Abstract
COOH-terminal (S3) domains are conserved within the MscL family of bacterial mechanosensitive channels, but their function remains unclear. The X-ray structure of MscL from Mycobacterium tuberculosis (TbMscL) revealed cytoplasmic domains forming a pentameric bundle (Chang, G., R.H. Spencer, A.T. Lee, M.T. Barclay, and D.C. Rees. 1998. Science. 282:2220–2226). The helices, however, have an unusual orientation in which hydrophobic sidechains face outside while charged residues face inside, possibly due to specific crystallization conditions. Based on the structure of pentameric cartilage protein , we modeled the COOH-terminal region of E. coli MscL to better satisfy the hydrophobicity criteria, with sidechains of conserved aliphatic residues all inside the bundle. Molecular dynamic simulations predicted higher stability for this conformation compared with one modeled after the crystal structure of TbMscL, and suggested distances for disulfide trapping experiments. The single cysteine mutants L121C and I125C formed dimers under ambient conditions and more so in the presence of an oxidant. The double-cysteine mutants, L121C/L122C and L128C/L129C, often cross-link into tetrameric and pentameric structures, consistent with the new model. Patch-clamp examination of these double mutants under moderately oxidizing or reducing conditions indicated that the bundle cross-linking neither prevents the channel from opening nor changes thermodynamic parameters of gating. Destabilization of the bundle by replacing conservative leucines with small polar residues, or complete removal of COOH-terminal domain (Δ110–136 mutation), increased the occupancy of subconducting states but did not change gating parameters substantially. The Δ110–136 truncation mutant was functional in in vivo osmotic shock assays; however, the amount of ATP released into the shock medium was considerably larger than in controls. The data strongly suggest that in contrast to previous gating models (Sukharev, S., M. Betanzos, C.S. Chiang, and H.R. Guy. 2001a. Nature. 409:720–724.), S3 domains are stably associated in both closed and open conformations. The bundle-like assembly of cytoplasmic helices provides stability to the open conformation, and may function as a size-exclusion filter at the cytoplasmic entrance to the MscL pore, preventing loss of essential metabolites.
Keywords: amphipathic helix, coiled coil, MD simulation, disulfide trapping, channel gating
INTROUCTION
Cytoplasmic domains of ion channels play multiple roles. The gating of the “simple” KcsA potassium channel, for instance, is associated with a pH-dependent conformational change in the cytoplasmic domains and the linkers connecting them to the transmembrane helices (Cortes et al., 2001). In ATP-dependent inward rectifier channels, COOH-terminal domains interact with phospholipids, providing modulation of the channel activity by phosphatidylinositol phosphates (Shyng et al., 2000). In more complex voltage- and cyclic nucleotide–gated channels, the cytoplasmic domains are involved in the channel assembly (Kreusch et al., 1998), mediate the attachment of auxiliary subunits (Sewing et al., 1996) and cytoskeleton (Nakahira et al., 1998), or provide binding sites for regulation by Ca2+ (Schreiber and Salkoff, 1997), calmodulin (Bian et al., 2001), or cyclic nucleotides (Matulef et al., 1999).
MscL, a large bacterial mechanosensitive channel apparently stands apart from these groups of channels. As a part of the turgor regulation system in bacteria, MscL acts as a “safety valve” for strong hypoosmotic shock (Levina et al., 1999). When tension in the membrane approaches the lytic limit (8–10 dyne/cm), MscL forms a pore of ∼3 nm in diameter in the inner membrane (Blount et al., 1996a; Sukharev et al., 1999b). The pore shows no ionic selectivity (Sukharev et al., 1993) and most likely is permeable to a variety of inorganic and small organic osmolytes that are expelled from the cell to avoid lysis (Martinac, 2001).
E. coli MscL (EcoMscL)* is a homomultimer of 136–amino acid polypeptide chains (Sukharev et al., 1994, 1999a). The crystal structure of the MscL homologue from M. tuberculosis (151 amino acids) revealed the pentameric channel in an apparently closed state (Chang et al., 1998). The core of the protein is formed by tightly packed M1 and M2 transmembrane helices from all five subunits constituting the complex. The COOH-terminal (S3) helices form a pentameric bundle in the cytoplasm. Each S3 helix is connected to the M2 transmembrane helix by a flexible linker. Detailed examination of the COOH-terminal bundle in the crystal structure reveals that hydrophilic and some charged side chains are packed inside the bundle, whereas many hydrophobic sidechains are exposed to the outside. This orientation may not be the natural one since the crystallization was achieved at low pH in the presence of a detergent with divalent cations bound between some of the carboxyl side chain groups of the S3 bundle. Molecular dynamic simulations of TbMscL have already shown that the crystal structure of S3 helices would be highly unstable at physiological pH (Elmore and Dougherty, 2001).
A model of TbMscL that better satisfies hydrophobicity criteria was recently proposed (Sukharev et al., 2001b). An inversion of S3 domains, which places the aliphatic side chains inside, is predicted to form a more stable conformation under physiological conditions. In parallel, a homology model of EcoMscL was built in an effort to relate the wealth of experimental observations collected mostly from E. coli MscL (EcoMscL) to the structural data obtained from TbMscL. EcoMscL was used because it is difficult to obtain patch-clamp recordings from TbMscL in E. coli spheroplasts (Moe et al., 2000).
