Abstract
SliK, a K+ channel encoded by the Streptomyces KcsA gene, was expressed, purified, and reconstituted in liposomes. A concentrative 86Rb+ flux assay was used to assess the ion transport properties of SliK. SliK-mediated ionic flux shows strong selectivity for K+ over Na+ and is inhibited by micromolar concentrations of Ba2+, mirroring the basic permeation characteristic of eukaryotic K+ channels studied by electrophysiological methods. 86Rb+ uptake kinetics and equilibrium measurements also demonstrate that the purified protein is fully active.
Keywords: liposome, conduction, selectivity, flux
introduction
Mechanistic insights about ion channels have traditionally emanated from physiological antecedents. Recent discoveries of molecular determinants of voltage-dependent gating, ligand binding, pore selectivity, and second messenger modulation connect directly to the essential neurobiological functions of ion channels. The combined use of high-resolution electrical recording techniques and recombinant DNA manipulation of ion channel sequences has yielded a hailstorm of information about the molecular foundations of these functions, but detailed understanding of these proteins will also require high-resolution structures. The K+ channels of electrically excitable cells, in particular, have been productive targets for functional analysis of mutated proteins (Miller, 1991; Jan and Jan, 1994), but they have proven exceedingly resistant to the biochemical manipulations required for structure determination (Klaiber et al., 1990; Spencer et al., 1997).
Prokaryotes provide a plausible route to K+ channel structure. Examination of eubacterial and archeal genome sequences is uncovering an unexpected class of genes with remarkable resemblance to K+ channels (Milkman, 1994; Schrempf et al., 1995; MacKinnon and Doyle, 1997). In particular, these genes all display transmembrane α-helical sequences flanking an ∼20-residue stretch similar to the highly conserved, pore-forming “P-region” of eukaryotic K+ channels. One of these, the KcsA gene from Streptomyces lividans, encodes a 160-residue protein, “SliK.” The predicted transmembrane topology of this protein is reminiscent of the inward rectifier subfamily of K+ channels—a P-region flanked by two putative membrane-spanning α-helices (Schrempf et al., 1995); moreover, a glutamate residue unique to inward rectifier P-regions is also found in SliK (E71). However, the P-region of SliK actually shows higher overall similarity to the pore-forming sequences of voltage-gated K+ channels than to inward rectifiers. In stark distinction to the biologically motivated studies on eukaryotic K+ channels, the physiological roles of these prokaryotic genes are entirely unknown.
When overexpressed in Escherichia coli (Schrempf et al., 1995; Cortes and Perozo, 1997; Heginbotham et al., 1997), SliK can be purified readily in quantities (1–5 mg/ liter culture) that make spectroscopic and biochemical studies possible (leCoutre et al., 1998; Tatulian et al., 1998); indeed, the structure of this channel has recently been determined at 3.5-Å resolution (Doyle et al., 1998). We were therefore motivated to verify that the SliK preparations used in such studies are functional. It is known that SliK forms a stable tetramer in detergent micelles (Cortes and Perozo, 1997; Heginbotham et al., 1997), as expected for a K+ channel (MacKinnon, 1991) and that it shows single-channel activity when reconstituted into planar lipid bilayer membranes (Schrempf et al., 1995; Cuello et al., 1998). However, single-channel recording, because of its extreme sensitivity and intrinsically anecdotal character, cannot by itself argue for the functional competence of a biochemical preparation. Here we describe an assay that permits a quantitative evaluation of SliK ion-transport activity. Reconstitution of SliK into liposomes renders these membrane vesicles selectively permeable to K+ and its close analogues, as expected of a K+ channel with properties similar to those of its well-studied eukaryotic homologues. The results demonstrate the functional competence of SliK protein purified from an E. coli heterologous expression system.
materials and methods
Materials
All chemicals were of reagent grade. E. coli phosphatidylethanolamine (PE),1 bovine brain phosphatidylcholine and phosphatidylserine, egg phosphatidylglycerol (PG), bovine heart cardiolipin (CL), and mixed E. coli lipids obtained from Avanti Polar Lipids (Alabaster, AL) were stored at −70°C in chloroform solutions under nitrogen. Mixed soy lipids (asolectin) were obtained ∼35 yr ago from Associated Concentrates (Woodside, NY). Dodecylmaltoside (C12M) was from Calbiochem Corp. (San Diego, CA), and CHAPS (3-[(3-cholamidopropyl)dimethyl-ammonio]- 1-propane-sulfonate) was from Pierce Chemical Co. (Rockford, IL). N-Methylglucamine (NMG) was obtained from Sigma Chemical Co. (St. Louis, MO) as the free base. NTA-agarose Ni2+ affinity matrix was obtained from QIAGEN Inc. (Chatsworth, CA). The cation-exchange matrix, Dowex 50 X-4-100 (H+ form) (Sigma Chemical Co., St. Louis, MO), was initially washed with MeOH, and then converted to either Tris+ or NMG+ form and thoroughly washed with water. 86Rb+ was obtained from New England Nuclear (Boston, MA). Polystyrene column bodies (No. 29920) used for the flux assay were obtained from Pierce Chemical Co. and were reused after washing.
Molar extinction coefficient at 280 nm of purified SliK, 38,500 ± 1,500 M−1 cm−1, was determined by quantitative amino acid analysis (W.M. Keck facility, Yale University, New Haven, CT) and is within 8% of the value calculated from the tryptophan + tyrosine content of the protein (Gill and von Hippel, 1989).
Purification and Reconstitution
SliK was expressed in E. coli and purified as described (Heginbotham et al., 1997), with minor modifications. Cells were grown in 30-liter cultures of “Terrific broth,” and expression was induced at A550 = 1.0. Bacterial membranes were prepared and stored at −70°C in aliquots in buffer A (100 mM NaPi, 5 mM NaCl, pH 7.0) containing 200 mM sucrose. For purification of SliK, an aliquot was thawed, extracted in 15 mM C12M, and loaded onto a Ni2+ affinity column in the presence of 30–35 mM imidazole. The purified protein was stored at 4°C at a concentration of 2–10 mg/ml in buffer A containing 1 mM C12M, with or without 400 mM imidazole. Activity measurements were performed 1–7 d after purification, with no obvious decrease in function over that time.
