Abstract
The Armadillo protein p120ctn associates with the cytoplasmic domain of cadherins and accumulates at cell–cell junctions. Particular Armadillo proteins such as β-catenin and plakophilins show a partly nuclear location, suggesting gene-regulatory activities. For different human E-cadherin-negative carcinoma cancer cell lines we found expression of endogenous p120ctn in the nucleus. Expression of E-cadherin directed p120ctn out of the nucleus. Previously, we reported that the human p120ctn gene might encode up to 32 protein isoforms as products of alternative splicing. Overexpression of p120ctn isoforms B in various cell lines resulted in cytoplasmic immunopositivity but never in nuclear staining. In contrast, upon expression of p120ctn cDNAs lacking exon B, the isoforms were detectable within both nuclei and cytoplasm. A putative nuclear export signal (NES) with a characteristic leucine-rich motif is encoded by exon B. This sequence element was shown to be required for nuclear export and to function autonomously when fused to a carrier protein and microinjected into cell nuclei. Moreover, the NES function of endogenously or exogenously expressed p120ctn isoforms B was sensitive to the nuclear export inhibitor leptomycin B. Expression of exogenous E-cadherin down-regulated nuclear p120ctn whereas activation of protein kinase C increased the level of nuclear p120ctn. These results reveal molecular mechanisms controlling the subcellular distribution of p120ctn.
The p120ctn protein originally was described as an efficient tyrosine kinase substrate implicated in both cell transformation by Src (1) and in ligand-induced receptor signaling through various tyrosine kinase receptors (2). It belongs to the Armadillo (Arm) family, which is characterized by a central Arm repeat domain in the member proteins (3). The p120ctn protein binds directly via its Arm domain to the cytoplasmic domain of E-cadherin and to other proteins (4–6). However, whereas the Arm proteins β-catenin and plakoglobin bind to sites nearby the E-cadherin carboxyl terminus (7), p120ctn binds to the juxtamembrane region (5, 6, 8). β-Catenin and plakoglobin also bind to αE-catenin, which links cadherin complexes to the actin cytoskeleton (9). In contrast, p120ctn does not appear to bind to α-catenin (4). Although its potential role in cell adhesion is still unclear, the juxtamembrane region of the cadherins plays a role in cadherin clustering, cell adhesion, and cell motility (8, 10–12).
For β-catenin an important function as signal transducer in the Wnt/wg signal pathway has been demonstrated (13, 14). When protected against proteolysis, either upon Wnt triggering or because of cancer-specific mutations, β-catenin can bind with its Arm domain to the DNA-binding LEF-1/Tcf transcription factors (15, 16). For this reason, β-catenin can be localized in the nucleus (13, 17).
Previously, we have shown detailed evidence for the possible occurrence of up to 32 human p120ctn isoforms on the basis of alternative splicing (18). Splicing events in the 5′ part of the p120ctn mRNA result in the use of four different start codons, yielding isoforms 1–4 (Fig. 1a). Additional variability is generated by combinations with alternatively used exons A and B near the end of the ORF and also of exon C within the Arm domain. Hence, the longest human isoform is designated 1ABC. Here we report the nuclear localization of both endogenously and exogenously expressed p120ctn in particular cell lines. Furthermore, we show that exon B of the p120ctn gene encodes a functional nuclear export signal (NES).
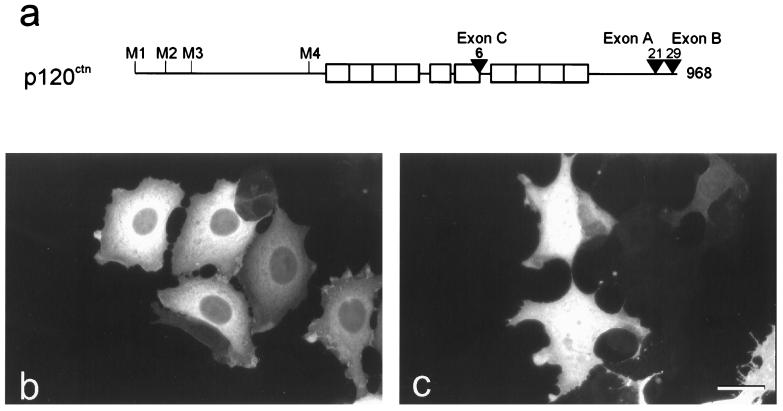
Figure 1.