Sequence analysis of >35 homologues from different bacteria (Spencer et al., 1999; Maurer et al., 2000; Sukharev et al., 2001b) revealed that the COOH termini may vary in length, but always contain a short (12 residue), highly conserved stretch that apparently folds as an amphipathic helix. Deletion of S3 in EcoMscL (Δ110–136) does not abolish gating, thus the domain was initially deemed “dispensable” (Blount et al., 1996b). An earlier analysis of the Δ110–136 mutant showed that the absence of the COOH-terminal domain increases the variability of the gating pattern and multiplicity of conducting states (Hase et al., 1997). In developing structural models in which a dramatic expansion of the transmembrane barrel is associated with substantial flattening of the transmembrane segments, we suggested that S3 helices may compensate for the flattening by interacting with lipids at the cytoplasmic surface of the membrane (Sukharev et al., 2001b; Betanzos et al., 2002). Therefore, it seems important to address the questions of whether S3 domains form a bundle-like structure at rest under physiological conditions, and whether the bundle remains stable or separates during channel opening.
In the present work we attempt to clarify the native conformation of the Eco MscL S3 bundle in the resting and open states. The sequence of the COOH terminus of MscL is similar to that of the oligomerization domain of the cartilage protein COMP, which has a fivefold coiled-coil structure stabilized by apolar interactions inside the bundle and by salt bridges on the periphery (Malashkevich et al., 1996) (see Fig. 1). We present a refined model of the EcoMscL S3 bundle that forms a left-handed coiled-coil analogous to the structure of COMP and compare it with the model built after the crystal structure of TbMscL. Molecular dynamic simulations were employed not only to test the stability of the modeled structures, but also to assess the dynamics and proximities of critical residues that were cysteine-substituted and used in disulfide-trapping experiments. Patch-clamp recordings were done on cysteine mutants that showed intersubunit coupling on gels, on mutants with disrupted S3 domain association, and on a truncated mutant completely lacking S3. In vivo experiments confirmed that the truncated MscL lacking S3 is functional but is more permeable to ATP. The combined analysis of structures, gating patterns, thermodynamic parameters, and permeabilities leads us to the conclusion that the bundle normally does not separate during the opening transition. We present a revised model of EcoMscL in the open state and discuss its functional implications.
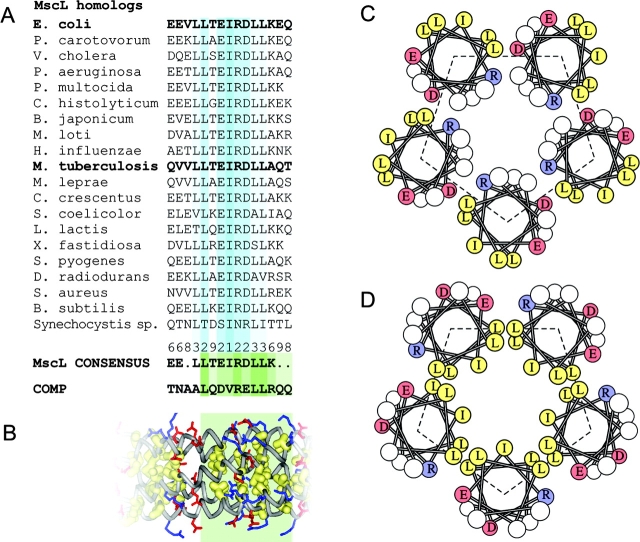
Figure 1.
Sequence analysis of conserved COOH-terminal regions of MscL and comparison with the cartilage protein COMP. Alignment of S3 domains (residues 118–132 for EcoMscL) for 19 representative MscL homologues from different bacterial species, the consensus and the sequence of the third heptad of the oligomerization domain of COMP (A). The fragment of the crystal structure of COMP representing the aligned sequence (green box), which shows all hydrophobic side chains packed inside the fivefold coiled-coil and the salt bridges between R48 or R52 (blue) and D46 (red) on different chains (B). Helical wheel representations of the packing of helices in the crystal structure of TbMscL (C) and in EcoMscL modeled after COMP (D).
MATERIALS AND METHODS
Modeling, Structure Analysis, and Molecular Dynamics Simulation
Models of the native conformation of EcoMscL have been reported previously (Sukharev et al., 2001b). An additional conformation of EcoMscL S3 bundle resembling the crystal structure of TbMscL was modeled by homology using the same tools (Sukharev et al., 2001b). The secondary structure of the entire COOH-terminal domain was assessed using web-based structure prediction tools such as PROF (Ouali and King, 2000), PSIPRED (Jones, 1999), and NNPREDICT (Kneller et al., 1990). The model for S3 termini with aliphatic sidechains inside, which was originally built almost as a parallel bundle, was refined; the helices were tilted to ∼19°, similar to the arrangement of helices in the coiled-coil structure of COMP (Malashkevich et al., 1996). The GETAREA web-based software (Fraczkiewicz and Braun, 2002) was used for analysis of water-accessible areas and hydration energies of assembled and disassembled bundles before or after the molecular dynamic (MD) simulation (below). The probe radius of 1.4 Å, atomic solvation parameters according to Wesson and Eisenberg (1992), or empirical potentials (Lomize et al., 2002) were used in energy computations.