All liposome reconstitution procedures were carried out at room temperature. Lipids were dried under a nitrogen stream, dissolved in pentane, redried, and resuspended at 10 mg/ml in buffer B (450 mM KCl, 10 mM HEPES, 4 mM NMG, pH 7.4) using a bath sonicator. CHAPS was added to a final concentration of 37 mM to solubilize the lipid, and the suspension was incubated for 2 h. SliK was added to the lipid at the desired protein: lipid ratio, and the mixture was incubated 20 min. Reconstituted liposomes were formed by centrifuging the detergent-solubilized lipid/SliK mixture through gel filtration columns. These were prepared from Sephadex G-50 (fine) swollen overnight in buffer B and poured into 5-ml disposable columns (1.5-ml bed volume). Columns were prespun in a clinical centrifuge at the highest speed setting (∼1,000 g) for 13 s; detergent-solubilized samples (95 μl) were loaded on the top of each column, and liposomes were recovered by spinning the columns at 700 g for 60 s. Liposomes were prepared 0.5–3 h before the flux assay and were stored at room temperature until use. Immediately before the flux measurement, a second G-50 spin column equilibrated with buffer C (400 mM sorbitol, 10 mM Hepes, 4 mM NMG, 30–50 μM KCl, pH 7.4) was used to exchange the extra-liposomal solution. Except for experiments reported in Fig. 5, mixed E. coli lipids were used for reconstitution.
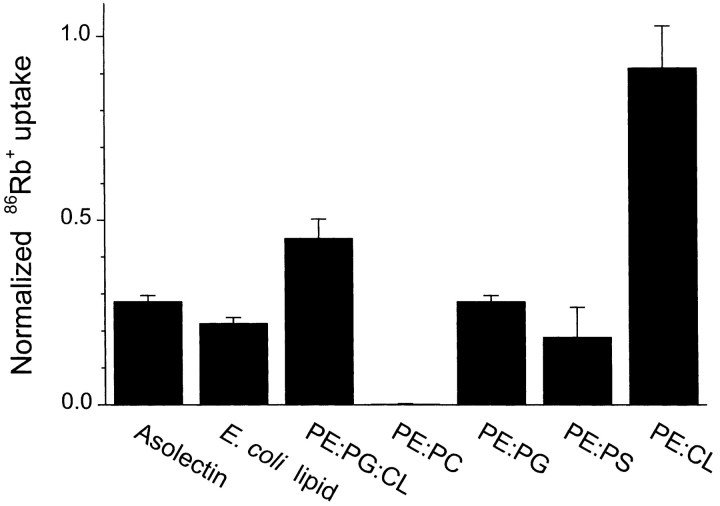
Figure 5.
Lipid dependence of SliK-mediated 86Rb+ uptake. Liposomes were made from the indicated lipid mixtures, incorporating SliK at a concentration of 2.5 μg/mg lipid. Values of 3.5-min 86Rb+ uptake were normalized to the equilibrium valinomycin- mediated uptake in the same vesicles. Bars reflect means ± SEM of three measurements. In mixtures of defined lipids, PE made up 75% of the lipid mass. For the PE:PG:CL mixture, the lipid composition was 75% PE, 15% PG, and 10% CL.
Flux Assay
86Rb+ uptake was initiated by mixing the liposomes with 1–3 vol flux buffer (buffer C containing 86Rb+ at a final concentration of ∼0.5 μCi/ml, 5 μM). Except as noted below, extra-liposomal 86Rb+ was removed at each time-point by passing an aliquot of this uptake mixture over a 1.5–ml Dowex cation exchange column in the Tris+ or NMG+ form. Typically, a 55–100-μl sample of uptake reaction mixture was loaded onto the column and eluted into scintillation vials with 1.5 ml 400 mM sucrose or sorbitol. Samples were mixed with 15 ml scintillation fluid and counted on a liquid scintillation counter (Beckman Instruments, Inc., Fullerton, CA). Nonspecific binding to the Dowex columns was minimized by prewashing each column with 2 ml of 5 mg/ml BSA in 400 mM sorbitol, followed by 2 ml 400 mM sorbitol.
In many experiments, we used valinomycin to assay equilibrium uptake into all the liposomes present. In such cases, valinomycin (100 μg/ml in EtOH) was added to the uptake reaction mixture, at a final concentration of ∼1 μg/ml (∼1,000-fold higher molar concentration than that of SliK), and samples were taken after a 2–6-min incubation. Under these conditions, valinomycin-mediated 86Rb+ uptake was complete within a minute.
We initially investigated the limits of the flux assay using control liposomes that contained valinomycin but no SliK. We found that in these controls, the 86Rb+ signal was sensitive to the ionic strength of the flux buffer, decreasing above 10 mM regardless of the salt used. We do not understand the reason for this sensitivity, but it imposes a practical limit on several of the experiments shown here. In particular, we could not measure uptake under standard “bi-ionic” conditions; i.e., with the internal and external solutions containing different ions at similar ionic strengths. This problem was associated with the Dowex beads and could be partially alleviated using Sephadex G-50 spin columns instead (a more time-consuming procedure). In such cases, 95-μl aliquots of flux uptake reaction were applied to G-50 columns swollen in buffer C, using the standard spin-column protocol described above.
In cases requiring variation of external ionic conditions, we divided the experiment into a series of “runs.” In a given run, a single batch of liposomes was aliquoted into flux buffers containing different test ions at the desired concentration, and 86Rb+ uptake was allowed to proceed. Uptake for a test condition was normalized to control samples from which test ions had been omitted. Data from two to three runs were averaged to generate final normalized results.
results
SliK labeled with a NH2-terminal hexahistidine tag may be purified in one step on a metal-affinity column and stored for weeks in C12M micellar solution without loss of homotetrameric structure (Heginbotham et al., 1997). Our initial attempts to examine SliK activity in liposomes failed (Heginbotham et al., 1997), but we have now developed a robust and quantitative assay for ionic flux catalyzed by purified SliK.
