Abstract
DNA replication origins (ORI) are regulatory regions from which the genome is replicated once every cell cycle. A widely used method for their identification in mammalian chromosomes relies on quantitative PCR of DNA nascent strands across candidate regions. We developed a new high-resolution PCR strategy to localize ORIs directly on total unfractionated human DNA. The increase in sensitivity provided by this approach has revealed that a short region of ∼200-base-pair overlapping well-characterized replication origins undergoes several rounds of replication, coinciding with their specific time of activation during S phase. This process generates a population of discrete dsDNA fragments detectable as free molecules in preparations of total DNA in normally proliferating cells. Overreplicated regions have precise boundaries at the edge of the nucleosome-free gap that encompasses the transcription initiation sites of CpG island promoters. By itself, active transcription does not induce overreplication but does stimulate it at ORIs associated with promoters. The coincidence in time and space between the overproduction of short DNA fragments and ORI activity predicts the precise localization of thousands of ORIs in the human genome and uncovers a previously unnoticed step in the initiation of DNA replication.
[Keywords: Replication origins, CpG islands, promoters, transcription, cell cycle, chromatin structure]
DNA replication origins (ORI) are regulatory regions that control the correct initiation of DNA replication during the cell cycle. Unscheduled ORI activity results in abnormal replication patterns and genomic instability. Despite their importance, to date only a small number of mammalian ORIs have been characterized, probably owing to the absence of a functional assay for their direct identification in complex populations of genomic fragments. However, three recently developed strategies based on subtractive hybridization of libraries of nascent DNA strands (Todorovic et al. 2005), direct isolation of restriction fragments containing replication bubbles (Mesner et al. 2006), and microarray hybridization of DNA nascent strands (Lucas et al. 2007) will likely contribute to expanding the catalog of mammalian ORIs. In contrast with the limited information about ORI localization, the protein complexes that regulate their activity are comparatively better known. ORIs are licensed for replication by the sequential recruitment of the origin recognition complex (ORC), Cdc6, Cdt1, and the MCM 2–7 hexamer to form the prereplication complex (pre-RC) (DePamphilis et al. 2006). Upon ORI firing, rereplication is prevented by the maintenance of high cyclin-dependent kinase activity and through the modulation of the stoichiometry of the complex formed by Cdt1 and its inhibitor geminin (Blow and Dutta 2005; Lutzmann et al. 2006).
How pre-RC complexes are targeted to ORIs is a major unresolved issue in the field of replication. In Xenopus egg extracts, transcription complexes have been shown to contribute to ORI positioning in plasmids (Danis et al. 2004), and recent studies in Drosophila and human cells (MacAlpine et al. 2004; White et al. 2004) have confirmed at genome-wide scale the link between transcription and early replication reported previously for a more limited number of examples (Goldman et al. 1984). Mammalian ORIs lack consensus sequence elements, and ORC shows no preferential affinity for ORI sequences, which suggests that epigenetic factors probably play an important role in ORI specification (Vashee et al. 2003; Antequera 2004).
Several of the mammalian ORIs identified to date are close to CpG island promoters (Giacca et al. 1994; Delgado et al. 1998; Keller et al. 2002; Gómez and Brockdorff 2004; Gray et al. 2007). However, the well-characterized human β-globin ORI (Kitsberg et al. 1993) does not colocalize with a CpG island, and examples of ORIs distant from known promoters have also been reported (Mesner and Hamlin 2005; Todorovic et al. 2005). In all these cases, and in another 28 human ORIs recently identified (Lucas et al. 2007), replication origins have been found to map to discrete genomic regions. In contrast, large regions containing multiple initiation sites have also been described downstream from the hamster DHFR gene (Hamlin and Dijkwel 1995), in the mouse immunoglobulin heavy chain locus (Norio et al. 2005), and in human primary keratinocytes (Lebofsky et al. 2006). We analyzed in detail the initiation of DNA replication at several human ORIs associated with promoters by a new quantitative PCR (Q-PCR) approach, and we report here that ORI firing during S phase is accompanied by the overproduction of short double-stranded fragments encompassing these regions.
Results
Overrepresentation of DNA fragments at DNA replication origins
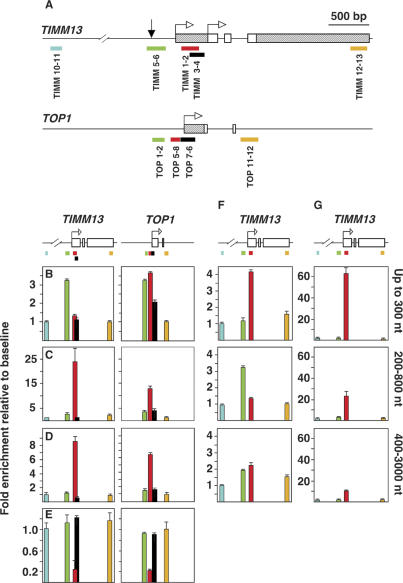
Q-PCR defines ORIs as regions preferentially amplified in relation to their flanking sequences in preparations of purified DNA nascent strands. Differences in PCR efficiency between different primer sets are corrected by normalization against total genomic DNA. However, since total genomic DNA usually contains replicating DNA molecules, the closer a region is to an ORI, the higher its abundance will be as a template. This implies that the efficiency of the primers covering these regions will be overestimated and, as a consequence of normalization, the amount of DNA nascent strands close to the ORI will be underestimated. We reasoned that the use of the cloned versions of the loci to be analyzed instead of total genomic DNA for normalization would circumvent this limitation and improve the accuracy of the measurements. We applied this strategy to two well-characterized human ORIs adjacent to the promoters of the TIMM13 gene (known as Lamin B2 ORI) and of the TOP1 gene (Fig. 1A). Total heat-denatured DNA from human HEK293 cells was run in a neutral sucrose gradient, and fractions enriched in nascent strands between 200 and 800 nucleotides (nt) long (Supplemental Fig. 1A, fractions 3,4) were pooled. They were then digested with λ-exonuclease to remove possible contaminating small nonreplicating DNA fragments without 5′ RNA primers prior to be used as input for Q-PCR (Bielinsky and Gerbi 1998). Primer efficiency was normalized against total sonicated HEK293 DNA. In agreement with previous observations, we detected a three- to fourfold enrichment of TIMM 5–6 and TOP 5–8 relative to the TIMM 10–11 and TOP 11–12 regions that were used as a baseline (Fig. 1B; Giacca et al. 1994; Keller et al. 2002). In contrast, when the same nascent strand-enriched fraction was used as a template and cloned DNA was used for normalization, two unexpected results were observed. First, overrepresentation of TIMM 1–2 and TOP 5–8 increased to ∼25-fold and 15-fold relative to TIMM 10–11 and TOP 11–12 (Fig. 1C), and second, the preferentially amplified fragment in the TIMM13 locus switched from TIMM 5–6 (Fig. 1B, green bar) to TIMM 1–2 (Fig. 1C, red bar), depending on whether genomic or cloned DNA was used for primer normalization, respectively. The striking differences between TIMM 1–2 and TIMM 3–4, which are so close to one another that they partially overlap, and between TOP 5–8 and TOP 7–6, which were only 42 base pairs (bp) apart (Fig. 1A), were difficult to reconcile with the expected smooth progression of replication forks after ORI firing.
