This is a wildly exciting period in the study of ion channels. Since the sequencing of Na+ and K+ channels was accomplished (Noda et al., 1984, Tempel et al., 1987), many of the functional parts of the channels have been located within the sequence. These include the pore through which ions travel (P region: Guy and Conti, 1990; Hartmann et al., 1991; Yellen et al., 1991), the voltage sensor (S4, the positively charged fourth transmembrane segment: Stühmer et al., 1989; Larsson et al., 1996; Yang et al., 1996) and the inactivation gate of both K and Na channels (NH2 terminus of Shaker B: Hoshi et al., 1990; for the Na channel, the linker between domains III and IV: Stühmer et al., 1989; West et al., 1992).
Missing from the list of identified functional parts is the activation gate. The paper by Holmgren, Smith, and Yellen in this issue of The Journal of General Physiology (Holmgren et al., 1997) and a preceding paper (Holmgren et al., 1996) give fascinating evidence about the location of this gate if not yet a complete answer.
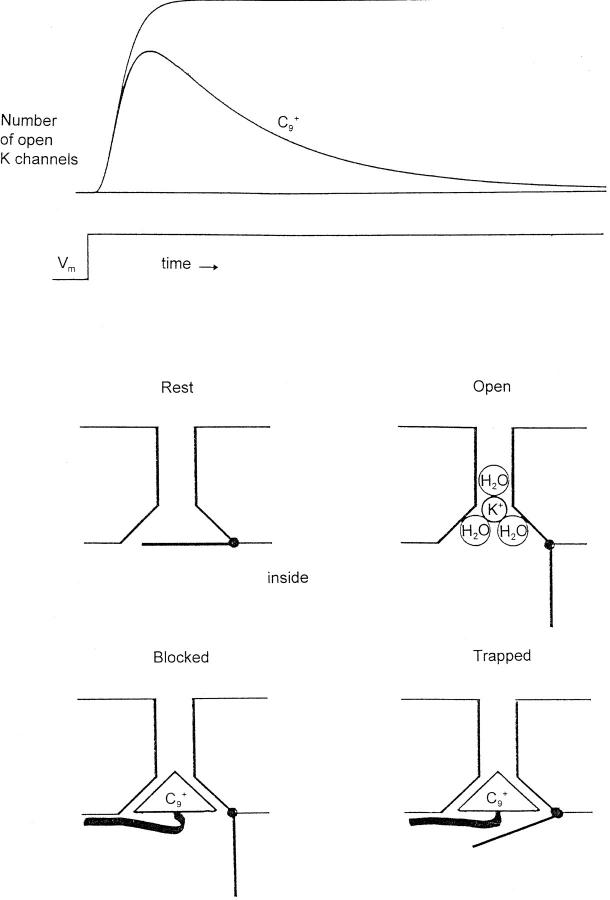
Where should one begin to look for the gate? As Holmgren et al. generously note, old experiments using TEA+ and analogues to block K channels (Armstrong, 1971) suggested that the activation gate is inside and that the search should be in the intracellular segments of the sequence. These experiments are summarized in Fig. 1. TEA+ and analogues block squid K channels only when applied inside. Further, they are “open channel blockers,” meaning they have access to their binding site in the pore only when the channel gate is open. During a voltage step the number of open K channels (measured as IK) normally rises to a steady level (Fig. 1, upper trace). If C9 +, an analogue of TEA+, is present inside the axon, the channels then “inactivate” as C9 + ions diffuse in and block them.
C9 + has a “head” group (the charged nitrogen and three surrounding ethyl groups) and a hydrophobic tail (nine carbons with their hydrogens). The head is similar in size to a K+ ion with one hydration shell, roughly 8 Å, and this is thought to make C9 + acceptable to the inner end of the pore. C9 + blocks because, unlike K+, which is thought to dehydrate partially as it moves into the narrow part of the pore (Hille, 1973), C9 + cannot shed its covalently linked ethyl groups. The hydrophobic tail increases binding of the blocker in the pore, the longer the tail the better, up to about 10 carbons (C10 +; Choi et al., 1993).
What happens when the gate tries to shut on repolarization? As might be expected, C9 + acts like a foot in the door, and hinders closing of the gate. At −60 mV few squid K channels close until C9 + has dissociated. At −100 mV, however, the closing pressure is very strong, and the gates of about one-third of the channels apparently close at least partially before C9 + has dissociated. The result is that C9 + is trapped in the channel and remains there for (relatively) a very long time. This trapping effect is easy to picture in terms of a gate at the inner end of the channel, as shown in the cartoon. Altogether, the experiments favor the idea of a stable channel with a gate at the inner end. The gate is the major moving part.
The gate thus seems to be inside, but where is it within the sequence? And where is the TEA+ binding site? Choi et al. showed that C10 + binding is affected not only by mutations in the P region but also by S6 (sixth transmembrane segment) mutations. The S6 mutation T469I, for example, greatly increases the affinity of the channel for C8 + and C10 +, almost certainly by improving interaction between the channel and the hydrophobic tail. Interestingly, the affinity increase is largest with C8 +, suggesting that C10 + is too large to fit well in the mutated channel.