SliK-mediated Ionic Fluxes in Reconstituted Liposomes
We employ a 86Rb+ flux measurement under “concentrative uptake” conditions, similar to that used for studying other ion channels in sealed membrane vesicles (Talvenheimo et al., 1982; Garty et al., 1983; Goldberg and Miller, 1991). Liposomes loaded with a high-KCl solution are incubated in low-KCl medium spiked with trace amounts of 86Rb+. Protein-free liposomes are impermeable to small inorganic ions and do not take up 86Rb+ on our experimental time scale. In contrast, liposomes reconstituted with a K+-conductive pathway are expected to take up 86Rb+ readily; moreover, 86Rb+ will be accumulated inside the liposomes at concentrations much higher than in the external medium. This concentration effect arises because the preestablished K+ gradient sets up and “clamps” a transmembrane electrical potential that 86Rb+, being at trace concentrations, must follow. If K+ and Rb+ are the only permeant species, then at equilibrium the 86Rb+ concentration ratio must be identical to that of K+:
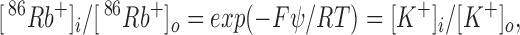 |
1 |
where ψ is the liposome membrane potential, and the subscripts refer to inside and outside concentrations.
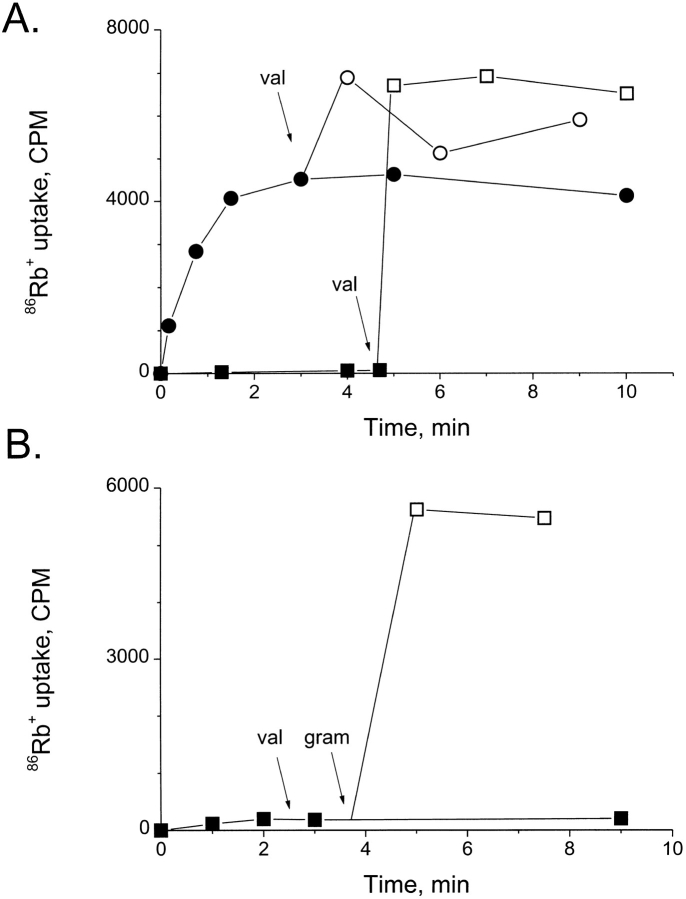
Fig. 1 A illustrates the 86Rb+ influx assay. In SliK-containing liposomes, 86Rb+ is taken up into the intraliposomal space and reaches a steady level in 3–5 min. Addition of the K+-selective ionophore valinomycin, which renders all the liposomes permeable to K+ and Rb+, increases the tracer uptake ∼30%. We attribute the initial, valinomycin-independent uptake to liposomes containing functional SliK, and the subsequent ionophore-stimulated uptake to a minor fraction of liposomes devoid of the channel. The figure also shows that in pure phospholipid liposomes, no 86Rb+ uptake is observed until valinomycin is added. Use of valinomycin in this way provides a convenient normalization procedure by which we could compare SliK-mediated 86Rb+ uptake in different preparations. The uptake observed in this flux assay represents a massive concentration (∼1,000-fold) of tracer inside the liposomes; in a typical experiment, in which the intraliposomal aqueous space represents 0.1–0.3% of the total sample volume, ∼50% of the 86Rb+ originally added to the external solution becomes trapped within the liposomes. Control experiments show that 86Rb+ uptake is abolished by removing the K+ gradient, by disrupting the liposome membrane with detergent, or by boiling the SliK protein before reconstitution (data not shown).
Figure 1.
Concentrative uptake of 86Rb+. (A) Time course of 86Rb+ uptake into liposomes reconstituted with 0.9 μg SliK/mg lipid (circles) or into control liposomes containing no protein (squares). Hollow points refer to samples taken after addition of valinomycin (arrows). Each point represents a single 60-ml sample containing 0.15 mg lipid, removed from the reaction mixture. External 86Rb+ concentration was ∼0.2 μCi/ml (104 cpm per 60-ml sample). (B) Liposomes were formed in the presence of SliK as in A, but were loaded with 450 mM NaCl instead of KCl. Where indicated, valinomycin was added, and at 3 min the sample was split into two parts: a control that was allowed to proceed without further addition (▪, at 9 min), and a test sample to which gramicidin A (10 μg/ml) was added (□).
The simple fact that 86Rb+ is concentrated in SliK- reconstituted liposomes shows that SliK strongly discriminates between K+ and Cl−, the only other abundant ion in the system; if, in addition to K+, Cl− were significantly permeant, then the transmembrane KCl gradient initially present would fail to establish a large membrane potential, and, indeed, both K+ and Cl− would quickly leak out of the liposomes and dissipate the gradient. SliK also discriminates between K+ and Na+; SliK-reconstituted liposomes loaded with Na+ instead of K+ fail to support 86Rb+ influx (Fig. 1 B) as expected if SliK, like eukaryotic K+ channels, selects against Na+; in such a case, 86Rb+, though itself permeant, cannot accumulate in the internal space since electroneutrality demands one-for-one exchange with the internal cation. As expected under these conditions, addition of valinomycin does not promote 86Rb+ uptake, while the liposomes do fill in the presence of gramicidin A, an ionophore that can transport all group IA cations (Fig. 1 B).