Exon B of p120ctn encodes a NES. (a) Schematic overview of the human p120ctn isoforms, generated by alternative splicing. Open boxes represent the sequences encoding the Arm repeats. Four different ATG start codons are designated M1–M4. In the C-terminal half of p120ctn, sequences C, A, and B are encoded by alternatively spliced exons. Numbers indicate the size of the longest isoform and of the inserts encoded by exons A–C. (b and c) In MDCK cells, overexpression of p120ctn isoforms 3AB and 3A was achieved by transient transfection of plasmids pEFBOSp120–3AB or pEFBOSp120–3A, respectively. Detection was done by immunofluorescence. Endogenous p120ctn in MDCK is hardly visible because of the short exposure time. (Bar = 18 μm.)
MATERIALS AND METHODS
Plasmids.
Various full-size p120ctn-expressing cDNA sequences were cloned into the pGEM-11Zf(+) vector (Promega) and the eukaryotic expression pEFBOS vector (19). The plasmids encoding isoform 3A were obtained by cloning a 2,877-bp EagI/PstI fragment, derived from plasmid pDR2Cas5 (18). The plasmid encoding isoform 3AB was generated by use of a 2,964-bp EagI/PstI fragment from pDR2Cas5B. The latter plasmid contained the cDNA sequence encoded by exon B and was derived from pDR2Cas5 by substituting a 426-bp PstI/ClaI fragment of a cloned reverse transcription–PCR (RT-PCR) product for the original 339-bp PstI/ClaI fragment. This RT-PCR was performed on cDNA of HT29 cells by using primer pair EX14F1+EX21R3 (18). Plasmids encoding p120ctn isoforms 3B or 3N were generated by substituting in, respectively, plasmids 3AB and 3A, a 162-bp ClaI/NdeI cDNA fragment without exon A for the original 249-bp fragment comprising exon A. The fragment lacking exon A was isolated by RT-PCR from the human cell line GLC34 by using primer set EX14F1+EX21R3 (18). Plasmids encoding the p120ctn isoform 3ABC or 3AC were generated by substituting in, respectively, plasmids 3AB and 3A, a 634-bp BstB1/AflII fragment with exon C for the original fragment without exon C. The fragment comprising exon C was isolated by RT-PCR from fetal brain, using primer set EX7F1+EX20R1 (18). For the construction of the pUHDp120 plasmids, EcoRI/XbaI fragments from the appropriate pGEM11 plasmids containing the complete cDNAs of the different human p120ctn isoforms were cloned into the EcoRI/XbaI-cut pUHD10–3 vector (20). For transfection of full-length murine E-cadherin cDNA, either plasmid pBATEM2 (21) or a derivative of the expression vector pPNT (22) was used. All plasmid sequences were verified by DNA sequencing.
Mutagenesis in human p120ctn 3AB of codon leucine-844 to alanine and codon leucine-846 to alanine was performed by the QuickChange Site-Directed Mutagenesis Kit (Stratagene) by using mutagenic primers 5′-GAAGAGTTGGATGTGGCGGTTGCGGATGATGAGGGGGGC-3′ and 5′-GCCCCCCTCATCCATCCGCAACCGCCACATCCAACTCTTC-3′.
Cell Culture and Transfection.
Human colon adenocarcinoma cells SW480 and human breast cancer cells SK-BR-3 and MDA-MB-231 were obtained from the American Tissue Culture Collection (Manassas, VA). Human breast carcinoma cells MCF7 were transfected with FuGENE 6 transfection reagent (Boehringer Mannheim), SW480 cells and canine kidney epithelial cells (MDCK) were transfected with Lipofectamine plus Reagent (GIBCO/BRL), and SK-BR-3 cells were transfected with PerFect Lipids (Invitrogen). RNA derived from mouse MO4 cells (18) was used for RT-PCR.
Mouse fibrosarcoma L929 cells were cotransfected by calcium phosphate coprecipitation with plasmid pUHD17–1, coding for the rtTA transactivator (23), plus the selection plasmid pSV2neo (24). G418-resistant transfectants were tested for rtTA expression by transient transfection with plasmid pUHC13–3 expressing the luciferase gene under control of the tetracycline-regulated promoter (20). A clone with tightly regulated, high-level luciferase expression was chosen for further studies (24). This clone was cotransfected further with pUHD10.3-derived plasmids (20), encoding p120ctn isoforms 3AB, 3AC, or 3A, plus pPHT. The latter is a derivative of pPNT (25) and confers resistance to hygromycin. Stable L929 transfectants were selected by G418 (400 μg/ml) or hygromycin-B (250 units/ml) for a period of 3 weeks. Cloned transfectants were induced for 24 or 48 h with 1 μg/ml of doxycycline (dox; Duchefa Biochemie, Haarlem, The Netherlands) and were assayed by immunofluorescence for p120ctn expression. Colonies with reliable and stable induction properties were selected.