The two models of EcoMscL S3 bundle, COMP-like and Tb-like, were taken as initial conformations for MD simulations. Hydrogens absent in the original model were added using PSFGEN, part of the NAMD package (Theoretical Biophysics Group and National Institutes of Health Resource for Macromolecular Modeling and Bioinformatics, Beckman Institute, UIUC, Urbana, IL). Residues 104–136 from all five subunits were placed in a preequilibrated water box. Only Cα atoms of R104 were harmonically restrained (with force constant k = 1 kcal/(mol*Å2)) in their initial positions at a distance of 21.7 Å from the axis of symmetry. All charged residues and COOH termini were in the ionized state (pH 7.0). The molecular dynamic simulation was performed using the NAMD2 program (version 2.3) (Kale et al., 1999) with CHARMM 22 force field (MacKerell et al., 1998). The complete system of 31,766 total atoms, 2,465 protein atoms, and 9,767 water molecules represented an NPT ensemble. Bad contacts were removed by energy minimization of the polypeptide assembly in the presence of water. The system was gradually heated to 310 K with temperature coupling over 3 ps with subsequent equilibration for 0.1 ns at 1 atm. The simulation was performed with full electrostatics using the particle mesh Ewald (PME) method (Darden et al., 1993) with a grid spacing of ∼1 Å in a flexible unit cell, under periodic boundary conditions. Other program settings were as in (Gullingsrud et al., 2001). The simulation time step was 1 fs. The system was usually kept at 310 K, 1 atm using the Langevin piston method (Feller et al., 1995). In one of the simulations the temperature was gradually raised to 360 K and equilibrated for 0.1 ns. The system was then simulated for one more nanosecond. Additional 2-ns simulations were performed in the steered mode, in which Cα atoms of residues 104 were “pulled” along the radii with the force constant k = 1 kcal/(mol*Å2) to new positions 36.5 Å away from the axis of symmetry, which corresponds to the open state of the barrel. The second pull was applied to Cα of residues 114 moving them to new positions 29.5 Å away from the axis, thus the stability of the bundle with highly strained linkers was examined. Results were visualized using VMD (Humphrey et al., 1996) and WebLab Viewer Pro (Molecular Simulations, Inc.). Trajectories were created using MS Excel. The traditional assessment of the structure stability was performed using RMS deviations (RMSD) of atomic coordinates relative to their initial positions. We also measured relative interatomic distances and their deviations (RMSDd) to exclude effects of movement of the helical assembly as a single unit. The presence of salt bridges was statistically assessed by the distance criterion, i.e., a bridge was scored if less than 4 Å separated the acidic oxygen and the hydrogen of a basic amino group in a frame, with the total of 500 frames analyzed.
Strains
All genetic manipulations and cloning steps were done in PB104 mscL −, recA− cells (Blount et al., 1996a). The Δ110–136 truncation mutant lacking COOH-terminal domain was briefly described previously (Blount et al., 1996b) and provided by Dr. Paul Blount (University of Texas, Dallas). The Δ110–136 mutant and WT MscL were expressed in the pB10a vector, behind the PlacUV5-inducible promoter (Sukharev et al., 1994). For patch clamp experiments, all mutant proteins (see below) were expressed in PB104 cells. For osmotic shock survival and ATP release experiments, the constructs were expressed in MJF455 (mscL −, mscS−) osmotically sensitive cells (Levina et al., 1999), donated by Dr. Ian Booth (University of Aberdeen, UK).
Site-directed Mutagenesis
Native EcoMscL contains no cysteines. Four single-cysteine (E119C, L121C, T123C, I125C), and two double-cysteine (L121C/ L122C and L128C/L129C) substitutions were generated using the standard PCR-based megaprimer technique as described previously (Yoshimura et al., 1999). The p5-2-2b plasmid (Moe et al., 1998) containing the wild-type mscL ORF was used as a template for both rounds of PCR performed with Pfu DNA polymerase (Stratagene). The amplified ORFs containing substitutions were trimmed and cloned as BglII-XhoI fragments into the pB10b vector, behind the LacPuv5-inducible promoter. Both double cysteine mutants were extended by six additional His (cat) codons on the 3′ end of the ORF as described (Blount et al., 1996a) to obtain COOH-terminally His-tagged versions of these proteins. All genetic modifications were confirmed by automated sequencing.
Disulfide Cross-linking
The proximity of engineered cysteines was tested using the disulfide trapping technique (Pakula and Simon, 1992) as modified (Sukharev et al., 2001a). pB104 mscL − cells carrying the corresponding expression vector were grown in 20 ml LB (plus ampicillin) to OD600 = 0.6. Before harvesting, the cells were induced by 1 mM isopropyl-b-D-thiogalactopyranoside (IPTG) for 45 min. The cells were washed once with the disruption buffer (25 mM KH2PO4, 25 mM K2HPO4, and 5 mM MgCl2, pH 7.2.), and French-pressed at 16,000 psi in the presence of 1 mM PMSF as a protease inhibitor. The membranes were collected by a 35-min centrifugation at 19,000 g. The resulting pellet was resuspended in a 100 mM NaCl, 30 mM K2HPO4 (pH 7.2) solution. The final volume of the suspension was separated into four equal fractions. One was exposed to ambient O2 in a shallow dish (control); the second sample was exposed to 10 mM DTT, 0.1 mM iodine (I2) was added to the third, and a solution of 0.3% hydrogen peroxide (H2O2) was added to the forth. All four samples were incubated for 15 min at room temperature. The samples, other than the control, were then quenched with 5 mM N-ethyl maleimide (NEM) and centrifuged at 19,000 g for 35 min. After the supernatant was discarded, the pellets were dried and 150 μl of nonreducing SDS sample buffer, containing 0.2 M sodium iodoacetate, was added. To analyze the cross-linking products, the solutions were heated to 70°C for 7 min and resolved in SDS-PAGE. Western blots were made, using purified antibodies directed toward the COOH terminus of MscL (Blount et al., 1996a) as the primary antibody and an anti-rabbit IgG AP conjugate (Promega) as the secondary antibody. Our preliminary trials showed that double-cysteine mutant proteins L121C/L122C and L128C/L129C were poorly recognized by the antibodies, apparently due to the close proximity of mutation sites to the epitope. These proteins, therefore, were tagged with six COOH-terminal histidines and were visualized with anti–6His-COOH monoclonal antibodies conjugated with alkaline phosphatase (Invitrogen). Western blots were developed using an NBT/BCIP reagent (Promega).