Ionic Selectivity of SliK
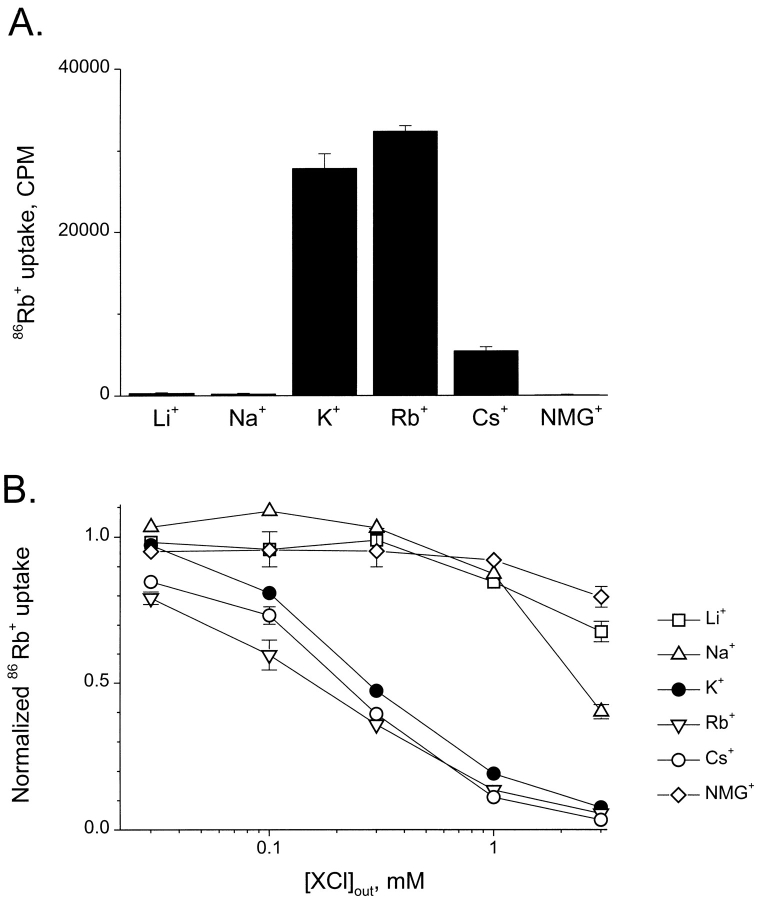
Eukaryotic K+ channels all display a characteristic selectivity among monovalent cations; K+, Rb+, and Cs+ permeate readily, while larger organic cations (i.e., NMG+) and smaller inorganic ones (Na+, Li+) do not (Hille, 1991; Heginbotham and MacKinnon, 1993). We investigated the relative permeabilities of these cations to SliK using two variations of the 86Rb+ flux assay. First, we examined which cations would support concentrative 86Rb+ uptake by loading SliK-containing liposomes with the Cl− salts of different test cations. Liposomes filled with K+ or Rb+ exhibited comparable levels of 86Rb+ accumulation. Uptake was lower with Cs+ and undetectable with Li+, Na+, and NMG+ as the internal cation (Fig. 2 A). Since tracer accumulation requires a large negative membrane potential, the results demonstrate that SliK is permeable to K+, Rb+, and Cs+. The absence of uptake observed with the other ions is consistent with the conventional explanation that SliK selects against these cations, but other possibilities should also be considered. For instance, since the process by which proteoliposomes form may be sensitive to ionic conditions, the efficiency of proper SliK incorporation may vary with different cations; furthermore, the functional integrity of the SliK protein, like some eukaryotic K+ channels, may be compromised by exposure to K+-free conditions (Almers and Armstrong, 1980; Korn and Ikeda, 1995; Khodakhah et al., 1997).
Figure 2.
Selectivity of SliK- dependent uptake. (A) Variation of internal cation. Liposomes were formed as in materials and methods except that the indicated cation was substituted for K+. Samples (60 μl, 0.12 mg lipid) were taken after 5 min of influx. Each bar represents samples reconstituted in triplicate. (B) Variation of external cation. 5-min uptake was determined in flux buffers to which test cations had been added at the indicated concentrations (on a background of 50 μM K+). Sephadex G-50 columns were used to terminate the uptake reaction, as in materials and methods. Samples were normalized to duplicate control samples containing 50 μM KCl but no added test ion. Points and error bars represent means and ranges of two separate runs.
To avoid these potential pitfalls, we examined the ionic selectivity of SliK using a second approach. Here, liposomes were loaded with high KCl, and influx of 86Rb+ was assayed in solutions containing different test cations at varying external concentrations. Any permeant test cation will depolarize the membrane and thus reduce tracer uptake, but an impermeant ion will have no effect. The experiments of Fig. 2 B show that accumulation of 86Rb+ is inhibited strongly by addition of external K+, Rb+, or Cs+. Half inhibition by these ions occurs at ∼300 μM. In contrast, addition of Li+ or NMG+ had no effect on 86Rb+ uptake. Na+ produced half inhibition at ∼3 mM; strictly interpreted, this result suggests that Na+ is 10% as permeant as K+, but we hesitate to assign this value unconditionally since the Na+ measurement was made at external ionic strength perilously close to the operational limits of the technique (see materials and methods). Thus, on the basis of this assay, SliK displays the permeability sequence familiar in eukaryotic K+ channels (Hille, 1991): K+ ≈ Rb+ ≈ Cs+ > Na+ >> Li+, NMG+.
The selectivity profiles derived from Fig. 2, A and B, are quite similar, but not identical. Cs+ appears to be more permeant when present externally than when loaded at high concentrations internally. This is not a disquieting result since Cs+ permeability of eukaryotic K+ channels is known to be sensitive to the ionic conditions used for its determination (Heginbotham and MacKinnon, 1993).
