one (Hsf1+/−) or both (Hsf1−/−) copies of the Hsf1 gene compared with wild-type mice (Hsf1+/+). (A) The voluntary locomotor activity was recorded as wheel-running activity for wild-type Hsf1+/+ mice, heterozygous Hsf1+/− mice, and homozygous Hsf1−/− mice in DD. Typical double-plot actograms obtained for two animals of each genotype are shown. In each actogram, the first few days were recorded under light–dark (LD) conditions (lights on at 7:00; lights off at 19:00). Time spans during which the lights were switched off are marked by gray shading. (B) Period length of wild-type, heterozygous, and homozygous mice in DD. At least 12 animals were analyzed per genotype. The lengths of the free-running periods in hours were 23.05 ± 0.32 (Hsf1+/+), 23.39 ± 0.33 (Hsf1+/−), and 23.68 ± 0.14 (Hsf1−/−). The Student’s t-test was used to examine the data, and the P values for the period length differences compared with Hsf1+/+ mice are 0.008 for Hsf1+/− mice and 2.7 × 10−7 for Hsf1−/− mice.
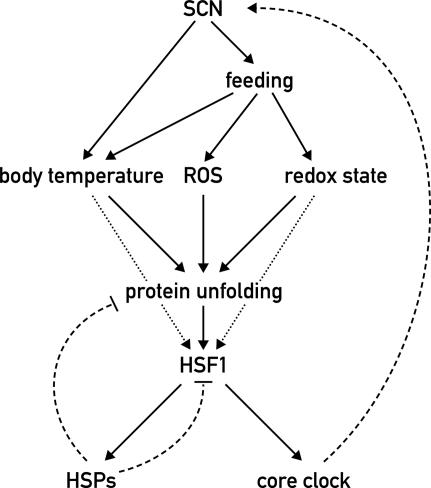
Taken together, our data suggest that circadian HSF1 activity plays a dual role for body homeostasis in mice: HSF1 exerts a cytoprotective function through the induction of heat-shock proteins and at the same time may assist the synchronization of the molecular clock to behavior and metabolism (see Fig. 7; Discussion).
Figure 7.
Model for the integration of HSF1 into the circadian network. In the mammalian body, the SCN regulate virtually all clock outputs, including temperature rhythms and feeding/fasting cycles. Elevated temperature directly triggers protein unfolding, whereas food processing leads to an increased production of ROS and changes in the cellular redox state. The oxidation of cysteine residues of polypeptides by ROS likewise promotes protein denaturation. Unfolded proteins induce HSF1 activity, which in turn promotes the expression of heat-shock proteins to counteract protein denaturation and to down-regulate HSF1 activity in a negative feedback loop (broken arrow). Additionally, HSF1 is itself a redox and temperature sensor under physiological conditions (dotted arrows) (Yan et al. 2002; Ahn and Thiele 2003). HSF1 also directly affects the core clock mechanism to synchronize circadian rhythms with animal behavior and physiology (dashed arrow).
Discussion
DDDP is an activity-based screen for differentially regulated transcription factors
The circadian clock network of mammals, like many other biological systems, is under active investigation by high-throughput genomics and proteomics methods. Transcriptional profiling has been repeatedly used to determine the circadian profile of mRNA levels in the SCN and in peripheral organs (see De Haro and Panda 2006 and references therein). In the tissues examined so far, ∼2%–10% of the genome seems to be under circadian control, while circadian transcription is at the same time highly tissue-specific; marked differences have been observed between the SCN and peripheral organs as well as between different peripheral tissues (Kornmann et al. 2001, 2007; Akhtar et al. 2002; Duffield et al. 2002; Panda et al. 2002; Storch et al. 2002; McCarthy et al. 2007; Miller et al. 2007). Many circadian output genes seem therefore to be regulated in a tissue-specific manner rather than exclusively by shared oscillator components.
