Abstract
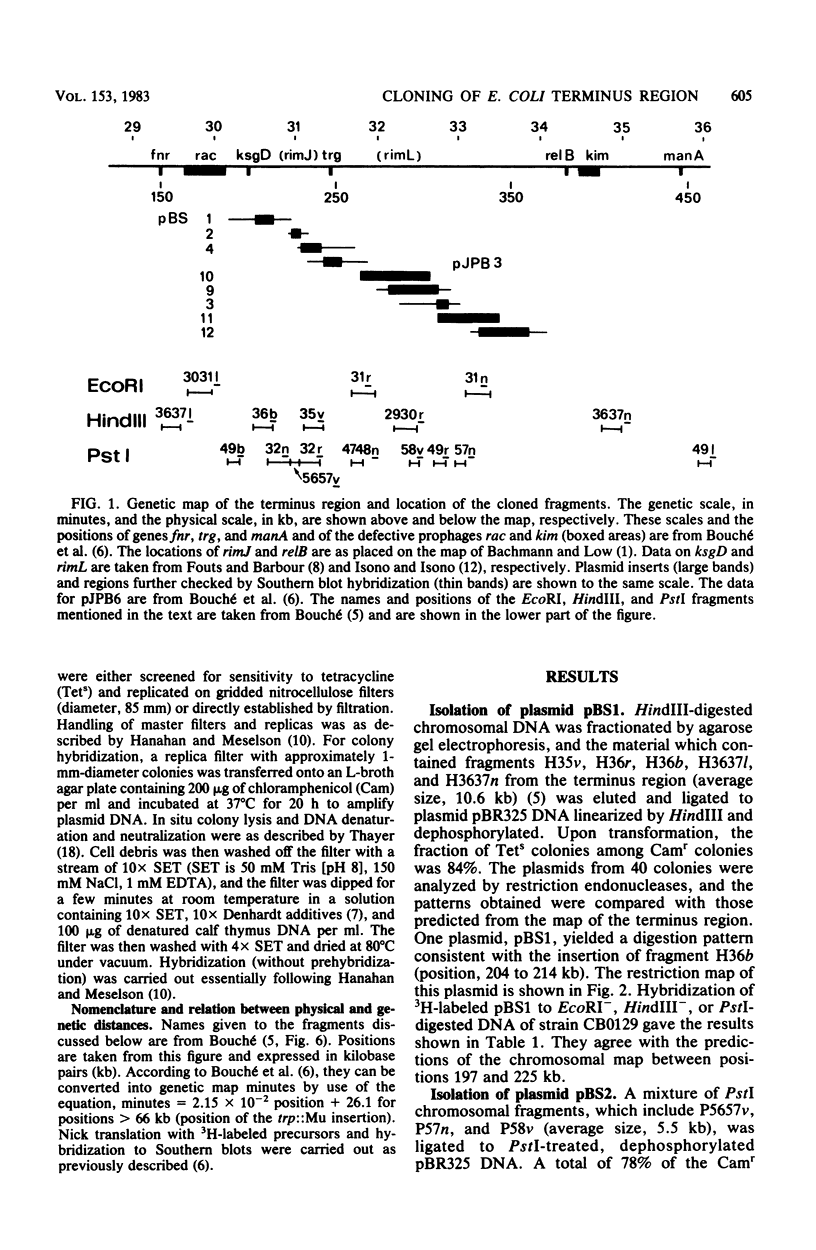
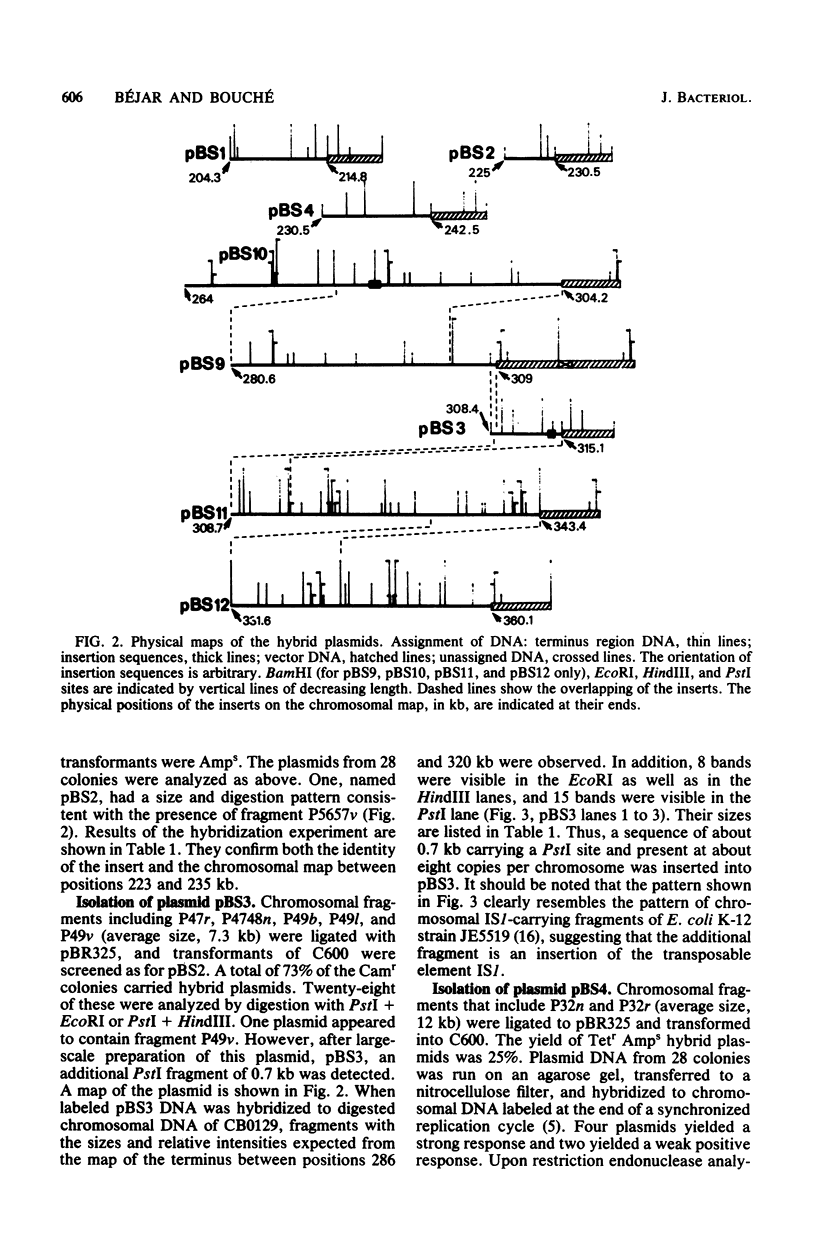
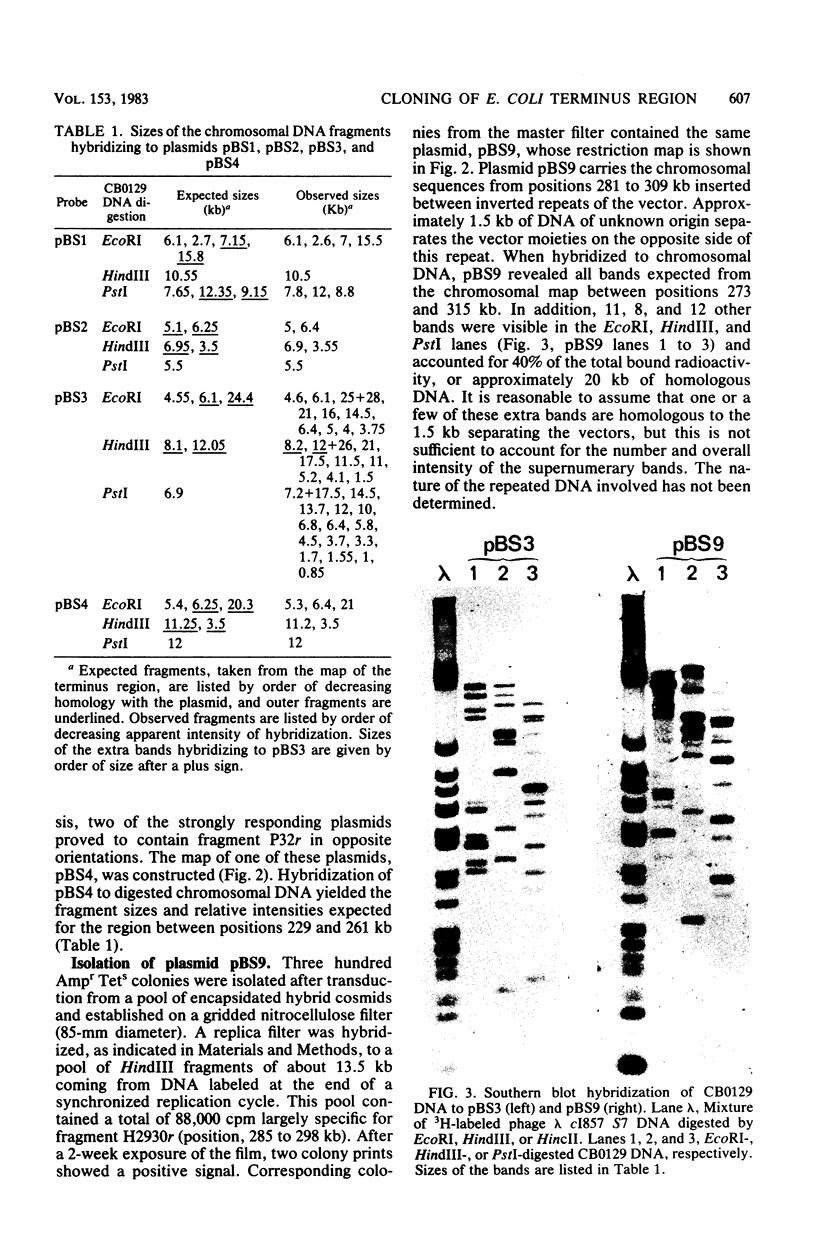
A series of plasmids have been isolated either by ligation of defined restriction fragments to plasmid pBR325 or by screening of a cosmid bank by in situ colony hybridization. Together with one previously isolated plasmid, they spanned 86% of the 30.5- to 34-min region of the genetic map of Escherichia coli K-12. Physical analysis of these plasmids and hybridizations to Southern blots confirmed the endonuclease map of this region, with the exception of a 9.3-kilobase pair inversion.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D., Germino J., Crosa J. H., Hale P. Molecular cloning of the replication terminus of the plasmid R6K. Gene. 1981 Jun-Jul;14(1-2):81–89. doi: 10.1016/0378-1119(81)90150-5. [DOI] [PubMed] [Google Scholar]
- Bitner R. M., Kuempel P. L. P1 transduction map spanning the replication terminus of Escherichia coli K12. Mol Gen Genet. 1981;184(2):208–212. doi: 10.1007/BF00272906. [DOI] [PubMed] [Google Scholar]
- Bouché J. P., Gélugne J. P., Louarn J., Louarn J. M., Kaiser K. Relationships between the physical and genetic maps of a 470 x 10(3) base-pair region around the terminus of Escherichia coli K12 DNA replication. J Mol Biol. 1982 Jan 5;154(1):21–32. doi: 10.1016/0022-2836(82)90414-4. [DOI] [PubMed] [Google Scholar]