Ba2+ Block
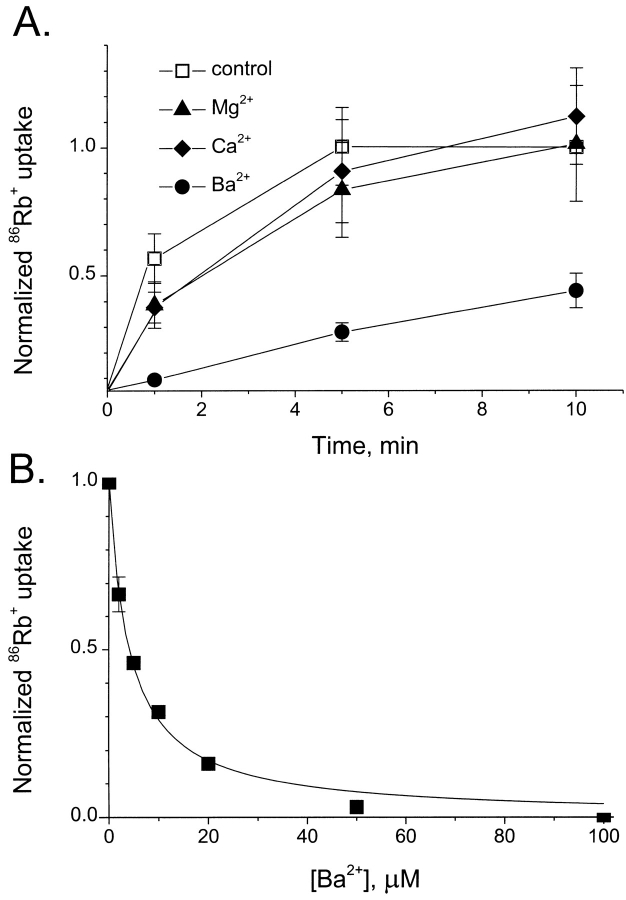
In addition to their trademark ionic selectivity, eukaryotic K+ channels share a second distinctive pore property: block by Ba2+ (Hille, 1991). Whereas many compounds (peptide toxins, alkylammonium and guanidinium derivatives, and various multivalent cations) act on subsets of K+ channels, Ba2+ is the only known universal inhibitor of K+ channels. This ion has been viewed, uniquely among the group IIA cations, as a “divalent analogue” of dehydrated K+ (Armstrong and Taylor, 1980; Eaton and Brodwick, 1980; Latorre and Miller, 1983) that prevents K+ conduction by binding tightly within the pore. We therefore tested Ba2+ as an inhibitor of SliK-mediated Rb+ uptake rates. The effects of divalent cations on kinetic profiles of 86Rb+ uptake are shown in Fig. 3 A. While 50 μM Ca2+ or Mg2+ has almost no effect on uptake, Ba2+ slows tracer flux dramatically. This effect is quantified in Fig. 3 B, where we examine the concentration dependence of Ba2+ inhibition. External Ba2+ reduces the initial rate of 86Rb+ uptake (20-s influx) according to a simple Langmuir curve with K i = 4 μM, mirroring Ba2+ block of eukaryotic K+ channels assayed under voltage clamp.
Figure 3.
Ba2+ block of SliK-mediated flux. Liposomes were reconstituted with SliK at 1 μg/mg. (A) Time courses of 86Rb+ influx were followed in the presence of 50 μM of the indicated divalent cations. Samples were normalized to 10-min control samples with no added divalent cation. (B) Concentration dependence of Ba2+ block. The indicated concentration of Ba2+ was added externally, and initial rate of 86Rb+ uptake was measured by taking samples at 20 s. Points and error bars represent means ± SEM of three separate runs.
Kinetics of 86Rb+ Uptake
The detailed characteristics of the 86Rb+ uptake time course depend on the concentration of SliK used in the reconstitution. At the submaximal concentration (0.5 μg SliK/mg lipid) shown in Fig. 4 A, 86Rb+ enters ∼35% of the liposomes (as measured by the fraction of uptake in the presence of valinomycin) with an approximately exponential time course (τ = 58 s). At higher SliK concentration, the influx speeds up (τ = 29 s), and the fraction of 86Rb+-accessible liposomes increases to ∼80% of the total internal space. This SliK dependence of the steady-level 86Rb+ uptake is shown in greater detail in Fig. 4 B. Uptake increases linearly at low SliK concentrations (<0.5 μg/mg lipid) and ultimately saturates, reaching 80–90% of the valinomycin-mediated uptake at high protein concentrations. As discussed later, a quantitative analysis of this concentration dependence permits an absolute estimate of the number of functional molecules in the purified SliK preparation.
Figure 4.
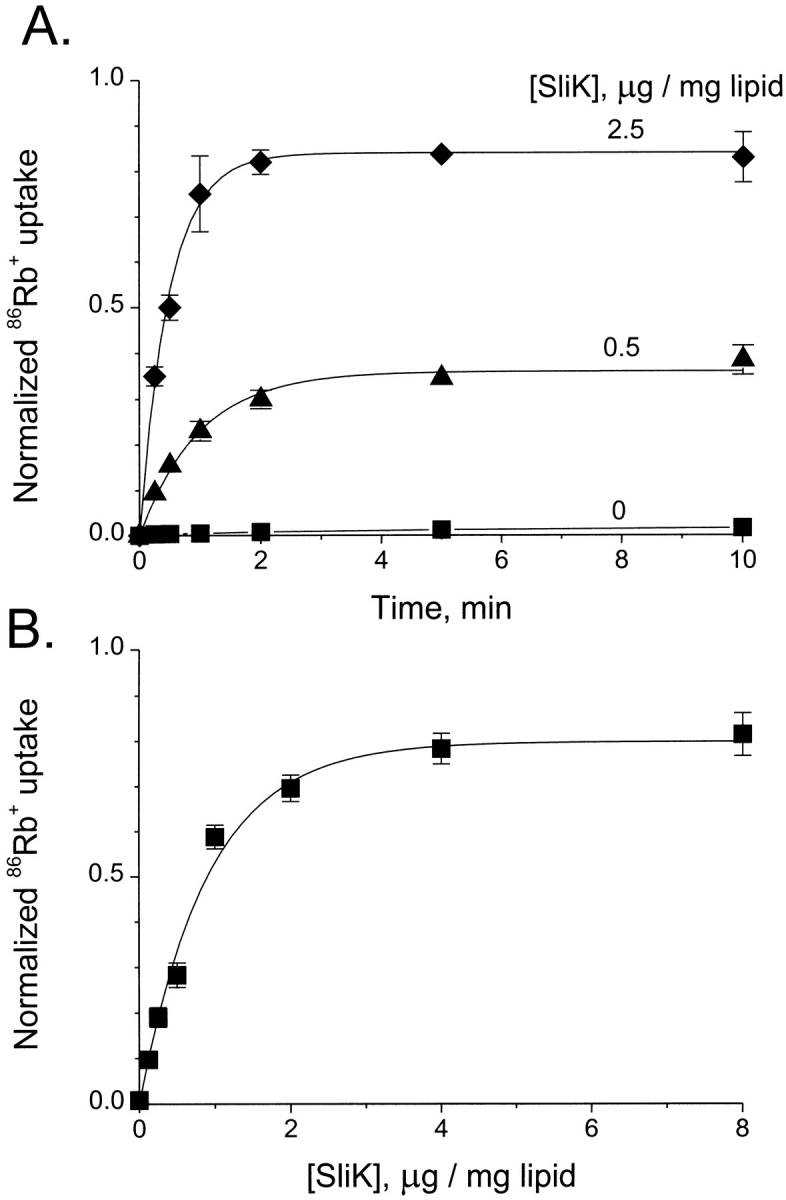
86Rb+ uptake at different SliK concentrations. (A) Time course of 86Rb+ uptake into vesicles containing 0, 0.5, or 2.5 μg SliK/mg lipid. Samples were removed from a common reaction mixture at the indicated times and normalized to valinomycin- induced uptake in the same vesicles. Solid curves are single exponentials with time constants of 58 and 29 s for 0.5 and 2.5 μg/mg SliK, respectively. (B) 86Rb+ uptake as a function of SliK concentration. Equilibrium 86Rb+ accumulation was measured after 10 min in liposomes containing SliK at the indicated concentrations. The solid curve is drawn according to Eq. 4, where r = 460 Å, φ = 1, θ = 0.20. Each point reflects mean ± SEM of three independent measurements.
Lipid Requirements