Figure 1.
Overrepresentation of DNA fragments at DNA replication origins. (A) All three exons of the TIMM13 gene and the first two exons of the TOP1 gene are represented by white boxes. Striped areas indicate nontranslated regions. Arrows indicate major transcription initiation sites, and color bars represent regions analyzed by Q-PCR. The black arrow points to a replication initiation site identified previously (Abdurashidova et al. 2000). (B) Relative abundance of the nine regions shown in A in a HEK293 DNA nascent strand fraction 200–800 nt long using total DNA for primer normalization. Values for the TIMM 10–11 and TOP 11–12 regions were considered as baseline. (C) Relative abundance in the same nascent strand preparation as in B but using the cloned TIMM13 and TOP1 regions for primer normalization. (D) Relative abundance in total unfractionated HEK293 DNA using the cloned TIMM13 and TOP1 regions for primer normalization. (E) Relative abundance in the cloned TIMM13 and TOP1 regions using total genomic DNA for primer normalization. (F) Relative abundance in three preparations of purified DNA nascent strands of the indicated sizes using total DNA for primer normalization. (G) Same as F but using the cloned TIMM13 region for primer normalization. All of the experiments were carried out in duplicate in at least two independent preparations of the standards and the samples used as a template. Standard deviation bars are indicated.
A radical implication of these results was that the overrepresented molecules should be directly detected on total genomic DNA without previous enrichment of nascent strands. We tested this possibility by Q-PCR across the TIMM13 and TOP1 ORI regions using total unfractionated HEK293 DNA as a template and primer normalization against the corresponding cloned regions and found that TIMM 1–2 and TOP 5–8 were indeed overrepresented ninefold and sevenfold relative to the baseline (Fig. 1D). As a control, we performed the reverse experiment using total genomic DNA as a reference for normalization and the cloned TIMM13 and TOP1 regions (where all sequences were equally represented) for quantitation. Figure 1E shows that both regions appeared to be underrepresented in the cloned molecules in a proportion comparable with their overrepresentation in Figure 1D. These results indicated that the usual normalization against genomic DNA greatly underestimates the number of short intermediates close to ORIs and suggested that the overrepresented molecules could have been generated through several rounds of replication without elongation into their neighboring sequences. Otherwise, the maximum possible overrepresentation of a replicated ORI region relative to their flanks in total unfractionated DNA would be twofold, even assuming the extreme possibility that ORI efficiency was 100% and that every cell in the population was in S phase (a highly unlikely situation in an exponential asynchronous culture).
The second unexpected result of the Q-PCR experiments was the switch from TIMM 5–6 to TIMM 1–2 as the preferentially amplified fragment using the same template but depending on whether genomic or cloned DNA was used for normalization (Fig. 1B,C, left panels, green and red bars). The TIMM13 ORI had originally been mapped to the TIMM 5–6 region using nascent strands ranging from 800 to 1200 nt long as input for Q-PCR (Giacca et al. 1994), and a site of replication initiation has also been mapped at nucleotide resolution in this region (Fig. 1A, black arrow; Abdurashidova et al. 2000). We hypothesized that preferential enrichment of TIMM 1–2 would have been detected if nascent strands shorter than 200–300 nt had not been excluded from that analysis. Figure 1F (top panel) shows that this prediction was correct and that TIMM 1–2 was indeed fourfold more abundant than TIMM 5–6 in fractions containing DNA fragments shorter than 300 nt, even when genomic DNA was used for normalization. However, the situation was reversed when nascent strands longer than 200 nt were used as a template, in agreement with Giacca et al. (1994), probably due to a depletion of short nascent strands containing TIMM 1–2 (Fig. 1F, middle panel). As expected, relative differences between the tested regions diminished when longer DNA nascent strands were used as a template (Fig. 1F, bottom panel). When cloned DNA was used for normalization, TIMM 1–2 reached a 60-fold overrepresentation in the fraction containing short replication intermediates, where it should be enriched, and was found to be the most abundant fragment in all fractions, regardless of their size (Fig. 1G).
It should be noted that the fraction containing nascent strands up to 300 nt should be enriched in Okazaki fragments. However, given that asynchronously growing cells were used in these experiments, Okazaki fragments were expected to derive from all the genome without a bias for particular loci. In addition, a higher abundance of the same short fragments was also clearly detectable in total unfractionated DNA (Fig. 1D), where overrepresentation of specific genomic regions cannot be attributed to any artificial enrichment.