Nevertheless, transcriptional regulatory proteins are often constantly expressed and rely heavily on post-translational mechanisms for activation. Hence, the identification of circadian transcription factors requires screens for rhythmic activity on the protein level. An efficient analysis by high-throughput proteomics methods is, however, complicated by the low abundance of these proteins and by the possibility that only a fraction of the cellular pool of a transcription factor might be converted to a transcriptionally active form. Indeed, a recently performed analysis of the circadian proteome of mouse liver by a combination of two-dimensional gel electrophoresis and mass spectrometry did not efficiently detect circadian transcription factors. Although the study clearly demonstrated extensive circadian regulation of the cellular proteome, the majority of analyzed proteins consisted of highly expressed, soluble proteins (Reddy et al. 2006).
We therefore decided to develop a novel method based on a functional assay that selectively screens for circadian DNA-binding activity and that is not easily confounded by proteins that are irrelevant in the context of gene regulation. For most known transcription factors, DNA binding goes hand in hand with transcriptional activity, although in some cases transcription factors can be regulated at a step subsequent to DNA binding (Calkhoven and Ab 1996). Nevertheless, a circadian pattern on an EMSA gel is likely to reflect the circadian activity of a transcription factor, regardless of the number and nature of regulatory steps that were required for its activation. Proof of principle for our experimental setup was provided by the identification of various circadian proteins that rely on entirely disparate modes of activation. Two regulatory extremes are exemplified by DBP on the one hand, and HSF1 on the other (see Figs. 3, 4). Circadian DBP oscillation has been shown previously to be the result of highly rhythmic transcription rates and mRNA accumulation (Wuarin and Schibler 1990, 1994). In contrast, we found the levels of Hsf1 mRNA to be constant throughout the day and the circadian formation of functionally active HSF1 complexes to be triggered exclusively by post-transcriptional mechanisms. EMSA has the additional advantage that it reveals certain properties of transcription factors in addition to their temporal accumulation. For example, the RF value of a DNA–protein complex may vary according to daytime, as was observed for CLOCK/BMAL1 heterodimers, indicating circadian oscillation of post-translational modifications and/or the association of additional cofactors.
From differentially bound DNA elements to novel circadian transcription factors
The initial round of DDDP screening resulted in a collection of circadian EMSA probes. These probes contained with high probability short sequence motifs that confer circadian transcriptional regulation onto target genes in vivo, either alone or in combination with other elements. The sequences of these probes became immediately accessible by sequencing the inserts of the corresponding clones of the random DNA library. In principle, this information can then be used in two ways. Firstly, knowledge of its DNA-binding sequence usually greatly accelerates the identification of an unknown transcription factor. Databases exist for several model organisms that list transcription factors according to their DNA consensus sites. Moreover, we facilitated the identification process further by establishing an easy two-step procedure that allows narrowing down any binding site obtained in the screen from 100 bp to 10–30 bp in a reasonably short time. A computational search for transcription factor-binding sites results in fewer false-positive candidates when a short sequence is analyzed, and for proteins that bind to more than one probe, sequence comparison will result in a consensus DNA-binding motif. Using this approach, we found HSEs in the sequences of three probes yielding circadian EMSA complexes, which quickly led to the identification of HSF1 as the corresponding DNA-binding activity. In cases in which no obvious candidate can be attributed to a binding sequence, the binding element could be used for the affinity purification of the cognate protein, which can then be identified by various mass spectrometry methods.
In our screen, we found several circadian probes to contain highly GC-rich sequence stretches that are typically bound by the transcription factor SP1. Although we were not yet able to obtain evidence for circadian regulation of SP1 target genes in vivo, a bioinformatics analysis of the GC-rich binding sequences might still provide clues about their circadian regulatory potential. In this context, it might be interesting to extend the search for circadian E-boxes to sequence motifs beyond the canonical E-box and to explore the contribution of flanking sequences. A long-standing question in the field of transcriptional regulation is how a short and statistically extremely abundant sequence motif such as the E-box can be selectively used by various transcription factors in different regulatory contexts. A CT-rich sequence element that has the ability to confer robust BMAL1/CLOCK responsiveness onto an adjacent E-Box has already been found in the mouse arginine vasopressin promoter (Munoz et al. 2006). In the course of our analysis of circadian E-box motifs found by DDDP, we identified a clear sequence preference in the flanking region that might help in selecting CLOCK/BMAL1 among the many E-box-binding proteins.