The wild-type Shaker channel, when exposed to a TEA+ analogue, shows all of the states seen in the cartoon of Fig. 1 except the trapped state: the gate cannot close until C10 + leaves the channel. Holmgren et al. have now discovered that I470C, an S6 mutation, confers the ability to trap C10 +. Presumably replacing isoleucine by a smaller cysteine residue makes it sterically possible for C10 + to reside in the inner vestibule of the closed channel.
Significantly, it is relatively easy (and easier than in squid) to close the gate of this mutant even with C10 + trapped inside: the closed state was destabilized by only ∼1.2 kcal/mol, less than the energy of a hydrogen bond. This implies that C10 + occupies a cavity that changes little when the gate closes. The smaller TEA+ ion actually stabilizes the closed state. Since TEA+ is similar in size to a hydrated K+ ion, does this mean that the channel normally closes with a K+ ion in the inner vestibule? Overall, these fascinating findings make it almost irresistible to think that one or more flaps fold over the inner end of a commodious vestibule that can accommodate C10 + without much strain.
What are the flaps? Since the channel is a tetramer, it is easiest to imagine there are four flaps. Where are they, and how are they linked to S4, the voltage sensor? One candidate is the S4-5 linker, an intracellular loop of 13 amino acids. This region was tested for this role in elegant fashion (Holmgren et al., 1996) by labeling cysteine mutants with MTS reagents (Akabas et al., 1992). They found that labeling rates at some positions differed 4- to 10-fold between the open and the closed state, showing that there is movement in this region during opening and closing. Perhaps the S4-5 linkers are the flaps, but conviction must await further experiments. We eagerly await the next chapter.
references
- Akabas MH, Stauffer DA, Xu M, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science (Wash DC) 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- Armstrong CM. Interaction of tetraethylammonium ion derivatives with the potassium channel of giant axons. J Gen Physiol. 1971;58:413–437. doi: 10.1085/jgp.58.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KL, Mossman C, Aubé J, Yellen G. The internal quaternary ammonium receptor site of Shakerpotassium channels. Neuron. 1993;10:533–541. doi: 10.1016/0896-6273(93)90340-w. [DOI] [PubMed] [Google Scholar]
- Guy HR, Conti F. Pursuing the structure and function of voltage-gated channels. Trends Neurosci. 1990;13:201–206. doi: 10.1016/0166-2236(90)90160-c. [DOI] [PubMed] [Google Scholar]
- Hartmann HA, Kirsch GE, Drewe JA, Taglialatela M, Joho RH, Brown AM. Exchange of conduction pathways between two related K+channels. Science (Wash DC) 1991;251:942–945. doi: 10.1126/science.2000495. [DOI] [PubMed] [Google Scholar]
- Hille B. Potassium channels in myelinated nerve. Selective permeability to small cations. J Gen Physiol. 1973;61:669–686. doi: 10.1085/jgp.61.6.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren M, Jurman ME, Yellen G. N-type inactivation and the S4-S5 region of the Shaker K+channel. J Gen Physiol. 1996;108:195–206. doi: 10.1085/jgp.108.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren M, Smith P, Yellen G. Trapping of organic blockers by closing of voltage-dependent K+channels: evidence for a trap door mechanism of activation gating. J Gen Physiol. 1997;109:527–535. doi: 10.1085/jgp.109.5.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shakerpotassium channel inactivation. Science (Wash DC) 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the Shaker K+channel S4. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- Noda M, Shimizu S, Tanabe T, Takai T, Kayano T, Ikeda T, Takahashi H, Nakayama H, Kanaoka Y, Minamino N, Kangawa K, et al. Primary structure of Electrophorous electricussodium channel deduced from cDNA sequence. Nature (Lond) 1984;312:121–127. doi: 10.1038/312121a0. [DOI] [PubMed] [Google Scholar]
- Stühmer WF, Conti, Suzuki H, Wang X, Noda M, Yahagi N, Kubo H, Numa S. Structural parts involved in activation and inactivation of the sodium channel. Nature (Lond) 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- Tempel BL, Papazian DM, Schwartz TL, Jan YN, Jan LY. Sequence of a probable potassium channel component encoded at Shaker locus of Drosophila. . Science (Wash DC) 1987;237:770–775. doi: 10.1126/science.2441471. [DOI] [PubMed] [Google Scholar]
- West JW, Patton DE, Scheuer T, Wang Y, Goldin AL, Catterall WA. A cluster of hydrophobic amino acid residues required for fast Na+-channel inactivation. Proc Natl Acad Sci USA. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, George AL, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- Yellen G, Jurman ME, Abramson T, MacKinnon R. Mutations affecting internal TEA blockade identify the probable pore-forming region of a K+channel. Science (Wash DC) 1991;251:939–941. doi: 10.1126/science.2000494. [DOI] [PubMed] [Google Scholar]