While developing this assay, we noticed that SliK activity is sensitive to the lipids used in the reconstitution mixture. Fig. 5 compares SliK-dependent 86Rb+ uptake in liposomes of different lipid compositions. Robust flux activity is observed in crude soy lipids (asolectin), native E. coli lipids, or in a reconstructed E. coli phospholipid blend of purified PE:PG:CL. A competent lipid cocktail could be further reduced to two components: PE and a negatively charged phospholipid. The negatively charged component appears to be necessary for activity, since SliK-mediated flux is undetectable in liposomes made from E. coli phosphatidylethanolamine: bovine brain phosphatidylcholine mixtures; all negatively charged headgroups tested, egg phosphatidylglycerol, bovine brain phosphatidylserine, and bovine heart cardiolipin, support flux activity. These results merely provide practical guidance, not mechanistic insight; we do not know whether the lipid affects the SliK protein itself or the process by which SliK is incorporated into the reconstituted liposome membranes.
discussion
In only four years, prokaryotic K+ channel homologues have progressed from oddities in genome sequence databases (Milkman, 1994) to proteins of known structure at atomic-level resolution (Doyle et al., 1998). The SliK protein, a target of particularly intense study, shows basic molecular features expected for a K+ channel: tetrameric architecture (Heginbotham et al., 1997; Doyle et al., 1998), high unitary ion transport rates (Schrempf et al., 1996; Cuello et al., 1998), and a multi-ion, single-file pore axially positioned in a tetrameric complex (Doyle et al., 1998). Largely due to the ease with which milligram quantities of SliK can be obtained, this channel provides a unique opportunity for structural analysis using direct protein-level techniques. We have therefore sought to quantify the functional integrity of purified SliK protein.
We have chosen a flux assay rather than an electrophysiological measurement to gauge the functional character of SliK. For the purpose of evaluating the competence of a purified channel preparation, single-channel recording suffers a fatal weakness: extraordinarily high sensitivity. A denumerable collection of single channel-like observations cannot validly argue for, or even qualitatively assess, the behavior of milligram quantities of protein molecules in a biochemical preparation. In contrast, liposome flux methods, although poor for detailed characterization of ion channels, are uniquely suited for surveying fundamental ion-transport properties in a molecular ensemble of purified proteins.
There are several aspects of our flux assay worth noting. First, we used 86Rb+ to avoid problems associated with the short-lived isotope 42K+; eukaryotic K+ channels are known to conduct Rb+ well (Hille, 1991). Second, since we are studying channel-mediated electrodiffusive transport rather than directional ion pumping, the orientation of the reconstituted protein within the liposome membrane is irrelevant to the basic features of the assay; 86Rb+ can enter any liposome containing active SliK, regardless of whether the protein is facing inward or outward. Third, 86Rb+ uptake was sensitive to ionic strength; the high values of uptake reported here were seen only below 10 mM salt, conditions that would be considered electrophysiologically repugnant, at least for eukaryotic cells. We do not understand the reason for this phenomenon, but we suspect that it accounts for ∼100-fold lower 86Rb+ flux activity described by Cuello et al. (1998) at 200 mM ionic strength.
Basic Transport Properties of SliK
The ion-transport properties of the SliK protein are precisely those expected for a K+ channel. SliK renders liposome membranes selectively permeable to K+ and its close analogues Rb+ and Cs+. Na+ is at most 10% as permeant as K+, and no permeability at all can be detected for Li+, organic cations, or Cl−. Moreover, SliK-mediated K+ flux is specifically blocked by Ba2+ in the micromolar range. These results are in harmony with those obtained by Cuello et al. (1998) in a similar assay; the quantitative differences between our two studies most likely arise from differences in the ionic conditions used.