A mechanism for circadian cytoprotection?
Clearly the most interesting result of our screen was the identification of HSF1 as a circadian transcription factor. Initially, sequence analysis of three EMSA probes that displayed maximal DNA-binding activity at the onset of the dark phase revealed a tripartite HSE. In a supershift experiment with anti-HSF1 antibodies, we identified HSF1 as the major binding factor for these elements, and in agreement with previous studies, we found post-transcriptional mechanisms to be responsible for its circadian DNA-binding activity. Furthermore, our results suggest that HSF1 confers circadian regulation to its target genes in vivo. As revealed by ChIP assays, the binding of HSF1 to recognition sites in the promoters of the heat-shock genes Hsp105 and Hspa1b is highly circadian and coincides with the peak of DNA binding in vitro. Importantly, the phase of HSF1 occupancy is in keeping with the phase of mRNA accumulation of various HSF1 target genes; e.g., Hsp105, Hspa1b, Hsp90, and Stip1 in mouse liver (Kornmann et al. 2007). Also the mRNA encoding CaMKII, which potentiates the transcriptional activity of HSF1 by phosphorylating it at Ser 230 (Holmberg et al. 2001), is diurnally expressed and in phase with maximal HSF1 activity (Kornmann et al. 2007). Taken together, the processes following the circadian activation of HSF1 highly resemble the ones during the classical heat-shock response. A likely explanation for this somewhat surprising finding is that HSF1 might be required to counteract proteotoxic side effects that accompany the circadian peak in body temperature at the onset of the dark phase. Interestingly, this increase in body temperature seems to be a regulated process that is connected to the circadian clock. Supporting evidence for this hypothesis was provided by Feillet et al. (2006), who found that Per2 knockout mice fail to display food anticipatory activity and lack the accompanying increase in core body temperature. Furthermore, feeding has been shown to induce HSF1 in the mouse liver and intestine (Katsuki et al. 2004). In keeping with these reports, we found temperature fluctuations in a range in which they occur in living animals to be able to entrain circadian HSF1 activation. In addition, restricted feeding strongly affects the phase of HSF1 activity, and the kinetics of body temperature fluctuations suggests that feeding works at least in part through a modulation of core body temperature.
The model in Figure 7 summarizes our conclusions about the role of HSF1 in the regulation of circadian physiology. An increase in the amount of unfolded proteins is considered to be the primary trigger for HSF1 activation. Several different physiological parameters that are themselves subjected to circadian regulation directly and indirectly affect the level of denatured proteins in the cell. An elevated temperature and an oxidizing environment lead to protein denaturation through protein unfolding and the oxidation of cysteines, respectively. In mice, feeding is accompanied by a rise in core body temperature and probably leads to increased levels of reactive oxygen species (ROS), which causes protein denaturation via the oxidation of amino acid side chains. HSF1 senses this proteotoxic stress and counteracts it by the up-regulation of protein chaperones and potentially other cytoprotective proteins. In addition, HSF1 might assist in the temperature-dependent synchronization of peripheral circadian oscillators (Brown et al. 2002; C. Saini, H. Reinke, and U. Schibler, unpubl.) by adjusting them according to physiological challenges leading to protein denaturation. In the SCN harboring the master pacemaker, this feedback mechanism may also be intertwined with the core oscillator mechanism, given that the disruption of Hsf1 leads to an altered period length in locomotor activity. One possible scenario explaining the function of HSF1 on the molecular level would be that HSF1-induced chaperones assist in the folding of core clock proteins, thereby leading to an interlocking of the HSF1–HSP cycle and circadian feedback loops (Fig. 7).