Single-channel Recording
Patch-clamp recording of mutant channels was performed in giant spheroplasts as originally described by Martinac et al. (1987). The channels were activated by applying steps of negative pressure to the pipette using a screw-driven syringe. The currents were recorded from excised patches in a 10 kHz bandwidth, digitized at 25,000 samples/s using a Digidata 1300 A/D converter and Clampex 8 Software (Axon Instruments, Inc.). After the baseline subtraction, Po was calculated as the mean current divided by the maximal current that could be achieved at saturating pressure. Prior to analysis, the pressures were converted into tensions using the pressure of MscS channel activation as a reference point (Blount et al., 1996b). According to a previous study (Sukharev, 2002), the MscS activation curve has the midpoint of 5.5 dyne/cm. Typically, 3–20 small channels were active in every patch. The dose–response curves for MscL were analyzed in the framework of two-state model (Sukharev et al., 1999b): Po/Pc = exp[−(ΔE − γΔA)/kT] where ΔE is the energy difference between the open and closed states in an unperturbed membrane, ΔA is the in-plane expansion of the protein upon gating transition, and γ is membrane tension. All-points histograms of single-channel traces and normalized probability density histograms reflecting relative occupancies of subconducting states were generated using the HISTAN program written in Matlab.
Osmotic Shock Survival/ATP Release Assays
Survival and ATP release into the osmotic shock medium were measured in the same experiment. Overnight cultures of MJF455 double knock-out (mscL −, mscS−) cells harboring one of the expression vectors, or the empty pB10a vector used as negative control, were grown in LB in the presence of 100 μg/ml ampicillin. The next morning the cultures were transferred to 50 ml of LB supplemented with 0.5 M NaCl (high-osmotic medium, 850 mOsm) and grown to OD600 of 0.7–0.8. Cells were induced with 0.8 mM IPTG 40 min before harvesting. Cells were pelleted, washed once with high-osmotic buffer (10 mM Tris, pH 7.4; adjusted to 850 mOsm with sorbitol) and finally resuspended in 1 ml of this buffer. 20 μl aliquots of the latter cell suspension were introduced in 1 ml of sterile Tris-sorbitol buffer of various osmolarity containing 20 μg/ml of Luciferase reagent (cat. #L 0633, Sigma-Aldrich). The samples were vortexed for 2–3 s and immediately counted in a TD-20/20 luminometer (Turner Designs). Calibration of the luminescent signal was done with K-ATP dissolved in the same Tris-sorbitol buffer. To account for the entire amount of ATP inside the cells, equal samples of cell suspension in the initial high-osmotic Tris-sorbitol buffer were French pressed, immediately mixed with Luciferase, and counted. The amount of ATP released during shocks was expressed as the fraction of the entire amount contained in that batch of culture. After shock and luminescent counting, the cell samples were diluted 104 or 105 times with the same shock buffer and plated on standard LB plates in triplicates. The colonies were counted manually and the mean and the standard error were determined.
RESULTS
Structure Analysis, Modeling, and Molecular Dynamic Simulation
According to the secondary structure prediction algorithms (PSIPRED, PROF, and NNPREDICT), the cytoplasmic part of EcoMscL (residues 104–136) can be depicted as a coil (R104-E118) followed by an α helix (E119 through Q132) predicted with a 95% confidence. Although the coiled linkers bear little sequence similarity, the latter helical (S3) domain is highly conserved among all identified MscL homologues (Spencer et al., 1999; Maurer et al., 2000; Sukharev et al., 2001b). The alignment of S3 domains for 20 MscL homologues is presented in Fig. 1 A. The consensus reveals the LLxEIRDLLK motif, which has a pattern of a strongly amphipathic helix. A similar sequence, xLQDVRELLR, occurs in the oligomerization domain of cartilage protein COMP, which forms a five-stranded coiled-coil structure as determined by the X-ray crystallography (Malashkevich et al., 1996). In this structure, all leucines and the central valine are packed inside the fivefold bundle. The bundle is additionally stabilized by salt bridges between the DVR segment aspartate and arginine sidechains from adjacent subunit (Fig. 1 B).