The unitary turnover rates of K+ channels are very rapid, 106–107 s−1 under physiological conditions (Latorre and Miller, 1983; Hille, 1991), and so we expect that in symmetric salt solution, a liposome containing only a single ion channel should equilibrate tracer on the millisecond time scale (Miller, 1984). Since SliK-mediated 86Rb+ influx occurs on the time scale of minutes (Figs. 1 and 4), it might seem that the SliK channel has an atypically low conductance. However, this expectation is mistaken because our measurements were done under highly asymmetric ionic conditions that produce concentrative uptake: at very low concentrations of external permeant ions; i.e., under Vmax/K m conditions (Latorre and Miller, 1983). Assuming simple ionic diffusion and constancy of external ionic concentrations, the time constant for influx, τ, into a spherical liposome of radius r containing a single open channel is:
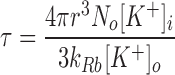 |
2 |
where k Rb is the second-order rate constant of 86Rb+ influx, and N 0 is Avogadro's number. Applying the parameters of the experiment (Fig. 4 A, 0.5 μg SliK/mg) and r = 400 Å (see below), we calculate that k Rb ≅ 3 × 107 M−1 s−1, similar to values found in other K+ channels (Latorre and Miller, 1983; Heginbotham and MacKinnon, 1993). This rate constant predicts a single-channel conductance on the order of 100 pS in symmetrical 100 mM Rb+, assuming that conductance varies linearly with concentration over this range and that the channel open probability is ∼0.1 (Heginbotham, L., and LeMasurier, data not shown). This estimate agrees remarkably well with the 90–140-pS conductance of the channels observed in planar lipid bilayers (Schrempf et al., 1996; Cuello et al., 1998).
The experiments demonstrating that SliK catalyzes high transport rates were performed at neutral pH. This result appears to contradict recent experiments (Cuello et al., 1998) reporting a sharp optimum for flux at pH 4 and no measurable activity at pH 7.0–7.5. Again, the different ionic conditions used in the two studies may provide a partial explanation for this apparent discrepancy. Our assays were all carried out in negatively charged liposomes at low (5–10 mM) ionic strength; under these conditions, the local pH near the external surface of the membrane will be substantially lower than in bulk solution—1.5–2 pH units under our conditions (McLaughlin, 1977). This effect of surface electrostatics would not be significant at the 20-fold higher ionic strength used by Cuello et al. (1998).
Functional Competence of the Purified SliK Preparation
The flux method used here allows us to address a central question about the purified SliK protein. What is the functional competence of this biochemical preparation; i.e., what proportion of the purified protein operates as an active channel, and how much is functionally compromised?
Any liposome containing at least one active SliK channel fills with 86Rb+ in several minutes; in contrast, liposomes without SliK do not accumulate 86Rb+ at all. Thus, the level of equilibrium 86Rb+ uptake in a given experiment is a direct measure of the fraction of liposomes that contain at least one active SliK channel. If SliK protein reconstitutes randomly into liposomes, this fraction, f, can be calculated from a Poisson distribution:
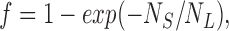 |
3 |
where N S and N L represent the number of SliK molecules and liposomes, respectively, in the sample. To avoid the intractability that would arise from a wholly realistic picture of the liposome population, we assume that all liposomes are spheres of radius r and that there is a fraction of liposomes, θ, incapable of reconstituting protein, as has been observed in other reconstitution studies (Eytan, 1982; Goldberg and Miller, 1991). Then, Eq. 3 leads to a prediction of the variation of equilibrium 86Rb+ uptake as a function of SliK concentration, S (grams SliK/grams lipid):
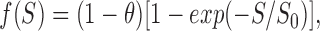 |
4 |
where the “characteristic concentration” S 0 is
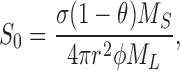 |
4A |
φ is the “competence coefficient,” the fraction of SliK protein that is functionally active, M S and M L represent molecular masses of SliK (74,000) and phospholipid (720), respectively, and σ is the area per lipid molecule in a PE-rich, fluid-phase lipid bilayer (30 Å2; Rand and Parsegian, 1989). Thus, we expect that the SliK titration curve (i.e., the 86Rb+-accessible space as a function of SliK) should follow a saturating exponential function, as is observed (Fig. 4 B). The parameters governing the SliK titration curve are known (M L, M S, σ, and r within a factor of ∼2) except for the crucial quantity to be determined, φ, the fraction of active protein. The data of Fig. 4 B tell us unambiguously that a large fraction of SliK is functionally active, with the solid curve drawn with φ = 1, r = 460 Å. Because our assumption of homogeneous liposome size is certainly unrealistic, we do not consider the fit value of r to be a precise measurement of liposome size; nevertheless, this value falls squarely in the range (150–1,600 Å, mean = 630 Å, n = 29) observed on our SliK-reconstituted liposomes in the electron microscope (data not shown), and on numerous preparations of unilamellar liposomes formed from detergent solutions under various conditions (Rhoden and Goldin, 1979; Zumbuel and Weder, 1981; Nozaki et al., 1982). We therefore conclude that SliK is fully active as a K+-selective channel.
Acknowledgments
We are grateful to Linda Melanson for electron microscopy and to Merritt Maduke, Michael Lawrence, and Fred Sigworth for gratuitous criticism of the manuscript.
This study was supported by National Institutes of Health grant GM-31768.