A major role for HSF1 outside the classical stress response emerged recently from work in Caenorhabditis elegans. The activity of two transcription factors, HSF1 and DAF-16, turned out to be essential in preventing the formation of intracellular protein aggregates that lead to neurodegeneration and aging (Cohen et al. 2006). However, under normal stress-free conditions, HSF1 is supposedly kept in the transcriptionally inactive form and is therefore unable to assist in this process. The circadian activation of HSF1 might provide a way to guarantee higher levels of the required target genes by turning on their expression at least once per day. Indeed, overexpression of a constantly active form of HSF1 can suppress polyglutamine aggregate formation in cultured cells and in mice (Fujimoto et al. 2005), and Hsf1-deficient mice have very recently been shown to display several symptoms that are classical hallmarks of degenerative diseases of the central nervous system (Homma et al. 2007).
Prospects and limitations of DDDP
Like any other screening method, DDDP also has its limitations. For several reasons, it should be difficult to perform DDDP in a saturating way. Some transcription factors may have special or even opposing requirements for in vitro DNA binding regarding salt concentration, temperature, cofactors, etc. Furthermore, the DNA library has to be limited to a manageable size. Our initial calculation showed a probability of >99% for the presence of a functional binding sequence for a typical eukaryotic transcription factor in <11,000 bp of random DNA sequence. Some transcription factors have more complex binding sites; other transcription factors might tolerate mismatches at only a limited number of positions in their binding sequence. Nevertheless, we would like to point out that our screen identified those circadian transcription factors that are known to cycle with high amplitude in peripheral tissues; e.g., CLOCK/BMAL1, REV-ERBα, and DBP. We speculate therefore that the number of strongly circadian transcription factors is limited since the remaining probes were either bound by HSF1 or fell into the group of probes containing GC-rich binding sequences. Given that most cyclically active genes were also expressed in a cell-type-specific manner (see above), it was surprising that our approach did not unveil liver-specific transcription factors. Conceivably, the circadian activity of hepatocyte-specific genes may involve the synergistic action between ubiquitously expressed circadian transcription factors (e.g., BMAL1, CLOCK, REV-ERBα, and DBP) and cell-type-specific transcription factors accumulating throughout the day (e.g., Hepatocyte Nuclear Factors 1, 3, and 4).
We believe that due to the easy accessibility of the required starting material, DDDP should be applicable to virtually every biological system that allows the preparation of concentrated protein extracts, preferably nuclear (and, perhaps, whole cell) extracts. We already demonstrated the presence of binding sites for SRF and NF-κB in the random DNA library, two transcription factors involved among other processes in muscle development and inflammation.
Materials and methods
DDDP
An oligonucleotide population with the sequence 5′-ATTGGC GCGCC-[N]50-AAGGGATCCCTT-3′ was dissolved in 100 mM NaCl and self-annealed by incubation in a beaker with 100 mL of boiling water that was cooled down to 4°C in the course of several hours. Ten micrograms of annealed oligonucleotide were incubated with 15 μL of Klenow fill-in buffer, 1.2 μL of 25 mM dNTPs, and 5 μL of 5 U/μL Klenow fragment in a volume of 150 μL for 30 min at room temperature. The resulting dsDNA fragment was cut with AscI, gel-purified, and cloned into the vector pUC19AscI, which had been generated by ligating the 5′-phosphorylated oligonucleotide 5′-TCGAGGCGCGCC-3′ into the SalI site of pUC19, thereby generating an AscI site in pUC19. Minipreps of 400 clones were prepared that constituted the random DNA library.