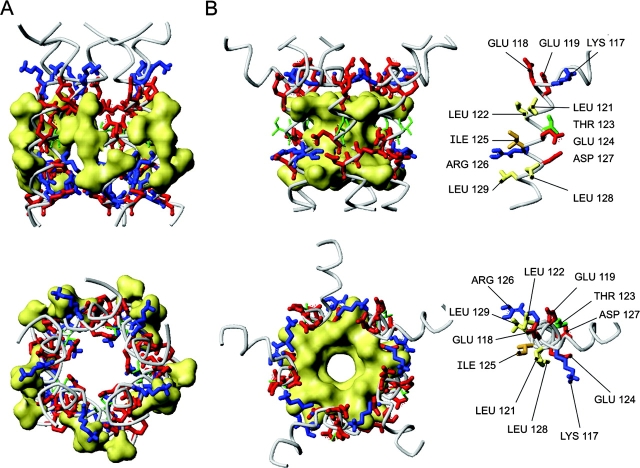
The crystal structure of TbMscL (Chang et al., 1998) was resolved for most of the molecule, from R11 at the NH2 terminus through R118 in the COOH-terminal (S3) segment. It includes most of the conserved amphipathic S3 cluster (residues L113-L121), which is in a helical conformation. These amphipathic helices, however, are in an unusual orientation. Represented as a helical wheel in Fig. 1 C, the structure indicates that the hydrophobic facet of each helix faces outside, while some of the polar and charged residues are buried inside the bundle. In all other coiled-coils of which we are aware, hydrophobic residues of the amphipathic helices face inwardly. It is possible that the crystallization conditions account for the atypical structure of the TbMscL coiled-coil. At low pH, aspartate 119 and other acidic sidechains can be protonated and therefore easily buried, at the same time the penalty of the water-exposed conformation for multiple leucines and isolucines could be low due to the presence of the detergent, dodecylmaltoside (Chang et al., 1998). Molecular dynamic simulations suggested that the crystal structure of COOH-terminal domains of TbMscL would be highly unstable at pH 7.0 in the absence of detergent (Elmore and Dougherty, 2001). A model of EcoMscL S3 domains in the TbMscL crystal–like conformation is shown in Fig. 2 A. Note, the last resolved residue in TbMscL structure was R118 homologous to R126 of EcoMscL, thus the remaining residues 127–136 were modeled as a continuous α helix. Aliphatic sidechains of conserved L121, L122, I125, L128, and L129, all shown by yellow surface, face outside. It seemed likely, therefore, that a simple reorientation of the helices, which would bury the large hydrophobic surface, would make the structure of the S3 bundle more stable.
Figure 2.
Models of the cytoplasmic bundle of EcoMscL. (A) A model built after the crystal structure of TbMscL as a left-handed bundle with sidechains of the conserved aliphatic residues L121, L122, I125, L128, and L129 all outside (shown by yellow surfaces). (B) A COMP-like model of S3 bundle. All five helices form a short left-handed coiled coil with a 19° tilt. All aliphatic side chains are inside, collectively shown by surface. Two rings of potential salt bridges were inferred. Positions of critical side chains on an individual S3 helix are shown on the right.
Fig. 2 B depicts a model in which the S3 helices form a left-handed pentameric coiled-coil closely resembling the analogous segment of COMP, with conformations of critical side chains on the monomer helix shown on the right side. The pairs of leucines and the centrally positioned isoleucine 125 are all packed inside the bundle. The assembly is stabilized by two rings of intersubunit salt bridges. The bridges were initially modeled as branched, K117 to E118 and E119 at the S3 NH2 terminus and R126 to E124 and D127 near the middle. The latter ring is more conserved among MscL homologues and is very similar to interactions in COMP.
The stability of assemblies of five fully hydrated 32-amino acid COOH-terminal fragments in both, Tb-like and COMP-like, conformations was further examined by molecular dynamic simulations. Only the α carbons of R104 of each subunit were harmonically restrained in their initial modeled positions. The Tb-like model of S3 was simulated for a total of 2 ns (Fig. 3 A). The first frame after heating and initial equilibration for 100 ps is shown in blue and the last frame after additional 900 ps of simulation is shown in red. Within the first 500 ps, the tight structure dilated such that average distances between equivalent α carbons increased by ∼15%, and water channels wedged between the helices connecting the interior and exterior of the bundle. About 70 water molecules were present inside the bundle after the initial equilibration, and the number increased to 130 by the end of the first nanosecond. The electrostatic conflict between E119 initially packed inside was resolved by rotations of sidechains and extensive hydration of the bundle interior. The association between the helices remained stable for at least 2 ns, apparently due to multiple electrostatic interactions. Out of 15 sterically possible salt bridges, an average of 10.5 were constantly present, with E131–R135 interactions being most stable.
Figure 3.