Abbreviations used in this paper
- CL
bovine heart cardiolipin
- NMG
N--methylglucamine
- PE
E. coli phosphatidylethanolamine
- PG
egg phosphatidylglycerol
references
- Almers W, Armstrong CM. Survival of K+ permeability and gating currents in squid axons perfused with K+-free media. J Gen Physiol. 1980;75:61–78. doi: 10.1085/jgp.75.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CM, Taylor SR. Interaction of barium ions with potassium channels in squid giant axons. Biophys J. 1980;30:473–488. doi: 10.1016/S0006-3495(80)85108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes DM, Perozo E. Structural dynamics of the Streptomyces lividans K+ channels (SCK+): oligomeric stoichiometry and stability. Biochemistry. 1997;36:10343–10352. doi: 10.1021/bi971018y. [DOI] [PubMed] [Google Scholar]
- Cuello LG, Romero JG, Cortes DM, Perozo E. pH-dependent gating in the Streptomyces lividans K+channel. Biochemistry. 1998;37:3229–3236. doi: 10.1021/bi972997x. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Cabral JM, Pfuetzner A, Kuo JM, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+conduction and selectivity. Science. 1998;280:69–76. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Eaton DC, Brodwick MS. Effect of barium on the potassium conductance of squid axons. J Gen Physiol. 1980;75:727–750. doi: 10.1085/jgp.75.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eytan GD. Use of liposomes for reconstitution of biological functions. Biochim Biophys Acta. 1982;694:185–202. doi: 10.1016/0304-4157(82)90024-7. [DOI] [PubMed] [Google Scholar]
- Garty H, Rudy B, Karlish SJD. A simple and sensitive procedure for measuring isotope fluxes through ion-specific channels in heterogeneous populations of membrane vesicles. J Biol Chem. 1983;258:13094–13099. [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Goldberg AFX, Miller C. Solubilization and functional reconstitution of a chloride channel from Torpedo californicaelectroplax. J Membr Biol. 1991;124:199–206. doi: 10.1007/BF01994354. [DOI] [PubMed] [Google Scholar]
- Heginbotham L, Odessey E, Miller C. Tetrameric stoichiometry of a prokaryotic K+channel. Biochemistry. 1997;36:10335–10342. doi: 10.1021/bi970988i. [DOI] [PubMed] [Google Scholar]
- Heginbotham L, MacKinnon R. Conduction properties of the cloned Shakerchannel. Biophys J. 1993;65:2089–2096. doi: 10.1016/S0006-3495(93)81244-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, B. 1991. Ionic Channels of Excitable Membranes. Sinauer Associates, Inc. Sunderland, MA.
- Jan LY, Jan YN. Potassium channels and their evolving gates. Nature. 1994;371:119–122. doi: 10.1038/371119a0. [DOI] [PubMed] [Google Scholar]
- Khodakhah K, Melishchuk A, Armstrong CM. Killing K channels with TEA+ . Proc Natl Acad Sci USA. 1997;94:13335–13338. doi: 10.1073/pnas.94.24.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaiber K, Williams N, Roberts TM, Papazian DM, Jan LY, Miller C. Functional expression of Shaker K+channels in a baculovirus-infected insect cell line. Neuron. 1990;5:221–226. doi: 10.1016/0896-6273(90)90311-3. [DOI] [PubMed] [Google Scholar]
- Korn SJ, Ikeda SR. Permeation selectivity by competition in a delayed rectifier potassium channel. Science. 1995;269:410–412. doi: 10.1126/science.7618108. [DOI] [PubMed] [Google Scholar]
- Latorre R, Miller C. Conduction and selectivity in potassium channels. J Membr Biol. 1983;71:11–30. doi: 10.1007/BF01870671. [DOI] [PubMed] [Google Scholar]
- leCoutre, J., L.R. Narasimhan, C.K.N. Patel, L. Heginbotham, and C. Miller. 1998. Fourier transform infrared spectroscopy reveals a rigid α-helical assembly for a tetrameric K+ channel from Streptomyces lividans. Proc. Natl. Acad. Sci. USA. In press. [DOI] [PMC free article] [PubMed]
- MacKinnon R. Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature. 1991;500:232–235. doi: 10.1038/350232a0. [DOI] [PubMed] [Google Scholar]
- MacKinnon R, Doyle DA. Prokaryotes offer hope for potassium channel structural studies. Nat Struct Biol. 1997;4:877–879. doi: 10.1038/nsb1197-877. [DOI] [PubMed] [Google Scholar]
- McLaughlin S. Electrostatic potentials at membrane-solution interfaces. Curr Top Membr Transport. 1977;9:71–144. [Google Scholar]
- Milkman R. An Escherichia colihomologue of eukaryotic potassium channel proteins. Proc Natl Acad Sci USA. 1994;91:3510–3514. doi: 10.1073/pnas.91.9.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. Ion channels in liposomes. Annu Rev Physiol. 1984;46:549–558. doi: 10.1146/annurev.ph.46.030184.003001. [DOI] [PubMed] [Google Scholar]
- Miller C. 1990: annus mirabilis of potassium channels. Science. 1991;252:1092–1096. doi: 10.1126/science.252.5009.1092. [DOI] [PubMed] [Google Scholar]
- Nozaki Y, Lasic DD, Tanford C, Reynolds JA. Size analysis of phospholipid vesicle preparations. Science. 1982;217:366–367. doi: 10.1126/science.7089571. [DOI] [PubMed] [Google Scholar]
- Rand RP, Parsegian AV. Hydration forces between lipid bilayers. Biochim Biophys Acta. 1989;988:351–376. [Google Scholar]
- Rhoden V, Goldin SM. Formation of unilamellar lipid vesicles of controllable dimensions by detergent dialysis. Biochemistry. 1979;18:4173–4176. doi: 10.1021/bi00586a020. [DOI] [PubMed] [Google Scholar]
- Schrempf H, Schmidt O, Kummerlin R, Hinnah S, Muller D, Betzler M, Steinkamp T, Wagner R. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. . EMBO (Eur Mol Biol Organ) J. 1995;14:5170–5178. doi: 10.1002/j.1460-2075.1995.tb00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer RH, Sokolov Y, Li H, Takenaka B, Milici AJ, Aiyar J, Nguyen A, Park H, Jap B, Hall JE, et al. Purification, visualization, and biophysical characterization of Kv1.3 tetramers. J Biol Chem. 1997;272:2389–2395. doi: 10.1074/jbc.272.4.2389. [DOI] [PubMed] [Google Scholar]
- Talvenheimo JA, Tamkun MM, Catterall WA. Reconstitution of neurotoxin stimulated sodium transport by the voltage-sensitive sodium channel purified from rat brain. J Biol Chem. 1982;257:11868–11871. [PubMed] [Google Scholar]
- Tatulian S, Cortes DM, Perozo E. Structural dynamics of the Streptomyces lividans K+channel: secondary structure characterization from FTIR spectroscopy. FEBS Lett. 1998;423:205–212. doi: 10.1016/s0014-5793(98)00091-x. [DOI] [PubMed] [Google Scholar]
- Zumbuel O, Weder HG. Liposomes of controllable size in the range of 40 to 180 nm by defined dialysis of lipid/detergent mixed micelles. Biochem Biophys Acta. 1981;640:252–262. doi: 10.1016/0005-2736(81)90550-2. [DOI] [PubMed] [Google Scholar]