EMSA probes were prepared from each clone by PCR amplification of the random DNA inserts with the primers rand100-A, 5′-AATTCGAGCTCGGTACCC-3′; and rand100-B, 5′ACCATGATTACGCCAAGC-3′. The PCR products were gel-purified and subsequently cleaved with the restriction enzymes AscI and BamHI. Three microliters of cleaved DNA fragments (10 ng/μL) were incubated with 0.4 μL of Klenow fill-in buffer, 0.2 μL of 5 mM dATP/dGTP/dTTP, 0.2 μL of 3000 Ci/mmol α32-dCTP, and 0.2 μL of 5 U/μL Klenow fragment for 15 min at room temperature. Master mixes without DNA were prepared on ice beforehand, and 1 μL of each was added to the DNA to avoid pipetting of minuscule volumes. Unincorporated radioactivity could largely be separated from the probes by precipitation with 95 μL of 75% ethanol at room temperature. After centrifugation for 15 min, the probes were resuspended in 8 μL of H2O.
Liver nuclei were prepared at different circadian time points according to the method described in Lavery and Schibler (1993). Nuclear proteins were extracted in NUN buffer containing 0.3 M NaCl, 1 M urea, and 1% Nonidet P-40 for 20 min on ice. Centrifugation for 10 min at 4°C left the core histones and histone H1 in the pellet and soluble nonhistone proteins in the supernatant. Nuclear extracts typically contained proteins at a concentration of 6–8 μg/μL.
EMSA reactions were performed with 5 μg of nuclear proteins in 25 mM HEPES-KOH (pH 7.6), 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 200 ng/μL sheared salmon sperm DNA, 50 ng/μL poly(dI-dC), and 1 μL of radioactive probe in a volume of 10 μL. One microliter of loading dye (15% Ficoll, 0.4% Orange G) was added, and the reaction mixes were loaded on 4% polyacrylamide gels. Gels were run for 3 h at room temperature (7.5 V/cm) and vacuum-dried for exposure on a film or PhosphorImager screen.
Supershift and DNA-competition experiments
EMSA experiments were essentially performed as described above. All oligonucleotide sequences are listed in Supplemental Table 2. For supershift experiments, 1 μL of antiserum or purified antibody was added immediately before the addition of the radioactive probe. The antibodies used were anti-SP1, anti-SP3 (G. Suske, University of Marburg; Upstate Biotechnology), anti-HSF1 (Assay Designs), anti-GR (Santa Cruz Biotechnology), anti-BMAL1, and anti-CLOCK (Ripperger et al. 2000).
In DNA competition experiments, cold competitor DNA was added before the addition of the radioactive probe and incubated for 10 min at room temperature.
Western blot experiments
Western blots were performed according to a standard protocol (Sambrook and Russel 2000) with antibodies against HSF1 (Assay Designs) and U2AF65 (Sigma-Aldrich). For an optimal resolution of phosphorylated HSF1-isoforms, 7% polyacrylamide gels were used.
Northern blot experiments
Mouse liver RNA was extracted as described before (Fonjallaz et al. 1996); 5 μg of RNA were used per gel lane, and blotting and hybridization were performed according to a standard protocol (Sambrook and Russel 2000).
RNA analysis by real-time quantitative PCR
Mouse liver RNA extraction and transcript quantification by TaqMan real-time PCR technology was performed as described in Preitner et al. (2002). Briefly, 1 μg of total RNA was reverse-transcribed using M-MLV Reverse Transcriptase (Invitrogen) and random hexamers. The resulting cDNA was PCR-amplified in an ABI PRISM 7700 Sequence Detection System from PE-Applied Biosystems (Heid et al. 1996). Primers and probes are listed in Supplemental Table 2.
ChIP analysis
ChIP analyses were performed as described in Ripperger and Schibler (2006). Immunoprecipitated DNA was quantitatively measured by the ABI PRISM 7700 Sequence Detection System using the primers and TaqMan probes listed in Supplemental Table 2.
Real-time bioluminescence and temperature monitoring of tissue culture cells
HSE-luciferase reporter plasmids were constructed by cloning either an oligonucleotide containing the HSE element found in the DDDP screen (HSE) or a mutated version of this element (HSEmut) (Supplemental Table 2) flanked by NheI and HindIII restriction sites into the vector pGL4.23 (Promega), which contains a minimal promoter upstream of the luc reporter gene.