- Bouché J. P. Physical map of a 470 x 10(3) base-pair region flanking the terminus of DNA replication in the Escherichia coli K12 genome. J Mol Biol. 1982 Jan 5;154(1):1–20. doi: 10.1016/0022-2836(82)90413-2. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Fouts K. E., Barbour S. D. Insertion of transposons through the major cotransduction gap of Escherichia coli K-12. J Bacteriol. 1982 Jan;149(1):106–113. doi: 10.1128/jb.149.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts K. E., Barbour S. D. Transductional mapping of ksgB and a new Tn5-induced kasugamycin resistance gene, ksgD, in Escherichia coli K-12. J Bacteriol. 1981 Feb;145(2):914–919. doi: 10.1128/jb.145.2.914-919.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Isono S., Isono K. Ribosomal protein modification in Escherichia coli. III. Studies of mutants lacking an acetylase activity specific for protein L12. Mol Gen Genet. 1981;183(3):473–477. doi: 10.1007/BF00268767. [DOI] [PubMed] [Google Scholar]
- Kuempel P. L., Duerr S. A., Seeley N. R. Terminus region of the chromosome in Escherichia coli inhibits replication forks. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3927–3931. doi: 10.1073/pnas.74.9.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louarn J., Patte J., Louarn J. M. Evidence for a fixed termination site of chromosome replication in Escherichia coli K12. J Mol Biol. 1977 Sep 25;115(3):295–314. doi: 10.1016/0022-2836(77)90156-5. [DOI] [PubMed] [Google Scholar]
- Louarn J., Patte J., Louarn J. M. Map position of the replication terminus on the Escherichia coli chromosome. Mol Gen Genet. 1979 Apr 17;172(1):7–11. doi: 10.1007/BF00276208. [DOI] [PubMed] [Google Scholar]
- Nyman K., Nakamura K., Ohtsubo H., Ohtsubo E. Distribution of the insertion sequence IS1 in gram-negative bacteria. Nature. 1981 Feb 12;289(5798):609–612. doi: 10.1038/289609a0. [DOI] [PubMed] [Google Scholar]
- Schoner B., Schoner R. G. Distribution of IS5 in bacteria. Gene. 1981 Dec;16(1-3):347–352. doi: 10.1016/0378-1119(81)90093-7. [DOI] [PubMed] [Google Scholar]
- Thayer R. E. An improved method for detecting foreign DNA in plasmids of Escherichia coli. Anal Biochem. 1979 Sep 15;98(1):60–63. doi: 10.1016/0003-2697(79)90705-x. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Virolle M. J., Gélugne J. P., Béjar S., Bouché J. P. Origin of Escherichia coli K-12 Hfr B7. J Bacteriol. 1983 Feb;153(2):610–615. doi: 10.1128/jb.153.2.610-615.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]