RNA Isolation and RT-PCR.
RNA isolation from various mouse tumor cell lines and RT-PCR were performed essentially as described previously (18). PCR primers were designed by using oligo 5.0 primer analysis software (National Biosciences, Plymouth, MN). Carboxyl-terminal mouse p120ctn-specific RT-PCR fragments were obtained by using primers 5′-AGGAGCTTCGGAAGCCACTG-3′ and 5′-GCGAAGAAAGGAAAAAAATC-3′.
Conjugation of Synthetic Peptides and Microinjection.
The NES peptide corresponding to part of exon B-encoded p120ctn and its mutated form (NESmut) were synthesized and then conjugated to ovalbumin (OV) by using the bifunctional cross-linking reagent sulfo-SMCC (Calbiochem) as described (26).
Mouse L929 cells were grown on glass coverslips and cultured for 5 days in DMEM supplemented with 10% newborn calf serum/100 μg/ml of streptomycin/250 units/ml of penicillin. Microinjection was performed by using an Eppendorf Micromanipulator/Transjector apparatus. A mixture of peptide-OV (1.0 mg/ml) and FITC-BSA (1.0 mg/ml; Nordic, Tilburg, The Netherlands) in PBS was injected into the cell nuclei.
Immunofluorescence Microscopy and Antibodies.
Monolayers prepared for fluorescent staining were grown on glass coverslips. Transfected, microinjected, or dox-treated cells were rinsed briefly with PBS and fixed with either ice-cold 100% methanol for 15 min at −20°C or with 3% paraformaldehyde in PBS for 25 min at room temperature. Immunostaining was performed as described before (27). Samples were examined with a Zeiss Axiophot photomicroscope or with a Zeiss LSM 410 confocal laser-scanning immunofluorescence microscope.
The following antibodies were used: two mouse mAbs, pp120 (Transduction Laboratories, Lexington, KY) and 12F4 (kindly provided by A. Reynolds, Department of Cell Biology, Vanderbilt University, Nashville, TN), both recognizing all p120ctn isoforms, rat mAb DECMA-1 against mouse E-cadherin (Sigma), and rabbit antiserum to OV (ICN/Cappel). Secondary antibodies used in immunofluorescence microscopy were Alexa594-coupled anti-rabbit Ig, Alexa594-coupled anti-mouse Ig, and Alexa488-coupled anti-rat Ig antibodies (Molecular Probes).
RESULTS
Subcellular Localization of Overexpressed p120ctn Isoforms.
We cloned various cDNA isoforms into a eukaryotic expression vector, followed by transient transfections into various fibroblastic or epithelial cell lines. Indirect immunofluorescent staining of p120ctn revealed staining for isoforms B in the cytoplasm but not in the nucleus (illustrated for isoform 3AB in Fig. 1b). In contrast, expression of isoforms lacking exon B-encoded amino acid residues reproducibly resulted in both cytoplasmic and nuclear staining (illustrated for isoform 3A in Fig. 1c). All cell lines used for these transfections displayed comparable expression patterns.
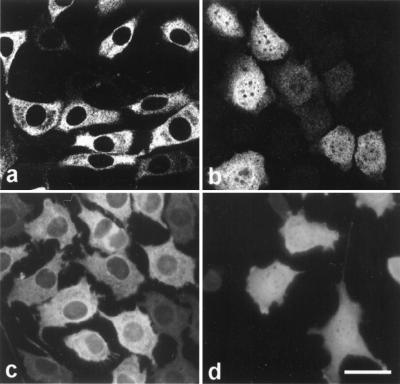
The same phenomena were observed in tetracycline-inducible cell lines. A L929 clone with high expression of the rtTA transactivator (24) was transfected with pUHD10.3-derived plasmids, encoding p120ctn isoforms 3AB, 3AC, or 3A. Upon induction with dox, expression of isoform 3AB was never observed in the nucleus (Fig. 2 a and c) in contrast to isoforms 3A (Fig. 2b) and 3AC (not shown). This lack of nuclear staining was abrogated by leptomycin B (LMB, Fig. 2d), a specific inhibitor of nuclear export mediated by a leucine-rich NES (28–30).
Figure 2.
LMB inhibits nuclear export of p120ctn in tetracycline-inducible L929 cells. L929 cells expressing the rtTA transactivator were transfected with pUHD10.3-derived plasmids encoding p120ctn isoform 3AB (a, c, and d) or isoform 3A (b). Expression of p120ctn was induced by dox treatment for 24 h. Cells were fixed by methanol and stained with mAb pp120. (d) During the last 3 h of dox treatment, LMB (5 ng/ml) was added to the culture medium; part of the overexpressed p120ctn isoform 3AB is retained in the nucleus. Images were obtained by laser-scanning confocal microscopy (a and b) or by standard fluorescence microscopy (c and d). (Bar = 22 μm.)