NIH-3T3 mouse fibroblasts were cultured in DMEM medium containing 10% fetal bovine serum (FBS) and 1% pigskin gelatin (PSG). Cells were transfected with the reporter constructs using FuGene6 Reagent (Roche), according to the manufacturer’s instructions. Two days post-transfection, the medium was changed to fresh phenol red-free DMEM medium, supplemented with 2% FBS and 0.1 mM luciferin, and cells were placed in a light-tight incubator to measure bioluminescence. Photon counts were integrated during 1 min, every 1 or 10 min, using Hamamatsu photomultiplier tube (PMT) detector assemblies (Hamamatsu).
A 1-h heat shock was performed in a separate incubator preheated at 42°C. Temperature cycles resembling those measured by telemetry in mice were generated using a homemade system. This computer-assisted, programmable incubator, which allows the generation of temporal temperature profiles with an accuracy of 0.1°C and the recording of bioluminescence in real time, was designed by André Liani and constructed by Yves-Alain Poget, both members of the mechanical workshop at the Department of Molecular Biology, University of Geneva (C. Saini, H. Reinke, A. Liani, Y.-A. Poget, and U. Schibler, unpubl.).
Animal care, monitoring of wheel-running activity, and real-time in vivo body temperature and activity measurements
The mice were housed and their wheel-running activity monitored as described in Lopez-Molina et al. (1997). Hsf1 knockout mice were first described in Xiao et al. (1999). Telemetric recording of body temperature and spontaneous activity was performed as described before (Brown et al. 2002) with software and PDT-4000 transmitter hardware from Minimitter Corporation.
Acknowledgments
We thank André Liani and Yves-Alain Poget for designing and constructing the bioluminescence recording systems, Guntram Suske (University of Marburg) for antibodies against SP1 and SP3, Jürgen Ripperger (University of Fribourg) for his gift of cross-linked chromatin and purified anti-CLOCK and anti-BMAL1 antibodies, Nicolas Roggli for the expert preparation of figures, and David Gatfield and Gad Asher for helpful comments on the manuscript. This research was supported by the Canton of Geneva, the Swiss National Science Foundation (through an individual research grant to U.S. and the National Center of Competence in Research program Frontiers in Genetics), the Louis Jeantet Foundation of Medicine, the BonizziTheler Stiftung, and the 6th European Framework Project EUCLOCK. H.R. received long-term fellowships from EMBO and HFSP.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.453808
References
- Ahn S.G., Thiele D.J. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes & Dev. 2003;17:516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar R.A., Reddy A.B., Maywood E.S., Clayton J.D., King V.M., Smith A.G., Gant T.W., Hastings M.H., Kyriacou C.P. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Anckar J., Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv. Exp. Med. Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Damiola F., Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Brown S.A., Marcacci L., Tronche F., Kellendonk C., Reichardt H.M., Schutz G., Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Berg J., Willmann S., Lassig M. Adaptive evolution of transcription factor binding sites. BMC Evol. Biol. 2004;4:42. doi: 10.1186/1471-2148-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Brown S.A., Zumbrunn G., Fleury-Olela F., Preitner N., Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr. Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- Calkhoven C.F., Ab G. Multiple steps in the regulation of transcription-factor level and activity. Biochem. J. 1996;317:329–342. doi: 10.1042/bj3170329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E., Bieschke J., Perciavalle R.M., Kelly J.W., Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Coulson J.M., Edgson J.L., Marshall-Jones Z.V., Mulgrew R., Quinn J.P., Woll P.J. Upstream stimulatory factor activates the vasopressin promoter via multiple motifs, including a non-canonical E-box. Biochem. J. 2003;369:549–561. doi: 10.1042/BJ20021176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haro L., Panda S. Systems biology of circadian rhythms: An outlook. J. Biol. Rhythms. 2006;21:507–518. doi: 10.1177/0748730406294767. [DOI] [PubMed] [Google Scholar]