Overrepresented fragments derive from a short region adjacent to the transcription initiation sites
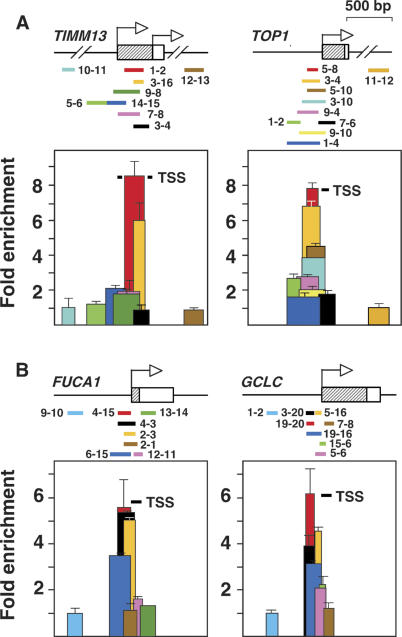
To define the size of the overrepresented molecules more precisely, we scanned the TIMM13 and TOP1 5′ regions by Q-PCR with closely spaced or overlapping pairs of primers using total unfractionated genomic DNA as input and primer normalization against the corresponding cloned regions as in Figure 1D. We found that overrepresented fragments derived from two narrow regions between primers TIMM 1 and TIMM 16 (214 bp) and TOP 3 and TOP 4 (203 bp) immediately adjacent to the transcription start sites (TSS) of both genes (Fig. 2A). Beyond these boundaries, the number of molecules fell sharply to the baseline level, consistent with results in Figure 1, suggesting that overrepresented molecules derived from short genomic regions with well-defined limits. TIMM13 and TOP1 are widely expressed genes whose 5′ ends lie within CpG islands, and because CpG islands are often associated with replication origins, we asked how general this phenomenon might be by examining two more examples. We chose the α-L-Fucosidase (FUCA1) and the glutamate-cysteine ligase (GCLC) genes at random from among the several thousand CpG island genes whose TSS have been accurately mapped in the DBTSS database (http://dbtss.hgc.jp). Neither of these two genes has been previously studied in the context of DNA replication. As shown in Figure 2B, a fragment of 187 bp between primers FUC 4 and FUC 3 in FUCA1 and another of 161 bp between primers GCLC 19 and GCLC 16 in GCLC immediately adjacent to their respective TSS were overrepresented in relation to their flanks in both cases in a proportion similar to that found in TIMM13 and TOP1. These experiments were done in HEK293 cells, and virtually identical results were obtained in HeLa, K562, HepG2 cells, exponential IMR90 primary human fibroblasts, and human blood (Supplemental Fig. 2). However, overrepresentation was reduced to one-third in senescent IMR90 cells, suggesting that the overproduction of short molecules requires active cell proliferation (Supplemental Fig. 2). We tested whether DNA nascent strands elongated from the overreplicated FUCA1 and GCLC regions by measuring the amount of replication intermediates of different sizes in two gradient fractions and normalizing primer efficiency against total DNA as previously done for the TIMM13 and TOP1 ORIs (Giacca et al. 1994; Keller et al. 2002). As shown in Supplemental Figure 3B, regions between primers FUC 2–11 and GCLC 19–20 were more abundant than their flanks in a gradient fraction containing replication intermediates up to 800 nt. However, the relative amount of their immediately flanking sequences increased when using intermediates between 400 and 3500 nt long (Supplemental Fig. 3C). These results were comparable with those obtained with TIMM13 and TOP1 and confirmed that overrepresented regions mark the position of ORIs from which DNA synthesis elongates to replicate the genome.
Figure 2.
Overrepresented fragments derive from a short region adjacent to the transcription initiation sites. Color bars span regions analyzed by Q-PCR across the promoter regions of the TIMM13 and TOP1 genes (A) and FUCA1 and GCLC genes (B). Black TSS bars indicate the transcription initiation region as annotated in the DBTSS database (http://dbtss.hgc.jp). Arrows indicate major transcription initiation sites. The two TSSs of the TIMM13 gene are indicated. Values for FUC 9–10 and GCLC 1–2 regions were considered as baseline. Other symbols are as in Figure 1. Total genomic DNA was used as input and the corresponding cloned loci for primer normalization. Duplicate experiments were carried out in at least two independent preparations of genomic DNA. Standard deviation bars are indicated. Primer sequences are shown in Supplemental Table 1. Sequence and genomic localization of overrepresented fragments are shown in Supplemental Table 2.
Overreplicated fragments are released from chromosomes as dsDNA molecules with 5′ RNA primers
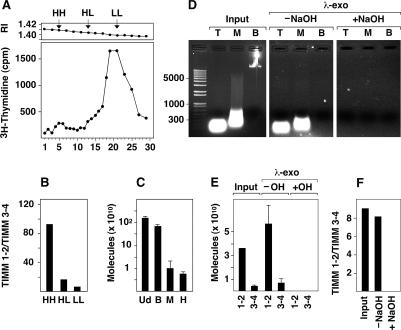
We next asked whether the overrereplicated fragments were single- or double-stranded and whether they could be detected as free molecules released from bulk DNA. We pulse-labeled exponential asynchronous HEK293 cells with bromodeoxyuridine (BrdU) and 3H-thymidine for 30 min and fractionated the total undigested DNA in a neutral cesium chloride (CsCl) gradient. The radioactivity profile displayed a small peak of heavy-heavy (HH) density, consistent with the presence of dsDNA molecules that had migrated independently from light-light (LL) DNA (Fig. 3A). HH fractions 3–7 were collected and fractionated again in a second CsCl gradient to further remove possible contamination from bulk DNA and were then used as input for Q-PCR. Figure 3B shows that TIMM 1–2 was ∼90-fold overrepresented relative to TIMM 3–4 in the HH fraction, and this overrepresentation declined to 15-fold and sixfold in the HL and LL fractions, respectively (Fig. 3B). These results indicated that both strands of TIMM 1–2 were synthesized during the 30-min pulse and that they were released from the chromosomes after their synthesis. End-labeling of fragments in the HH fraction with Klenow revealed that their size ranged between ∼100 and 200 bp (Supplemental Fig. 1B), consistent with the size of the overrepresented fragments shown in Figure 2. To further ascertain the double-stranded nature of these molecules, we digested total HEK293 DNA prior to Q-PCR with MspI or HpaII (which cut inside the TIMM 1–2 and TOP 5–8 fragments) or with BamHI (which cuts outside them). In both cases, digestion with MspI or HpaII resulted in an ∼100-fold reduction in the number of amplified molecules relative to undigested or BamHI-digested DNA, further supporting the notion that the overrepresented molecules were mostly double-stranded. The results for TIMM 1–2 are shown in Figure 3C.
Figure 3.
Overreplicated fragments are released from chromosomes. (A) Refraction index (RI) and 3H-thymidine incorporation across the CsCl gradient fractions. HH, HL, and LL indicate the positions of heavy-heavy, heavy-light, and light-light DNA, respectively. (B) DNA recovered from the HH, HL, and LL fractions was used as input for Q-PCR to measure the enrichment of TIMM 1–2 relative to TIMM 3–4. (C) Number of amplified TIMM 1–2 molecules (logarithmic scale) detected by Q-PCR using as input the same amount of total genomic DNA digested previously with BamHI (B), MspI (M) or HpaII (H). (Ud) Undigested control. (D) Total HEK293 DNA was fractionated in a neutral sucrose gradient, and three fractions (top [T]; medium [M], and bottom [B]) were collected. DNA from each fraction was electrophoresed directly (input) or after λ-exonuclease digestion (λ-exo) without or with previous incubation with alkali (− and + NaOH). Because all high-molecular-weight DNA migrates to the bottom of the gradient, the equivalent volume of the B fraction loaded in the gel was 4 orders of magnitude lower than that of the T and M fractions. (E) DNA from the top fraction after each treatment was used as input for Q-PCR with the TIMM 1–2 and TIMM 3–4 pairs of primers. The amount of amplified molecules after incubation with alkali and λ-exo was below the level of detection established for the range of the standards. TIMM 3–4 fragments were sensitive to restriction enzymes when digested prior to Q-PCR, indicating that they were mostly double-stranded (data not shown). Histograms represent the average of two quantitations, and the bars represent the range of results. Cloned DNA was used for primer normalization. (F) Overrepresentation of TIMM 1–2 relative to TIMM 3–4 in the three cases.