A NES Sequence in p120ctn Isoforms B.
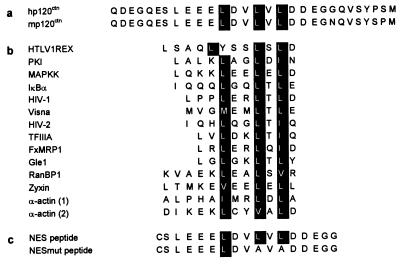
Exon B in human p120ctn encodes a remarkably negatively charged peptide with 10 acidic amino acids of 29 (Fig. 3). An exon B homologue has not yet been reported for mouse p120ctn mRNAs. By using appropriate mouse cDNA primers, flanking the predicted position of a putative exon B, we cloned the latter sequence from transcripts of murine MO4 cells (GenBank accession no. AF140220; Fig. 3a). The exon B-encoded sequences share key features with NES sequences that were shown previously to trigger the rapid, active export of proteins and RNA–protein complexes from the nucleus to the cytoplasm (Fig. 3b; reviewed in ref. 31). The key hydrophobic residues for NES function in other proteins also were found to be present in both human and mouse sequences encoded by exon B (Fig. 3a).
Figure 3.
Exon B-encoded sequence in p120ctn displays conservation of the key hydrophobic residues (highlighted) required for NES functionality. (a) Exon B-encoded protein sequences for human and mouse p120ctn. (b) NES sequence from a variety of proteins (31, 51, 52). (c) A peptide corresponding to part of the exon B-encoded NES and a mutated derivative peptide (NESmut) were synthesized and used for microinjection experiments (Fig. 4).
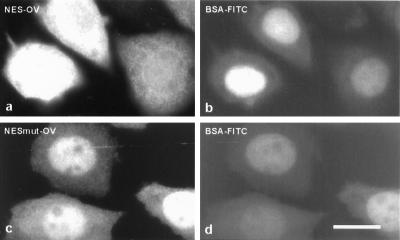
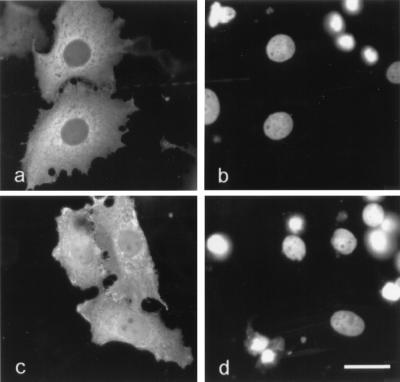
To test whether the leucine-rich sequence in human p120ctn can act as NES, a corresponding peptide (Fig. 3c) was chemically conjugated to OV. Nuclei of L929 cells were coinjected with FITC-labeled BSA and NES-OV. Within 20 min after injection, part of the latter conjugate was excluded from the nucleus (Fig. 4a). In contrast, coinjected BSA-FITC remained in the nucleus (Fig. 4b). The nuclear export from NES-OV was inhibited completely by LMB (data not shown). A mutant peptide, in which two conserved leucine residues were replaced by alanine residues (NESmut; Fig. 3c), also was conjugated to OV. It has been established previously that this type of mutation makes NES nonfunctional (32, 33). After injection into the nucleus, the NESmut-OV was unable to cross the nuclear envelope as coinjected BSA-FITC (Fig. 4 c and d). Taken together, these results indicate that exon B of p120ctn encodes a typical, functional NES.
Figure 4.
The exon B-encoded, leucine-rich sequence of p120ctn is necessary and sufficient to direct export of microinjected OV from the nucleus to the cytoplasm. A mixture of peptide-conjugated OV and FITC-BSA was injected into the nuclei of L929 cells. Either a wild-type NES sequence (a and b) or a mutated NES sequence (c and d) was conjugated to OV. At 20 min after injection, cells were fixed with 3% paraformaldehyde and stained with anti-OV antibody and Alexa594-conjugated secondary antibody. Dual-channel fluorescence was used to reveal both OV (a and c) and FITC-BSA (b and d) in the injected cells. (Bar = 10 μm.)
To confirm that the nuclear export of full-size p120ctn proteins was mediated by this exon B-encoded NES sequence, we overexpressed a mutated p120ctn isoform 3AB, in which the two critical leucine residues within the NES were replaced by alanine residues. Wild-type and mutated cDNA expression plasmids were transfected in epithelial and fibroblastic cell lines. Fig. 5 shows representative results in MCF7 cells. Overexpressed wild-type isoform 3AB localized to the cytoplasm (Fig. 5 a and b). The 3AB protein with mutated NES was partly retained in the nucleus (Fig. 5 c and d).