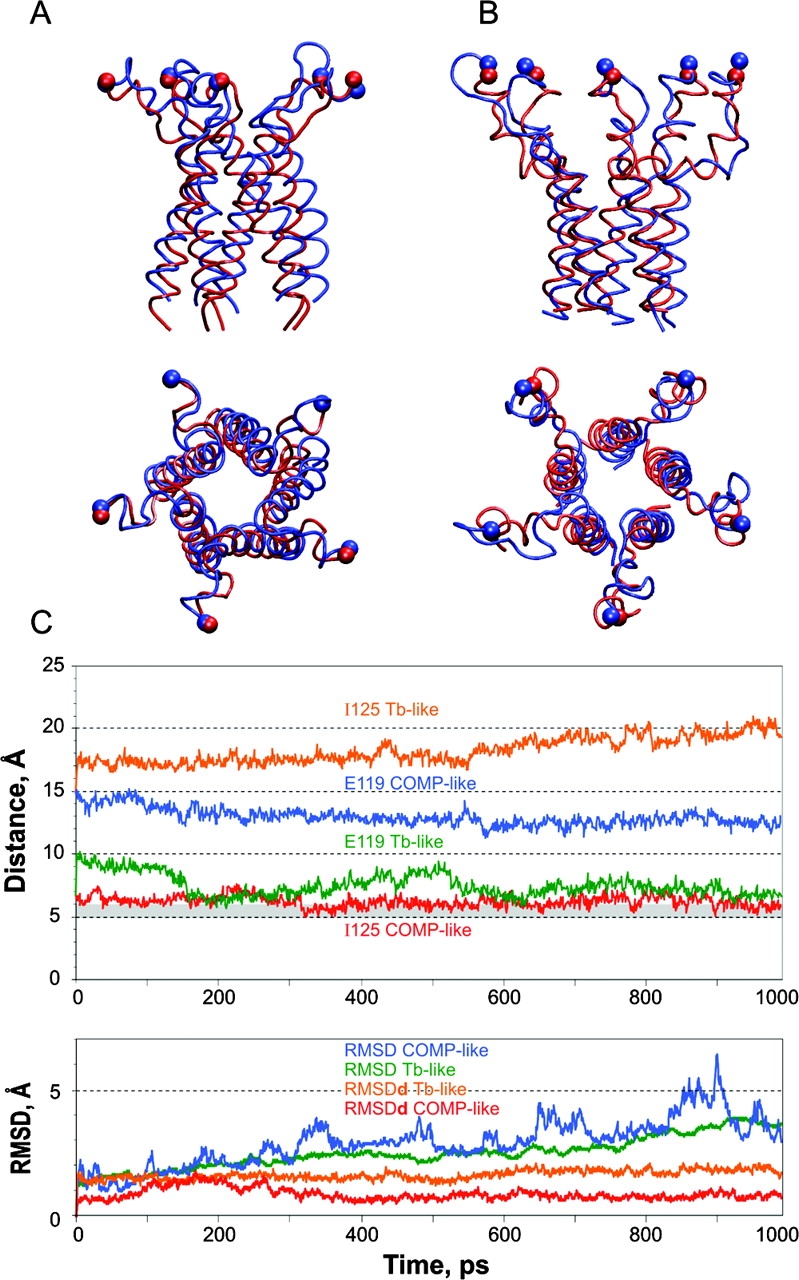
Molecular simulations of the COOH-terminal domains of EcoMscL. The results of 1-ns simulation of (A) Tb-like model, and (B) COMP-like model at 310 K. The initial conformations are represented as red tube; the final frames are shown as blue tube. (C) Distance trajectories for representative pairs of equivalent residues on the hydrophobic (I125) and polar (E119) sides of adjacent S3 helixes. The bottom panel shows root mean square deviations of absolute coordinates (RMSD) and of relative distances (RMSDd) derived from the 310K trajectories of Cβ atoms of the residues K106, P109, and A112 (linkers) and L121, I125, and L129 (bundle).
The minimization of energy in water did not significantly change the backbone position (RMSD 0.7 Å for α carbons and 1 Å for all atoms) for the COMP-like structure. A string of water molecules initially trapped inside the bundle during the system assembly was expelled in the course of heating, and water never entered the interior of the bundle again. The bundle retained its structure throughout the simulation at 310 K. The initial and the final frames of the 1-ns simulation are shown in Fig. 3 B. The conserved hydrophobic residues always remained inside the bundle. The salt bridges initially modeled as branched turned into single ionic pairs and the average number of intersubunit bridges in the equilibrated state was 8.5 per pentamer. The most stable associations were between K117 and E118 and between R126 and E124. The former were also H-bonded 36% of the time during last 500 ps of simulation at 310 K. Upon completion of the 1-ns simulation, we heated the system by 50 K and continued for one additional nanosecond. Although more distorted, the bundle remained stable at 360 K (87°C) for two more nanoseconds, without letting water inside.
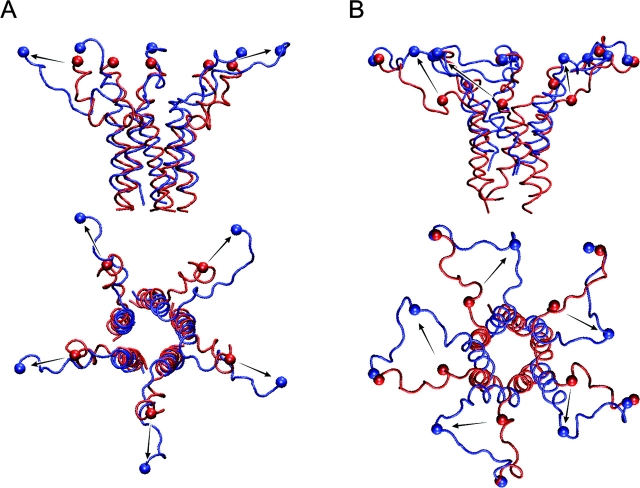
Another test for stability of the bundle was performed by pulling the proximal ends of M2-S3 linkers radially, simulating their movement during channel opening. The simulation started with the last conformation obtained in the previous simulation at 310 K. α-Carbons of arginines 104 previously restrained at the distance of r = 21.7 Å from the channel axis were directed with a harmonic force to a new position r = 35.6 Å, corresponding to the modeled open conformation 12 with the pore of 31 Å in diameter (Sukharev et al., 2001b). The ends of linkers arrived to their destination point within 150 ps. The simulation was continued for a total of 2 ns, during which the bundle was stable and “dry” inside, while the linkers remained partially coiled (Fig. 4 A). To further explore the limits of the bundle stability, an additional pull was applied to Cα of alanines 114, moving them to the final destination 29.5A away from the central axis (Fig. 4 B). The strain uncoiled the linkers and partially separated the upper portion of the bundle. The assembly yet remained stable for additional 2 ns, not letting water inside.