To confirm by an independent technique that short overreplicated molecules were indeed released from bulk DNA and to test whether they contained primers of RNA at their 5′ ends, we pelleted total nondenatured, non-RNAse-treated DNA from HEK293 cells through a neutral sucrose gradient and collected the top fraction (T), which contained fragments <400 bp (Fig. 3D). An aliquot (input) was used directly for Q-PCR using TIMM 1–2 and TIMM 3–4 primers and cloned DNA for normalization. Another two aliquots of the same volume were digested with λ-exonuclease with or without previous treatment with NaOH under conditions that remove 5′ RNA primers (Abdurashidova et al. 2000). The results in Figure 3, E and F, show that the amount and proportion of both fragments were unaffected by treatment with λ-exonuclease under conditions in which bulk DNA was completely digested (Fig. 3D, middle panel). In contrast, no amplification with any pair of primers was detected in the sample pretreated with NaOH. These results supported the idea that overreplicated fragments are independent of bulk DNA, in agreement with their behavior in the CsCl gradient (Fig. 3A), and they are consistent with the presence of RNA primers at the 5′ ends. We also tested the possibility that some fragments might adopt the conformation of extrachromosomal circular DNA described for several genomic regions in mammalian cells under conditions of stress and genomic instability (Cohen et al. 2006). Standard PCR using divergent primers from the TIMM 1–2 region failed to amplify any product in parallel reactions in which a circular 3-kb plasmid containing this region was readily amplified (data not shown).
Reiterative synthesis of short DNA molecules coincides with the time of activation of specific ORIs during S phase
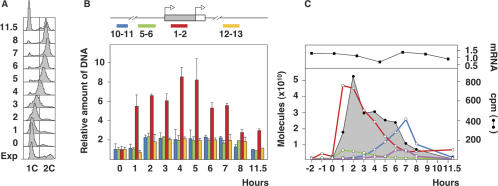
We asked next whether the mechanism that generated the overrepresented molecules was constitutive or cell cycle-regulated and whether it occurred simultaneously to the time of activation of ORIs during DNA replication. To address this point, HeLa cells were arrested very early in S phase by a double-thymidine block, and synchronization across S phase was monitored by FACS analysis and 3H-thymidine incorporation (Fig. 4A,C). We analyzed the replication profile of four regions of TIMM13 (Fig. 4B) by Q-PCR using total unfractionated DNA as a template and cloned DNA for normalization. The data in Figure 4B show that TIMM 10–11, TIMM 5–6, and TIMM 12–13 duplicated in the first 2 h after release into S phase, as expected for sequences close to an early firing ORI (Giacca et al. 1994). Their levels remained constant until 11.5 h, when it was reduced to half because most cells have gone through mitosis at that time (Fig. 4A). In contrast, the replication profile of TIMM 1–2 raised sharply immediately after release, reached about eightfold in the 4–5-h samples, and declined to a level comparable with that of the other three regions in the 8-h sample (Fig. 4B). This result indicated that the TIMM 1–2 region must have undergone several rounds of synthesis during a single S phase.
Figure 4.
Overreplication at early and late origin regions during S phase. (A) HeLa cells arrested in early S phase by a double-thymidine block (0-h sample) were released in thymidine-free medium, and their progress through S phase was monitored by FACS analysis. The profile of an exponential asynchronous culture is also shown (Exp). (B) Total DNA samples from an equivalent number of cells were taken at the indicated times after release, and the amount of the four TIMM13 regions shown relative to the 0-h sample was measured by Q-PCR. Total genomic DNA was used as input and cloned DNA was used for primer normalization. Histograms represent the average of two measurements, and the bars indicate the range of results. (C, top panel) Level of TIMM13 mRNA during S phase relative to that in an asynchronous exponentially growing culture. (Bottom panel) Incorporation of 3H-thymidine (black line) and amount of amplified molecules after immunoprecipitation of TIMM 1–2 (red), TIMM 5–6 (green), HBB 5–14 (blue), and HBB 7–8 (purple) fragments measured by Q-PCR across the S phase.
To compare the replication rate of TIMM 1–2 and TIMM 5–6 per unit of time, we pulse-labeled cells with BrdU for 30 min at 1-h intervals, and the amount of each BrdU-immunoprecipitated fragment was measured by Q-PCR. Results in Figure 4C show that both fragments were virtually undetectable during the last 2 h before release from the thymidine block and started incorporating BrdU immediately after entry into S phase. Maximum incorporation occurred in the 1-h sample and declined gradually to almost a background level at the end of S phase (Fig. 4C, red and green lines). A striking eightfold difference, however, was detected in the amount of immunoprecipitated TIMM 1–2 region relative to TIMM 5–6. This result was consistent with those in Figures 1D, 2A, and 4B and suggested that a burst of TIMM 1–2 synthesis was associated with the activation of the TIMM13 ORI at the onset of S phase. The different profile of TIMM 1–2 in Figure 4, B and C, indicates that fragments generated at the beginning of S phase (Fig. 4C) were not immediately degraded and accumulated toward the middle of S phase (Fig. 4B) before reaching a background level toward the end of the S phase.
We also asked whether overrepresentation of TIMM 1–2 correlated with a differential transcriptional rate of TIMM13 during S phase. Figure 4C (top) shows that TIMM13 mRNA remained constant at all tested times and at a level similar to that in asynchronously growing cells. This results rules out that the overrepresentation of short fragments could be due to DNA repair associated with a higher rate of TIMM13 transcription at the beginning of the S phase.
Overreplication at the β-globin ORI
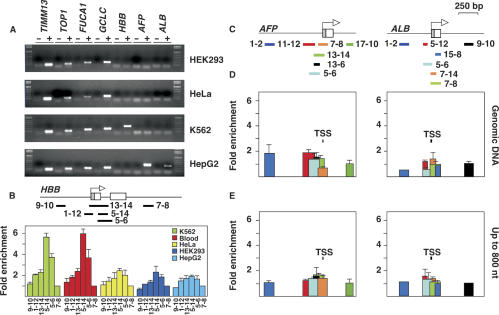
The TIMM13, TOP1, FUCA1, and GCLC genes were active in all cell lines tested (Fig. 5A), and their promoters colocalized with CpG islands and ORIs. The coincidence of these three features raised the questions of whether overreplication occurred only in ORIs associated with CpG island promoters and whether it depended on active transcription. To address these issues, we selected the β-globin (HBB) gene, which is not associated with a CpG island and is specifically expressed in erythroid cells such as K562 (Fig. 5A). In addition, an ORI that is active even in cells where the gene is not expressed has been located previously in a 1200-bp region encompassing the promoter and first exon of the gene (Kitsberg et al. 1993; Aladjem et al. 1995). We analyzed by Q-PCR the six regions of the HBB gene shown in the diagram of Figure 5B using total unfractionated DNA as a template and the cloned regions for normalization. Results revealed that a 209-bp fragment spanning the 5–14 and 5–6 regions, where the initiation region has been mapped, was overrepresented in expressing K562 cells at a level comparable with that found in the four ORIs associated with CpG islands (Fig. 5B, green histograms). An almost identical result was obtained when total blood DNA was used (Fig. 5B, red histograms), indicating that this phenomenon occurs in the organism and is not restricted to cells in culture. Overrepresentation of the 5–14 fragment was also detected in HeLa, HEK293, and HepG2 cell lines although at a lower rate than in expressing cells (Fig. 5B). This indicated that transcription was not required to drive replication initiation, but it stimulated overreplication, as shown in K562 and blood cells. Given that transcription of the HBB gene is associated with early replication, it is also possible that a higher concentration of replication initiation factors at the beginning of the S phase could contribute to overreplication.