Figure 5.
Mutation of exon B-encoded NES interferes with nuclear export of p120ctn. MCF7 cells were transiently transfected with pEFBOSp120–3AB expressing the wild-type p120ctn isoform 3AB (a and b) or with pEFBOSp120–3ABNESmut expressing p120ctn isoform 3AB with a mutated NES (c and d). (a and c) Two days after transfection, the subcellular distribution of p120ctn was determined by staining with mAb pp120. (b and d) Nuclear DNA staining of the same cells as shown in a and c. (Bar = 20 μm.)
LMB Induces Nuclear Accumulation of Endogenous p120ctn.
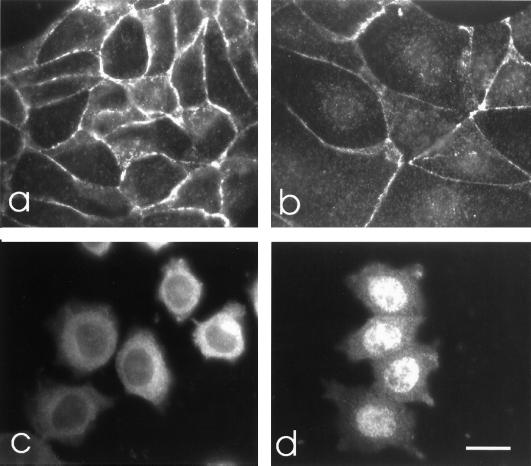
To check whether the subcellular distribution of endogenous p120ctn isoforms B also is regulated by its NES sequence, E-cadherin-positive MDCK and E-cadherin-negative L929 cells were treated with LMB. Immunofluorescent staining showed that endogenous p120ctn was present almost exclusively at sites of cell–cell contact in untreated MDCK cells (Fig. 6a). Upon LMB treatment, p120ctn became partly localized in the nucleus (Fig. 6b). In untreated L929 cells, p120ctn was present in the cytoplasm (Fig. 6c), but LMB treatment induced a complete shift to the nucleus (Fig. 6d).
Figure 6.
Effect of LMB on the subcellular distribution of endogenous p120ctn. E-cadherin-positive MDCK cells (a and b) and E-cadherin-negative L929 cells (c and d) were incubated for 18 h in the absence (a and c) or presence of LMB (5 ng/ml; b and d). Cells then were fixed with paraformaldehyde and stained with mAb pp120. The weak nuclear localization of p120ctn in LMB-treated MDCK cells (b) was confirmed by DNA counterstaining. (Bar = 12 μm.)
Expression Patterns of Endogenous p120ctn.
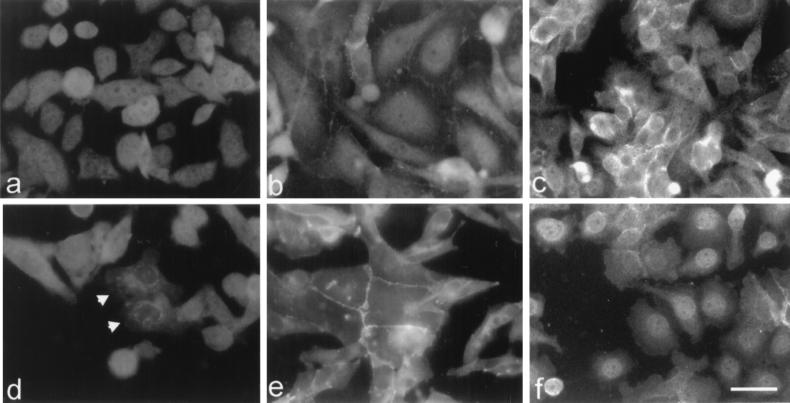
The presence of a functional NES in p120ctn suggests that endogenous p120ctn might actually be able to shuttle between the nucleus and the cytoplasm or plasma membrane. By immunofluorescence using two p120ctn-specific mAbs, we checked a large collection of human cell lines for nuclear localization of endogenous p120ctn. Nuclear p120ctn was detectable in the E-cadherin-negative breast carcinoma cell lines SK-BR-3 and MDA-MB-231 (Fig. 7 a and b). In SW480 colon carcinoma cells, which show negligible E-cadherin expression (34), p120ctn is present at the plasma membrane, in the cytoplasm, but also in the nucleus (Fig. 7c). Treatment of SW480 cells with the phorbol ester 12-O-tetradecanoyl-phorbol-13-acetate induced cell spreading, relocalization of E-cadherin to the cytoplasm, and increased amounts of both β-catenin and p120ctn in the nucleus (illustrated for p120ctn in Fig. 7f).