Figure 4.
The virtual “pull” experiment in which Cα atoms of residues R104 (A) or residues A114 (B) were steered to new positions shown by arrows. A harmonic force with the constant of 1 kcal/mole*Å2 was applied to those atoms shown as balls. The initial positions are depicted in red; the final frames after 2 ns simulation are shown in blue. The bundle remained stable during both simulations.
To characterize the dynamics of each type of assembly we scored the distances between β carbons (Cβ) for several identical residues on separate subunits. Fig. 3 C shows distance trajectories for two representative residues, one on the hydrophobic facet of the helix (I125) and one on the opposite side (E119) from two adjacent subunits, during 1-ns simulations of COMP-like and Tb-like models. To separate the relative motions of individual chains from the motions of the entire assembly, we scored the deviations of absolute coordinates from the initial position (RMSDs) and the deviations of relative distances between several pairs of identical atoms on different subunits (RMSDds). The trajectories presented in Fig. 3 C (bottom) and the data in Table I show that the RMSD is higher for the COMP-like assembly, which is most likely due to a higher flexibility of the linkers (residues 104–115) in that particularly conformation. The RMSDd parameter for the helical part (residues 119–136), denoted as bundle, however, is half as large (0.8 Å), which indicates that the COMP-like bundle is moving strictly as one unit. The motions of the Tb-like bundle are apparently less concerted.
TABLE I.
The Dynamics of COMP-like and Tb-like Structures
| COMP-like model | Tb-like model | |
|---|---|---|
| Å | Å | |
| RMSD bundle | 3.5 ± 0.73 | 2.96 ± 0.5 |
| RMSD linkers | 4.34 ± 0.22 | 3.65 ± 0.29 |
| RMSDd bundle | 0.8 ± 0.11 | 1.75 ± 0.17 |
| RMSDd linkers | 4.14 ± 0.51 | 2.67 ± 0.22 |
The dynamics of COMP-like and Tb-like structures as represented by mean deviations of absolute coordinates (RMSD) and relative distances (RMSDd) for groups of identical residues on the linkers and S3 helices. The parameters represent distances between all five beta carbons of residues 106, 109, and 112 in the linkers, or residues 121, 125, and 129 forming the core of the bundle. Based on 500 frames separated by 1-ps time intervals.
The distances and standard deviations were computed for residues E119, L122, I125, T123, L129, and L129 and for the adjacent of L121-L122 and L128-L129 pairs for each of the models (Table II) . To evaluate the probabilities of disulfide cross-linking between cysteines in corresponding positions, we scored the fraction of time that the pairs of β carbons spent within 6 Å of each other. The histograms for the COMP-like conformation (unpublished data) revealed that β carbons of adjacent residues at positions 121, 125, and 128 spend 3–17% of time in close proximity and have a good chance of being trapped by a disulfide bond without substantially distorting the structure of the bundle. The adjacent L121-L122 and L128-L129 pairs spend 30% and 88% of time within 6 Å. Note that E119 and T123, which are located on the periphery of the COMP-like bundle, were chosen as negative controls for cross-linking experiments because during the simulation they were separated by 14 Å most of the time and never closer than 11 Å.
TABLE II.
Statistical Assessment of Distances between Beta Carbons of Selected Residues during an MD Simulation of the S3 Domain of MscL
| COMP-like model | TbMscL-like model | |||
|---|---|---|---|---|
| Residue pairs | Distance (mean ± SD) | Occurrences within 6 Å | Distance (mean ± SD) | Occurrences within 6 Å |
| Å | % of time | Å | % of time | |
| L121-L121 | 7.0 ± 0.7 | 5.2 | 14.6 ± 1.7 | 0 |
| L122-L122 | 11 ± 0.6 | 0 | 11.5 ± 1.8 | 0 |
| L121-L122a | 6.4 ± 0.8 | 30.5 | 11.5 ± 1.6 | 0 |
| I125-I125 | 6.9 ± 0.8 | 16.6 | 16.3 ± 1.6 | 0 |
| L128-L128 | 6.9 ± 0.5 | 3.0 | 17.8 ± 1.5 | 0 |
| L129-L129 | 10.2 ± 0.5 | 0 | 14.8 ± 0.9 | 0 |
| L128-L129a | 5.3 ± 0.5 | 87.8 | 13.7 ± 1.4 | 0 |
| E119-E119 | 14 ± 1.1 | 0 | 9.1 ± 1.4 | 0.16 |
| T123-T123 | 14.3 ± 0.6 | 0 | 8.8 ± 1.2 | 0.04 |
Distribution was calculated over the last 500-ps periods of 1-ns simulations. The distances between Cb of all five neighboring residues are included. The fraction of occurrences within 6 Å was calculated based on the distances in 500 frames.
Only the distances between L122 of one subunit and L121 of the next clockwise subunit (looking from the cytoplasmic side) are considered. The distances measured in counterclockwise direction or across the bundle axis (e.g. subunits 1–2 or 1–4) are above 11 Å at any simulation step.