Figure 5.
Transcription does not drive replication or induce rereplication. (A) Expression analysis by RT–PCR using an equal amount of total RNA from the indicated cell lines. Plus (+) and minus (−) indicate cDNA synthesis with or without RT. The primers used are described in Supplemental Table 3. (B) Q-PCR analysis across the HBB ORI region in total genomic DNA from K562, blood, HeLa, HEK293, and HepG2 cells. The cloned version of the locus was used for normalization. Values for the GLO 7–8 region were considered as baseline. (C) First exons of the AFP and ALB genes are represented by boxes. Striped areas indicate nontranslated regions. Arrows indicate major transcription initiation sites, and color bars represent regions analyzed by Q-PCR. (D) Relative abundance of the indicated fragments across the AFP and ALB promoter regions using total genomic HepG2 DNA as input and the corresponding cloned regions for primer normalization. Black TSS bars indicate transcription initiation regions. (E) Relative abundance using a preparation of nascent strands up to 800 nt long from HepG2 cells as input DNA and total genomic DNA for primer normalization. Histograms represent the average of two measurements, and the bars indicate the range of results. Values for the AFP 17–10 and ALB 9–10 regions were considered as baseline.
To test whether overreplication of the 5–14 region was coincident with the activation of the β-globin ORI, we measured the rate of incorporation of BrdU in the HBB 5–14 and HBB 7–8 fragments during a synchronous S phase in HeLa cells. Figure 4C shows that both regions replicated at the end of the S phase in agreement with previous observations that this ORI fires late in nonexpressing cells (Kitsberg et al. 1993). In addition, there is a fivefold difference between the immunoprecipitated amount of the 5–14 fragment (Fig. 4C, blue line) relative to 7–8 (Fig. 4C, purple line), consistent with the differences found in asynchronous HeLa cell cultures (Fig. 5B, yellow histograms). These results, together with those of the TIMM13 ORI in Figure 4C, show that the overproduction of short fragments is strictly associated with the activation of ORIs regardless of their time of activation during S phase or their colocalization with CpG islands.
Transcription per se is not sufficient to drive replication or induce overreplication
We asked next whether transcription would be enough to drive replication initiation or overreplication at other tissue-specific promoters not tested previously for ORI activity. We selected the α-fetoprotein (AFP) and serum albumin (ALB) genes (Fig. 5C) that are liver-specific and are not associated with CpG islands. We tested whether some fragments adjacent to the TSS region of these genes were overrepresented in total DNA in liver HepG2 cells, where both genes were transcribed (Fig. 5A). Figure 5D shows that the Q-PCR scan analysis failed to detect a relative enrichment of any of the fragments analyzed when total HepG2 DNA was used as input and the primers were normalized against the cloned loci.
Despite this result, it was still possible that replication could initiate at these promoters but without the overproduction of any fragment. We addressed this possibility by using a purified DNA nascent strands fraction up to 800 nt from HepG2 cells as input for Q-PCR and total genomic DNA for primer normalization. No enrichment of any of the analyzed fragments was found either (Fig. 5E), indicating that replication initiation was undetectable at these promoters even when the genes were actively transcribed. The nascent strands fraction used in this experiment was the same as in Supplemental Figure 3, where replication initiation at FUCA1 and GCLC was readily detected. The results obtained with the ALB and AFP promoter regions indicated that active transcription per se is not sufficient to trigger replication or to induce rereplication in the regions adjacent to the TSS.
Overreplicated regions overlap with clusters of DNAse hypersensitive sites (HS)
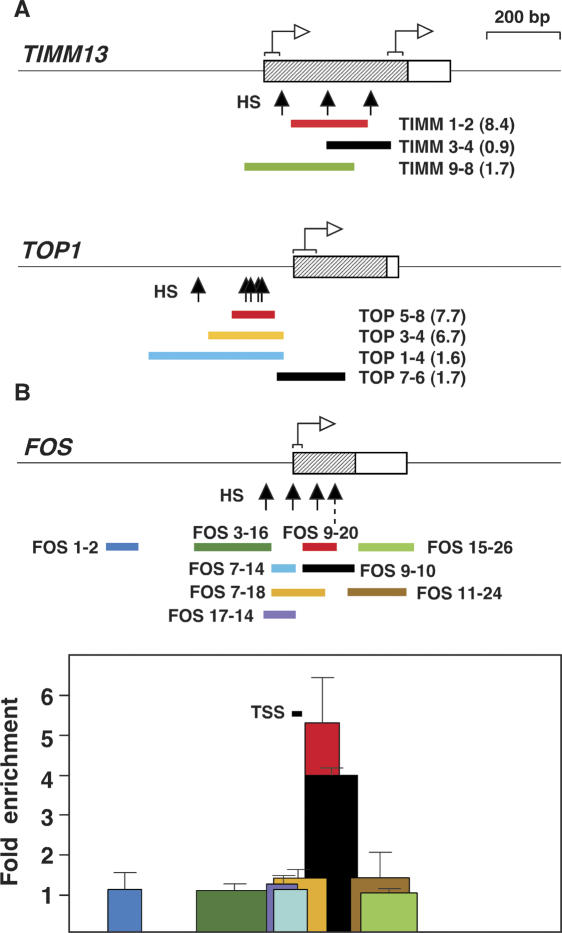
The five overreplicated regions described above did not show any significant sequence homology between them. However, their similar size of ∼200 bp and their location immediately adjacent to the TSS suggested that they could derive from genomic regions sharing a distinctive chromatin structure. We tested their possible relationship with the nucleosome-free gap that often surrounds the TSS of many active genes by searching for the presence of DNase I HS in a DNase HS database derived from human primary CD4+ cells (Crawford et al. 2004; http://research.nhgri.nih.gov/DNaseHS/May2005). We found that the promoters of the TIMM13 and TOP1 genes contained several clustered DNase HS sites that defined a hypersensitive domain precisely overlapping the TIMM 1–2 and TOP 5–8/TOP 3–4 overreplicated regions (Fig. 6A). This coincidence was further reinforced by the observation that the TIMM 3–4, TIMM 9–8, TOP 1–4, and TOP 7–6 fragments, which stretched beyond the boundaries of the HS cluster, gave a background level of replication intermediates in the Q-PCR assay shown in Figure 2A. Based on these observations, we hypothesized that overreplication would also occur at other genomic DNase I hypersensitive regions close to the TSS of active CpG island genes. We selected the FOS promoter because it is contained in a CpG island, and a nucleosome-free region encompassing the TSS has been mapped in detail in HeLa cells (Tazi and Bird 1990). Q-PCR analysis using total unfractionated HeLa DNA as a template and the cloned region for normalization confirmed our prediction and showed that the FOS 9–20 region, located precisely between the TSS and the nucleosome boundary, was five- to sixfold overrepresented relative to the remaining fragments tested (Fig. 6B). FOS 9–10 encompasses a site of partial nucleosomal protection (Fig. 6B, dashed arrow; Tazi and Bird 1990). This means that primer FOS-10 lies at the edge of a nucleosome that is not precisely positioned. As a consequence, the FOS 9–20 region is free of nucleosomes in every cell, while FOS 9–10 would be accessible only in some of them. The fact that the level of FOS 9–20 was higher than that of FOS 9–10 and that beyond them no enrichment above background was detected suggests that nucleosomes mark the boundaries of the overreplicated regions. A similar situation could apply to some of the other regions we analyzed (Figs. 2, 5), where a slight displacement of the primers that detects the highest amount of overreplication still detects an intermediate level before reaching the baseline.