Figure 7.
Expression of endogenous p120ctn in the nucleus and influences by E-cadherin and phorbol ester. Human breast cancer cells SK-BR-3 (a) and MDA-MB-231 (b) express p120ctn both within and outside the nucleus. Human colon cancer SW480 cells (c) express p120ctn mainly at cell–cell contacts but also in cytoplasm and nucleus. SK-BR-3 cells were transfected transiently with E-cadherin cDNA and stained for E-cadherin (not shown) and p120ctn (d). Two cells that were positive for E-cadherin show accumulation of p120ctn outside the nucleus (arrows). An MDA-MB-231 transfectant, which was stably expressing E-cadherin cDNA, shows colocalization of E-cadherin (not shown) and p120ctn at cell–cell contacts only (e). Upon treatment of SW480 cells with 250 ng/ml of phorbol ester for 3 h, p120ctn relocalized from extranuclear sites to the nucleus (f). (Bar = 18 μm.)
To study the effect of E-cadherin on the nuclear localization of endogenous p120ctn, E-cadherin cDNA was transfected into the MDA-MB-231, SK-BR-3, and SW480 cell lines. Immunofluorescence showed colocalization of exogenous E-cadherin with endogenous p120ctn at the plasma membrane or in the cytoplasm, whereas nuclear p120ctn expression clearly was down-regulated (illustrated for SK-BR-3 and MDA-MB-231 cells in Fig. 7 d and e).
DISCUSSION
The p120ctn protein is a prominent substrate for the oncoprotein Src, and its tyrosine phosphorylation correlates with phenotypic transformation (1). In ras-transformed breast epithelial cell lines the association of tyrosine phosphorylated p120ctn with E-cadherin is elevated (35). Altered p120ctn staining patterns or complete loss of p120ctn expression have been reported for colonic carcinoma (36, 37), adenomatous polyps of colon (38), bladder cancer (39), and breast cancer (40). To better understand the role and regulation of the p120ctn protein in embryonic development, morphogenesis, and cancer, we initiated an analysis of the expression patterns of the numerous p120ctn isoforms (18). By transient transfection and inducible expression we have demonstrated that human and mouse p120ctn isoforms B contain a functional NES. Overexpression of isoforms B results in expression of p120ctn outside the nucleus. Absence or mutation of the exon B-encoded NES leads to accumulation in both cytoplasm and nucleus. Moreover, this p120ctn-derived NES sequence turned out to be sufficient for induction of nuclear export of unrelated proteins.
The alternative use of exon B was observed regularly in human transcripts of p120ctn (18). Human mesenchymal cell types tend to express isoform A transcripts lacking exon B, whereas most epithelial cells express variable amounts of isoform AB. Despite the general expression of p120ctn isoforms without an NES, usually p120ctn cannot be detected in the nucleus. In contrast, partial nuclear localization has been described for several Armadillo proteins, such as β-catenin (13, 17), plakoglobin (41), plakophilin-1 (42), plakophilin-2 (43), and plakophilin-3 (44). Nuclear accumulation also is regulated by the efficiency of nuclear import. Some sequences of p120ctn and other Armadillo proteins bear resemblance to previously characterized nuclear localization signal motifs (45). In spite of such a sequence, β-catenin is imported into the nucleus by binding directly to the nuclear pore machinery (46). The general lack of nuclear p120ctn may be explained by inhibition of p120ctn import because of binding to E-cadherin or other extranuclear proteins.
Nevertheless, we found that the human carcinoma cells SW480, SK-BR-3, and MDA-MB-231 express p120ctn in the nucleus. These carcinoma cells express low to undetectable levels of E-cadherin (4, 34, 47). Upon expression of exogenous E-cadherin, p120ctn is no longer detectable in the nucleus. Apparently, binding of p120ctn to E-cadherin at the plasma membrane or in the cytoplasm restrains p120ctn from entering the nucleus. Recently, a similar correlation was reported for β-catenin in SW480 cells, as expression of exogenous N-cadherin directed β-catenin from the nucleus to the plasma membrane and cytoplasm (48). Another observation is in line with such an influence of E-cadherin on p120ctn localization. Treatment of SW480 cells with phorbol ester resulted in increased nuclear levels of both p120ctn and β-catenin, whereas E-cadherin was concomitantly decreased at the plasma membrane. Further, inhibition of nuclear import of p120ctn by E-cadherin is suggested by the LMB effects. LMB treatment of E-cadherin-positive cells (e.g., MDCK) hardly induced nuclear expression of p120ctn in contrast to the situation in LMB-treated, E-cadherin-negative cell types (e.g., L929 cells).