- Duffield G.E., Best J.D., Meurers B.H., Bittner A., Loros J.J., Dunlap J.C. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Eisner V., Quiroga C., Criollo A., Eltit J.M., Chiong M., Parra V., Hidalgo K., Toro B., Diaz-Araya G., Lavandero S. Hyperosmotic stress activates p65/RelB NFκB in cultured cardiomyocytes with dichotomic actions on caspase activation and cell death. FEBS Lett. 2006;580:3469–3476. doi: 10.1016/j.febslet.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Ellis P.D., Martin K.M., Rickman C., Metcalfe J.C., Kemp P.R. Increased actin polymerization reduces the inhibition of serum response factor activity by Yin Yang 1. Biochem. J. 2002;364:547–554. doi: 10.1042/BJ20020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvey E., Marcacci L., Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: A single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol. Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- Feillet C.A., Ripperger J.A., Magnone M.C., Dulloo A., Albrecht U., Challet E. Lack of food anticipation in Per2 mutant mice. Curr. Biol. 2006;16:2016–2022. doi: 10.1016/j.cub.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Fonjallaz P., Ossipow V., Wanner G., Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- Forman B.M., Chen J., Blumberg B., Kliewer S.A., Henshaw R., Ong E.S., Evans R.M. Cross-talk among ROR α 1 and the Rev-erb family of orphan nuclear receptors. Mol. Endocrinol. 1994;8:1253–1261. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Takaki E., Hayashi T., Kitaura Y., Tanaka Y., Inouye S., Nakai A. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J. Biol. Chem. 2005;280:34908–34916. doi: 10.1074/jbc.M506288200. [DOI] [PubMed] [Google Scholar]
- Gachon F., Fonjallaz P., Damiola F., Gos P., Kodama T., Zakany J., Duboule D., Petit B., Tafti M., Schibler U. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes & Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F., Olela F.F., Schaad O., Descombes P., Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Gallego M., Virshup D.M. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Hardin P.E. Essential and expendable features of the circadian timekeeping mechanism. Curr. Opin. Neurobiol. 2006;16:686–692. doi: 10.1016/j.conb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Hardin P.E., Hall J.C., Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Harris V.K., Coticchia C.M., List H.J., Wellstein A., Riegel A.T. Mitogen-induced expression of the fibroblast growth factor-binding protein is transcriptionally repressed through a non-canonical E-box element. J. Biol. Chem. 2000;275:28539–28548. doi: 10.1074/jbc.M001677200. [DOI] [PubMed] [Google Scholar]
- Heid C.A., Stevens J., Livak K.J., Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Holmberg C.I., Hietakangas V., Mikhailov A., Rantanen J.O., Kallio M., Meinander A., Hellman J., Morrice N., MacKintosh C., Morimoto R.I., et al. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 2001;20:3800–3810. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma S., Jin X., Wang G., Tu N., Min J., Yanasak N., Mivechi N.F. Demyelination, astrogliosis, and accumulation of ubiquitinated proteins, hallmarks of CNS disease in hsf1-deficient mice. J. Neurosci. 2007;27:7974–7986. doi: 10.1523/JNEUROSCI.0006-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuki K., Fujimoto M., Zhang X.Y., Izu H., Takaki E., Tanizawa Y., Inouye S., Nakai A. Feeding induces expression of heat shock proteins that reduce oxidative stress. FEBS Lett. 2004;571:187–191. doi: 10.1016/j.febslet.2004.06.087. [DOI] [PubMed] [Google Scholar]
- Kornmann B., Preitner N., Rifat D., Fleury-Olela F., Schibler U. Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res. 2001;29:e51. doi: 10.1093/nar/29.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Schaad O., Bujard H., Takahashi J.S., Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery D.J., Schibler U. Circadian transcription of the cholesterol 7 α hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes & Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- Lee C., Etchegaray J.P., Cagampang F.R., Loudon A.S., Reppert S.M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Liu A.C., Welsh D.K., Ko C.H., Tran H.G., Zhang E.E., Priest A.A., Buhr E.D., Singer O., Meeker K., Verma I.M., et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Conquet F., Dubois-Dauphin M., Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell B.B., Spiegelman B.M. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- Luisi B.F., Xu W.X., Otwinowski Z., Freedman L.P., Yamamoto K.R., Sigler P.B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Makoto M., Takuji N. Mersenne twister: A 623-dimensionally equidistributed uniform pseudo-random number generator. ACM Trans. Model. Comput. Simul. 1998;8:3–30. [Google Scholar]