Figure 6.
Overreplicated regions overlap with clusters of DNase HS. (A) Black arrows point to HS mapped by DNase I digestion across the TIMM13 and TOP1 promoters. Color bars represent regions analyzed by Q-PCR in Figure 2 and figures in parentheses refer to their enrichment relative to the baseline. White arrows with brackets indicate transcription initiation regions. (B) HS arrows indicate chromatin HS mapped by HhaI restriction enzyme digestion (Tazi and Bird 1990). The dashed arrow points to a site of partial protection. Colored bars span regions analyzed by Q-PCR across the human FOS promoter region using total genomic HeLa DNA as input and the cloned locus for primer normalization. TSS bar marks the transcription start sites annotated in the DBTSS. Duplicate experiments were carried out in at least two independent preparations of genomic DNA. Standard deviation bars are indicated. Values for FOS 1–2 were considered as baseline.
Discussion
Q-PCR is a widely used method for ORI mapping in mammalian cells. It relies on the preferential amplification of regions close to ORIs using as a template nascent DNA strands of between ∼500 and 1500 nt or longer, previously enriched in sucrose gradients. The reliability of this approach for ORI identification is supported by the fact that several mammalian ORIs that were initially identified by a variety of non-PCR approaches have been validated later by Q-PCR. Examples include the preferential initiation sites within the disperse initiation region downstream from the hamster DHFR gene identified by two-dimensional electrophoresis (Pelizon et al. 1996; Kobayashi et al. 1998), the human GM-CSF Ori1 and Ori2 identified by subtractive hybridization for origin capture (Todorovic et al. 2005), and several human ORIs recently identified by high-throughput array hybridization (Lucas et al. 2007). Conversely, ORIs initially identified by Q-PCR have been shown to bind ORC and MCM proteins as in the case of the human Lamin B2 (Abdurashidova et al. 2003), c-Myc (Ghosh et al. 2006), and FMR1 (Gray et al. 2007). In most cases reported in the literature, overrepresentation of fragments defined as ORIs range between fourfold and 10-fold relative to their flanks. This relatively modest enrichment could be due to a general inefficiency of mammalian ORIs or to a limited sensitivity of the technique. Primers for Q-PCR are routinely normalized against total genomic DNA on the assumption that replication intermediates present in that sample do not significantly affect normalization.
As a starting point for our work, we hypothesized that cloned versions of the regions to be analyzed would be a more reliable reference for primer normalization than total DNA. The increase in resolution provided by this new approach has uncovered a population of molecules generated by overreplication of short nucleosome-free regions encompassing the transcription initiation sites of promoters associated with replication origins. This phenomenon has hitherto remained unnoticed because genomic DNA has always been used for primer normalization, and also because short DNA molecules have been routinely excluded from the assays. A comparison of the results obtained with either method of normalization using the same population of nascent strands is shown in Figure 1, F and G. The improvement in sensitivity reported here is compatible with the previous identification of ORIs by Q-PCR. In fact, generation of short fragments takes place at well-characterized ORIs, such as TIMM13 (Lamin B2) TOP1 and β-globin (Figs. 1, 2, 5).
An important question is whether the overproduction of short molecules plays some role in the activation of ORIs or whether it is a byproduct of the replication initiation event. In the first case, some of the short molecules could be prevented from extrusion and allowed to elongate to generate daughter leading and lagging strands. This is consistent with the results for TOP1 and β-globin ORIs, where overabundant molecules derive precisely from regions where Orc2 binds and the polarity of nascent strand switches in these two ORIs, respectively (Figs. 2, 5; Aladjem et al. 1995; Keller et al. 2002). Alternatively, the generation of short molecules could take place somewhere in the vicinity of the replication initiation sites and might not participate in the generation of the mature daughter strands. In this context, a site of replication initiation has been reported within the TIMM 5–6 region (Abdurashidova et al. 2000) at ∼350 bp upstream of the overreplicated TIMM 1–2 region (Fig. 1A). So far this is the only replication initiation point reported in mammalian cells at nucleotide resolution, and Abdurashidova et al. (2000) contemplated that additional initiation sites might lie adjacent to the region they studied. Consistent with this possibility, several sites of initiation have been found by template polarity assays within a 2.4-kb region in the human c-myc ORI (Waltz et al. 1996). As regards the nature of the overrepresented molecules, given that they are double-stranded and contain 5′ RNA primers (Fig. 3), they could be generated by a process of repeated extrusion of the two complementary newly synthesized DNA strands by fork reversal due to the instability of small replication bubbles (Zannis-Hadjopoulos et al. 1981).