Until recently, cadherins were the only proteins known to associate directly with p120ctn (49). By a yeast two-hybrid approach using p120ctn as bait, Daniel and Reynolds (50) showed a novel BTB/POZ domain zinc finger protein to be a specific interaction partner of p120ctn. This putative transcription factor, termed Kaiso, was expressed in the nucleus. Because the interactions with both E-cadherin and Kaiso require the Arm domain of p120ctn, it is likely that p120ctn forms mutually exclusive complexes with these two proteins. Further analysis of Kaiso and other p120ctn-associated proteins may reveal the functions of the many p120ctn isoforms at diverse subcellular sites including the nucleus. Interestingly, in the related Armadillo proteins ARVCF, p0071, and δ-catenin a similar NES can be predicted on the basis of sequence comparisons (data not shown). Additional experiments are needed to prove that these NES sequences are functional. If consolidated, the appearance of a NES in many members of this subfamily of Armadillo proteins may point at peculiar shared functions.
Acknowledgments
We thank K. Strumane for providing the E-cadherin transfected MDA-MB-231 cell line, Dr. M. Yoshida for a kind gift of LMB, Dr. A. Reynolds for supplying mAb 12F4, and Dr. J. Vandekerckhove for the preparation of the synthetic peptides. We thank Barbara Gilbert and Petra Vermassen for expert technical assistance. J.v.H. and F.v.R. are, respectively, Postdoctoral Fellow and Research Director with the Fund for Scientific Research–Flanders (FSRF). The present research is supported by the FSRF, Geconcerteerde Onderzoekacties (University of Ghent), Sportvereniging tegen Kanker (Belgium), Centrum voor Studie en Behandeling van Gezwelziekten (Flanders), and Algemene Spaar en Lijfrentekas (Belgium).
ABBREVIATIONS
- Arm
armadillo
- dox
doxycycline
- LMB
leptomycin B
- NES
nuclear export signal
- OV
ovalbumin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF140220 (exon B of murine p120ctn)].
References
- 1.Reynolds A B, Roesel D J, Kanner S B, Parsons J T. Mol Cell Biol. 1989;9:629–638. doi: 10.1128/mcb.9.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanner S B, Reynolds A B, Parsons J T. Mol Cell Biol. 1991;11:713–720. doi: 10.1128/mcb.11.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riggleman B, Wieschaus E, Schedl P. Gene Dev. 1989;3:96–113. doi: 10.1101/gad.3.1.96. [DOI] [PubMed] [Google Scholar]
- 4.Daniel J M, Reynolds A B. Mol Cell Biol. 1995;15:4819–4824. doi: 10.1128/mcb.15.9.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finnemann S, Mitrik I, Hess M, Otto G, Wedlich D. J Biol Chem. 1997;272:11856–11862. doi: 10.1074/jbc.272.18.11856. [DOI] [PubMed] [Google Scholar]
- 6.Lampugnani M G, Corada M, Andriopoulou P, Esser S, Risau W, Dejana E. J Cell Sci. 1997;110:2065–2077. doi: 10.1242/jcs.110.17.2065. [DOI] [PubMed] [Google Scholar]
- 7.Jou T S, Stewart D B, Stappert J, Nelson W J, Marrs J A. Proc Natl Acad Sci USA. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yap A S, Niessen C M, Gumbiner B M. J Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrenknecht K, Ozawa M, Eckerskorn C, Lottspeich F, Lenter M, Kemler R. Proc Natl Acad Sci USA. 1991;88:9156–9160. doi: 10.1073/pnas.88.20.9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kintner C. Cell. 1992;69:225–236. doi: 10.1016/0092-8674(92)90404-z. [DOI] [PubMed] [Google Scholar]
- 11.Chen H Y, Paradies N E, Fedor-Chaiken M, Brackenbury R. J Cell Sci. 1997;110:345–356. doi: 10.1242/jcs.110.3.345. [DOI] [PubMed] [Google Scholar]
- 12.Navarro P, Caveda L, Breviario F, Mandoteanu I, Lampugnani M G, Dejana E. J Biol Chem. 1995;270:30965–30972. doi: 10.1074/jbc.270.52.30965. [DOI] [PubMed] [Google Scholar]
- 13.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 14.Gumbiner B M. Curr Opin Genet Dev. 1998;8:430–435. doi: 10.1016/s0959-437x(98)80114-7. [DOI] [PubMed] [Google Scholar]