- McCarthy J.J., Andrews J.L., McDearmon E.L., Campbell K.S., Barber B.K., Miller B.H., Walker J.R., Hogenesch J.B., Takahashi J.S., Esser K.A. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B.H., McDearmon E.L., Panda S., Hayes K.R., Zhang J., Andrews J.L., Antoch M.P., Walker J.R., Esser K.A., Hogenesch J.B., et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz E., Brewer M., Baler R. Modulation of BMAL/CLOCK/E-box complex activity by a CT-rich cis-acting element. Mol. Cell. Endocrinol. 2006;252:74–81. doi: 10.1016/j.mce.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Panda S., Antoch M.P., Miller B.H., Su A.I., Schook A.B., Straume M., Schultz P.G., Kay S.A., Takahashi J.S., Hogenesch J.B. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Panda S., Sato T.K., Hampton G.M., Hogenesch J.B. An array of insights: Application of DNA chip technology in the study of cell biology. Trends Cell Biol. 2003;13:151–156. doi: 10.1016/s0962-8924(03)00006-0. [DOI] [PubMed] [Google Scholar]
- Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Reddy A.B., Karp N.A., Maywood E.S., Sage E.A., Deery M., O’Neill J.S., Wong G.K., Chesham J., Odell M., Lilley K.S., et al. Circadian orchestration of the hepatic proteome. Curr. Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Refinetti R. Comparison of the body temperature rhythms of diurnal and nocturnal rodents. J. Exp. Zool. 1996;275:67–70. doi: 10.1002/(SICI)1097-010X(19960501)275:1<67::AID-JEZ10>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Reppert S.M., Weaver D.R. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Ripperger J.A., Schibler U. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Ripperger J.A., Shearman L.P., Reppert S.M., Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes & Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Takemori Y. Interaction between heat shock transcription factors (HSFs) and divergent binding sequences: Binding specificities of yeast HSFs and human HSF1. J. Biol. Chem. 2007;282:13334–13341. doi: 10.1074/jbc.M611801200. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Saper C.B., Lu J., Chou T.C., Gooley J. The hypothalamic integrator for circadian rhythms. Trends Neurosci. 2005;28:152–157. doi: 10.1016/j.tins.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Shimomura K., Low-Zeddies S.S., King D.P., Steeves T.D., Whiteley A., Kushla J., Zemenides P.D., Lin A., Vitaterna M.H., Churchill G.A., et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11:959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- Storch K.F., Lipan O., Leykin I., Viswanathan N., Davis F.C., Wong W.H., Weitz C.J. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Ueda H.R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Voellmy R. Feedback regulation of the heat shock response. Handb. Exp. Pharmacol. 2006;172:43–68. doi: 10.1007/3-540-29717-0_2. [DOI] [PubMed] [Google Scholar]
- Welsh D.K., Yoo S.H., Liu A.C., Takahashi J.S., Kay S.A. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J., Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63:1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]
- Wuarin J. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: Evidence for cotranscriptional splicing. Mol. Cell. Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Zuo X., Davis A.A., McMillan D.R., Curry B.B., Richardson J.A., Benjamin I.J. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Numano R., Abe M., Hida A., Takahashi R., Ueda M., Block G.D., Sakaki Y., Menaker M., Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yan L.J., Christians E.S., Liu L., Xiao X., Sohal R.S., Benjamin I.J. Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO J. 2002;21:5164–5172. doi: 10.1093/emboj/cdf528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.H., Ko C.H., Lowrey P.L., Buhr E.D., Song E.J., Chang S., Yoo O.J., Yamazaki S., Lee C., Takahashi J.S. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]