It is possible that overproduction of short fragments could be related to the repeated loading of multiple Mcm2–7 complexes at ORIs through several cycles of ATP hydrolysis by Cdc6 and ORC (Randell et al. 2006). In fact, overreplicated molecules could be generated by a mechanism similar to that suggested to operate in Xenopus egg extracts under conditions of Cdt1 overexpression. This model proposes that successive loading of Mcm2–7 could generate replication forks that would run into the previous ones and result in the release of small, newly synthesized double-stranded fragments (Davidson et al. 2006). However, a crucial difference with the overproduction of short molecules reported here is that it occurs in cells proliferating under physiological conditions. This implies that this limited overreplication falls below the level of detection by the ATM/ATR checkpoint and that it is tolerated as a constituent step that takes place during the normal activation of ORIs. It is not clear at this stage whether overreplication is mechanistically required for ORI activity. However, the physical proximity between overreplicated regions and replication origins, together with the coincidence of the burst of overreplication with the time of firing of specific ORIs during S phase (Fig. 4C), suggests that this phenomenon is intimately associated with the activation of properly licensed mammalian ORIs. Accordingly, overreplicated molecules could be good predictors for replication initiation regions in the genome. On the other hand, it should be noted that all ORIs included in our study were associated with promoters, either constitutive or tissue-specific (Fig. 5A). We did not explore whether the same phenomenon takes place at ORIs dispersed across large regions, such as the one downstream from the DHFR gene in hamster CHO cells (Hamlin and Dijkwel 1995), or at loci containing tightly clustered ORIs, such as the mouse immunoglobulin heavy-chain region (Norio et al. 2005).
The physical overlapping between overreplicated regions and transcription initiation sites raises the possibility of a mechanistic link between both processes. However, we showed that active transcription of the ALB and AFP genes is not sufficient to drive detectable replication initiation or overreplication (Fig. 5D,E). Also, the continuous rate of transcription of the TIMM 13 gene during S phase (Fig. 4C) indicates that the generation of short DNA molecules is unlikely to be due to DNA repair associated with active transcription. In ORIs close to promoters, however, active transcription enhances overreplication, as shown for the β-globin ORI in expressing relative to nonexpressing cells (Fig. 5B). This process could be facilitated by the presence of a short region of unwound ssDNA generated by the DNA-scrunching mechanism of transcription initiation (Kapanidis et al. 2006; Revyakin et al. 2006).
The coincidence of the overreplicated regions with HS (Fig. 6) is consistent with the idea of nucleosomes acting as a boundary to prevent elongation, and it suggests that chromatin remodeling factors, such as the FACT heterodimer, might not be present or operative at that stage. This complex facilitates transcription elongation in the chromatin context (Orphanides et al. 1998) and has recently been shown to be recruited to ORIs in vivo through binding to the MCM complex and to be required for efficient helicase activity and fork progression across nucleosomes (Tan et al. 2006).
The frequency of histone H3 modifications such as Lys 4 methylation and Lys 9/14 acetylation peaks at regions surrounding the transcription initiation sites (Liang et al. 2004; Roh et al. 2005; Barski et al. 2007), and it is interesting that many promoters associated with these modifications initiate transcription that in many cases does not produce full-length mRNAs (Guenther et al. 2007). We speculate that in addition to the distinctive chromatin structure of these regions, the generation of short RNA and DNA molecules that do not elongate into mature transcripts or replicons might be used as a dynamic signaling mechanism to highlight and perhaps contribute to maintaining these essential regulatory regions in the genome.
Materials and methods
Cell culture
Human cell lines were grown as recommended by the American Type Culture Collection and were harvested from exponential cultures. Senescent IMR90 cells were harvested after a month in culture without detectable proliferation.
Nascent strand purification
Total human genomic DNA was extracted and nascent strands were obtained essentially as described in Abdurashidova et al. (2000). Purified DNAs were denatured by heating and size-fractionated by centrifugation at 24,000 rpm in a Beckman SW-40 rotor for 20 h at 20°C in a seven-step sucrose gradient (5%–20% in steps of 2.5%) made up in 10 mM Tris (pH 8.0), 1 mM EDTA, and 100 mM NaCl. Twelve fractions of 1 mL were collected and aliquots were electrophoresed in alkaline agarose gels to monitor correct fractionation. The top three fractions containing DNA up to 300 nt, 200–800 nt, and 400–3000 nt, respectively, were enriched in RNA-primed nascent DNA by λ-exonuclease treatment (Bielinsky and Gerbi 1998). Sucrose gradients in Figure 3 were run without previous heat denaturation of DNA. Alkaline treatment of DNA to remove 5′ RNA primers was as in Abdurashidova et al. (2000).
Quantitative real-time PCR
Quantitative real-time PCR was performed using an Abi Prism 7000 Detection System (Applied Biosystems) with HotStar Taq polymerase (Qiagen) and SYBR Green (Molecular Probes). Four to five serial 10-fold dilutions of sonicated genomic DNA or the corresponding cloned fragments were amplified using the same reaction mixture as the samples to construct the standard curves. The regions studied were PCR-amplified with the primer pairs and conditions indicated in Supplemental Table 1 and cloned into pGEM-T (Promega). All real-time reactions were performed in duplicate, and a reaction mix without input DNA was always included as a control. Amplifications were run for 40 cycles in a final volume of 30 μL with 1 μL of input DNA, 3 μL of 10× Qiagen HotStar Taq buffer, 1.8 μL of 25 mM MgCl, 0.6 μL of 10 mM dNTPs, 0.75 μL of SYBR Green (commercial stock), 0.75 U of polymerase, and 2.25 μL of primers (5 μM each). After amplification, melting curve analyses were performed to control the specificity of the reactions. Quantitative analyses were carried out using the Abi Prism 7000 SDS Software.
Cell synchronization and immunoprecipitation with anti-BrdU antibodies
A double-thymidine block was performed by growing HeLa cells for 13 h in DMEM medium containing 2.5 mM thymidine followed by 9 h of incubation in fresh medium and further incubation for 13 h in the presence of 2.5 mM thymidine. Progression through S phase was checked every hour after block removal by FACS analysis and by 30-min labeling with 100 μM BrdU and 5 μCi/mL 3H-thymidine prior to cell harvest. Purification of replicated DNA at each time point was performed by immunoprecipitation with monoclonal anti-BrdU antibodies (Becton Dickinson) as described previously (Hansen et al. 1993).
Density substitution
HEK293 cells were pulse-labeled for 30 min with 100 μM BrdU and 5 μCi/mL 3H-thymidine. Purified DNAs were mixed with CsCl made up in 15 mM NaCl and 15 mM EDTA to reach a final refraction index of 1.402. Samples were centrifuged at 35,000 rpm in a 70.1Ti rotor (Beckman) for 62 h at 20°C. Thirty fractions of 400 μL were collected from bottom to top, and equal aliquots from each fraction were used to measure the refraction index and the radioactivity incorporated.