- 15.Behrens J, von Kries J P, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 16.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Els J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 17.Inomata M, Ochiai A, Akimoto S, Kitano S, Hirohashi S. Cancer Res. 1996;56:2213–2217. [PubMed] [Google Scholar]
- 18.Keirsebilck A, Bonné S, Staes K, van Hengel J, Nollet F, Reynolds A, van Roy F. Genomics. 1998;50:129–146. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagafuchi A, Shirayoshi Y, Okazaki K, Yasuda K, Takeichi M. Nature (London) 1987;329:341–343. doi: 10.1038/329341a0. [DOI] [PubMed] [Google Scholar]
- 22.Keirsebilck A, Van Hoorde L, Gao Y, De Bruyne G, Bruyneel E, Vermassen P, Mareel M, van Roy F. Invasion Metastasis. 1998;18:44–56. doi: 10.1159/000024498. [DOI] [PubMed] [Google Scholar]
- 23.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 24.Vanhoenacker P, Gommeren W, Luyten W H M L, Leysen J E, Haegeman G. Gene Ther Mol Biol. 1998;3:301–310. [Google Scholar]
- 25.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M, Gotoh I, Gotoh Y, Nishida E. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 27.van Hengel J, Gohon L, Bruyneel E, Vermeulen S, Cornelissen M, Mareel M, van Roy F. J Cell Biol. 1997;137:1103–1116. doi: 10.1083/jcb.137.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 30.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 31.Hope T J. Chem Biol. 1997;4:335–344. doi: 10.1016/s1074-5521(97)90124-1. [DOI] [PubMed] [Google Scholar]
- 32.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 33.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 34.Daniel C W, Strickland P, Friedmann Y. Dev Biol. 1995;169:511–519. doi: 10.1006/dbio.1995.1165. [DOI] [PubMed] [Google Scholar]
- 35.Kinch M S, Clark G J, Der C J, Burridge K. J Cell Biol. 1995;130:461–471. doi: 10.1083/jcb.130.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skoudy A, Gomez S, Fabre M, de Herreros A G. Int J Cancer. 1996;68:14–20. doi: 10.1002/(SICI)1097-0215(19960927)68:1<14::AID-IJC3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 37.Gold J S, Reynolds A B, Rimm D L. Cancer Lett. 1998;132:193–201. doi: 10.1016/s0304-3835(98)00190-6. [DOI] [PubMed] [Google Scholar]
- 38.Valizadeh A, Karayiannakis A J, El Hariry I, Kmiot W, Pignatelli M. Am J Pathol. 1997;150:1977–1984. [PMC free article] [PubMed] [Google Scholar]
- 39.Shimazui T, Schalken J A, Giroldi L A, Jansen C F J, Akaza H, Koiso K, Debruyne F M J, Bringuier P P. Cancer Res. 1996;56:4154–4158. [PubMed] [Google Scholar]
- 40.Dillon D A, D’Aquila T, Reynolds A B, Faeron E R, Rimm D L. Am J Pathol. 1998;152:75–82. [PMC free article] [PubMed] [Google Scholar]
- 41.Karnovsky A, Klymkowsky M W. Proc Natl Acad Sci USA. 1995;92:4522–4526. doi: 10.1073/pnas.92.10.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt A, Langbein L, Rode M, Pratzel S, Zimbelmann R, Franke W W. Cell Tissue Res. 1997;290:481–499. doi: 10.1007/s004410050956. [DOI] [PubMed] [Google Scholar]
- 43.Mertens C, Kuhn C, Franke W W. J Cell Biol. 1996;135:1009–1025. doi: 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonné, S., van Hengel, J., Nollet, F., Kools, P. & van Roy, F. (1999) J. Cell Sci., in press. [DOI] [PubMed]
- 45.Reynolds A B, Daniel J M. In: Cytoskeletal-Membrane Interactions and Signal Transduction. Cowin P, Klymkowsky M W, editors. Austin, TX: Landes Company and Chapman & Hall; 1997. pp. 1–48. [Google Scholar]
- 46.Fagotto F, Gluck U, Gumbiner B M. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 47.Pierceall W E, Woodard A S, Morrow J S, Rimm D, Fearon E R. Oncogene. 1995;11:1319–1326. [PubMed] [Google Scholar]
- 48.Sadot E, Simcha I, Shtutman M, Ben-Ze’ev A, Geiger B. Proc Natl Acad Sci USA. 1998;95:15339–15344. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reynolds A B, Daniel J, McCrea P D, Wheelock M J, Wu J, Zhang Z. Mol Cell Biol. 1994;14:8333–8342. doi: 10.1128/mcb.14.12.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniel J M, Reynolds A B. Mol Cell Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wada A, Fukuda M, Mishima M, Nishida E. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nix D A, Beckerle M C. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]