Transcriptase reverse reaction and PCR
Total RNA was extracted with a guanidium isothyocianate solution, and cDNA was synthesized using oligo-dT and SuperScript II Reverse Transcriptase (Invitrogen). The amount of cDNA synthesized from 200 ng of total RNA (− and + SuperScript II) was used as template for each PCR reaction, using the primer pairs and annealing temperatures listed in Supplemental Table 3. PCR reactions were performed with 34 cycles of amplification with HotStar Taq polymerase (Qiagen), and all primer pairs were designed to amplify from adjacent exons.
Acknowledgments
We thank members of the M.G. and F.A. laboratories for helpful discussions and advice and Karim Labib and Juan Méndez for advice and a critical reading of the manuscript. We are also grateful to three anonymous reviewers for excellent constructive criticism. This work was supported by the Programa Ramón y Cajal from the Spanish Ministry of Education and Science and grants from the Spanish Ministry of Education and Science (BFU2004-02595) and Junta de Castilla y León (CSI03B05) to M.G. and the Spanish Ministry of Education and Science (BFU2005-01959) to F.A.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.445608
References
- Abdurashidova G., Deganuto M., Klima R., Riva S., Biamonti G., Giacca M., Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- Abdurashidova G., Danailov M.B., Ochem A., Triolo G., Djeliova V., Radulescu S., Vindigni A., Riva S., Falaschi A. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 2003;22:4294–4303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem M.I., Groudine M., Brody L.L., Dieken E.S., Fournier R.E., Wahl G.M., Epner E.M. Participation of the human β-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- Antequera F. Genomic specification and epigenetic regulation of eukaryotic DNA replication origins. EMBO J. 2004;23:4365–4370. doi: 10.1038/sj.emboj.7600450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;4:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bielinsky A.K., Gerbi S.A. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- Blow J.J., Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Z., Bacharach E., Lavi S. Mouse major satellite DNA is prone to eccDNA formation via DNA Ligase IV-dependent pathway. Oncogene. 2006;25:4515–4524. doi: 10.1038/sj.onc.1209485. [DOI] [PubMed] [Google Scholar]
- Crawford G.E., Holt I.E., Mullikin J.C., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis E., Brodolin K., Menut S., Maiorano D., Girard-Reydet C., Mechali M. Specification of a DNA replication origin by a transcription complex. Nat. Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- Davidson I.F., Li A., Blow J.J. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol. Cell. 2006;24:433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S., Gómez M., Bird A., Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M.L., Blow J.J., Ghosh S., Saha T., Noguchi K., Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Ghosh M., Kemp M., Liu G., Ritzi M., Schepers A., Leffak M. Differential binding of replication proteins across the human c-myc replicator. Mol. Cell. Biol. 2006;26:5270–5283. doi: 10.1128/MCB.02137-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacca M., Zentilin L., Norio P., Diviacco S., Dimitrova D., Contreas G., Biamonti G., Perini G., Weighardt F., Riva S., et al. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. 1994;91:7119–7123. doi: 10.1073/pnas.91.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M.A., Holmquist G.P., Gray M.C., Caston L.A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Gómez M., Brockdorff N. Heterochromatin on the inactive X chromosome delays replication timing without affecting origin usage. Proc. Natl. Acad. Sci. 2004;101:6923–6928. doi: 10.1073/pnas.0401854101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S.J., Gerhardt J., Doerfler W., Small L.E., Fanning E. An origin of DNA replication in the promoter region of the human fragile X mental retardation (FMR1) gene. Mol. Cell. Biol. 2007;27:426–437. doi: 10.1128/MCB.01382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin J.L., Dijkwel P.A. On the nature of the replication origins in higher eukaryotes. Curr. Opin. Genet. Dev. 1995;5:153–161. doi: 10.1016/0959-437x(95)80002-6. [DOI] [PubMed] [Google Scholar]
- Hansen R.S., Canfield T.K., Lamb M.M., Gartler S.M., Laird C.D. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- Kapanidis A.N., Margeat E., Ho S.O., Kortkhonjia E., Weiss S., Ebright R.H. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C., Ladenburger E.M., Kremer M., Knippers R. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 2002;277:31430–31440. doi: 10.1074/jbc.M202165200. [DOI] [PubMed] [Google Scholar]
- Kitsberg D., Selig S., Keshet I., Cedar H. Replication structure of the human β-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Rein T., DePamphilis M.L. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 1998;18:3266–3277. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R., Heilig R., Sonnleitner M., Weissenbach J., Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol. Biol. Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Lin J.C., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. 2004;19:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I., Palakodeti A., Jiang Y., Young D.J., Jiang N., Fernald A.A., Le Beau M.M. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Maiorano D., Mechali M. A Cdt1–geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus. EMBO J. 2006;25:5764–5774. doi: 10.1038/sj.emboj.7601436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine D.M., Rodriguez H.K., Bell S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes & Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner L.D., Hamlin J.L. Specific signals at the 3′ end of the DHFR gene define one boundary of the downstream origin of replication. Genes & Dev. 2005;19:1053–1066. doi: 10.1101/gad.1307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner L.D., Crawford E.L., Hamlin J.L. Isolating apparently pure libraries of replication origins from complex genomes. Mol. Cell. 2006;21:719–726. doi: 10.1016/j.molcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Norio P., Kosiyatrakul S., Yang Q., Guan Z., Brown N.M., Thomas S., Riblet R., Schildkraut C.L. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell. 2005;20:575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C.H., Luse D.S., Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Pelizon C., Diviacco S., Falaschi A., Giacca M. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol. 1996;16:5358–5364. doi: 10.1128/mcb.16.10.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell J.C., Bowers J.L., Rodriguez H.K., Bell S.P. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol. Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Revyakin A., Liu C., Ebright R.H., Strick T.R. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev. 2005;5:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B.C., Chien C.T., Hirose S., Lee S.C. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006;25:3975–3985. doi: 10.1038/sj.emboj.7601271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Todorovic V., Giadrossi S., Pelizon C., Mendoza-Maldonado R., Masai H., Giacca M. Human origins of DNA replication selected from a library of nascent DNA. Mol. Cell. 2005;19:567–575. doi: 10.1016/j.molcel.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Vashee S., Cvetic C., Lu W., Simancek P., Kelly T.J., Walter J.C. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes & Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltz S.E., Trivedi A.A., Leffak M. DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res. 1996;10:1887–1894. doi: 10.1093/nar/24.10.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E.J., Emanuelsson O., Scalzo D., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., et al. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc. Natl. Acad. Sci. 2004;101:17771–17776. doi: 10.1073/pnas.0408170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Persico M., Martin R.G. The remarkable instability of replication loops provides a general method for the isolation of origins of DNA replication. Cell. 1981;27:155–163. doi: 10.1016/0092-8674(81)90369-x. [DOI] [PubMed] [Google Scholar]