For the Tb-like model, the β carbons of all equivalent aliphatic residues were >11 Å apart at any time. The neighboring pairs of L121 and L122 never occurred within 8.8 Å, whereas pairs of L128 and L129 were never closer than 10.8 Å in the equilibrated state. The residues E119 and T123, which were distant in the COMP-like conformation, are now in the core of the bundle only 8–10 Å apart. From the table one can see that neighboring E119 pairs spend 0.2% and T123 pairs 0.04% of time within 6 Å. Given the RMSDds of 1.2–1.4 Å, cysteine substitutions at these positions would be expected to cross-link monomers under oxidizing conditions.
Cysteine Cross-linking
We introduced single cysteine substitutions for residues E119, L121, T123, and I125. Additionally, double cysteine mutants L121C/L122C and L128C/L129C were generated. As was mentioned, the antibodies generated against the COOH-terminal epitope poorly recognized the double cysteine mutants. For this reason, the double mutants were tagged with six COOH-terminal histidines. Correspondingly, those cross-linking products were visualized with anti-6His monoclonal antibodies (see materials and methods).
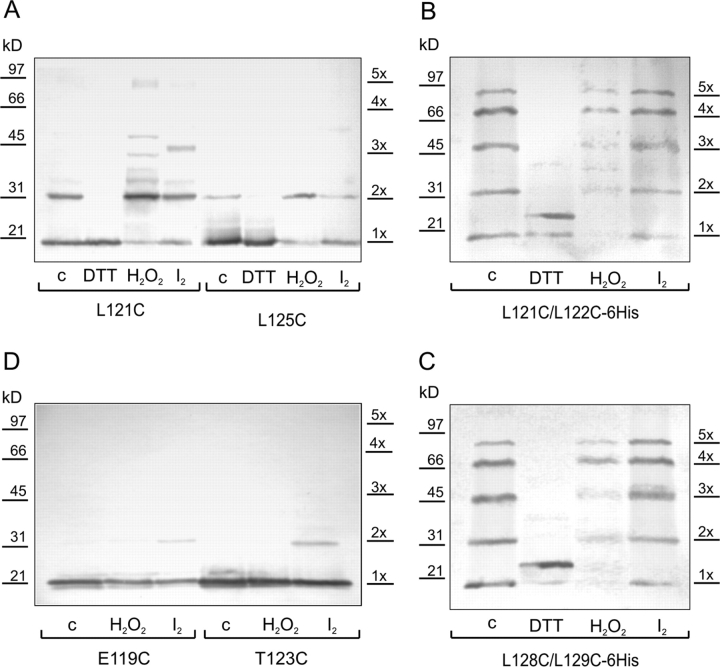
The Western blot for the cross-linking experiment with the L121C mutant (Fig. 5 A) exhibits subunit dimerization under ambient conditions (15 min exposure to atmospheric oxygen). While monomer bands are still present, darker dimer bands appear upon oxidation with H2O2 or I2. Under reducing conditions with DTT, only a monomer is present. The cross-linking patterns for double mutants L121C/L122C-6His and L128C/L129C-6His under ambient oxygen show ladders of products from monomers to complete pentamers (Fig. 5, B and C). Stronger oxidation with H2O2 reduced the total amount of visible protein with relatively more tetramers and pentamers and fewer monomer. Oxidation with iodine produced similar patterns. Reduction with DTT results in a monomeric form, which for an unknown reason migrates in the gel as a double band.
Figure 5.
Disulfide cross-linking in single and double cysteine MscL mutants. The patterns were visualized with the Western blot technique. Cross-linking conditions are denoted as c (control) - ambient oxygen, DTT (10 mM), H2O2 (0.3%), I2 (0.1 mM), duration of all treatments was 15 min.
The cysteines in E119C and T123C are located on the opposite side of the helix relative to L121, I125, or L129. One can observe that the monomeric form prevails for all three types of oxidative conditions for both mutants with a light dimer band occurring in T123C in the presence of I2 (Fig. 5 D). One can conclude that even under strongly oxidizing conditions the cysteines at position 119 do not cross-link and those at positions 123 rarely cross-link. The cross-linking data therefore strongly favors the COMP-like conformation of S3 helices.
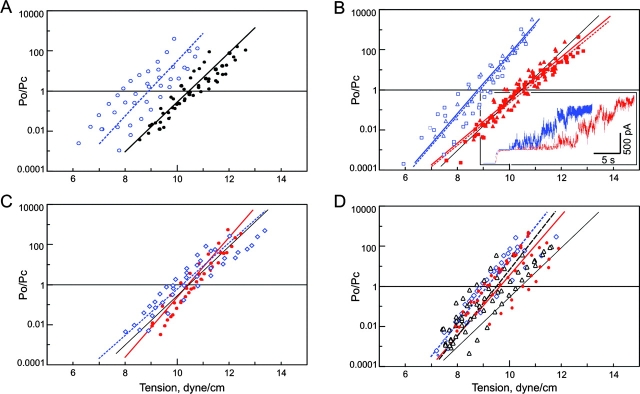
Effects of Cross-linking and Destabilizing Mutations in S3 on Single-channel Currents and Dose–response Curves
If S3 domains change their conformation substantially during the gating process, then their immobilization via covalent cross-linking should affect single-channel currents and the dose–response curves. Conversely, mutations that destabilize the bundle may have an opposite effect on the gating process. Therefore, we first studied the effects of disulfide bonds formation or reduction on L121C/L122C and L128C/L129C channels. Then we characterized the channels in which these highly conserved aliphatic amino acids were replaced by small-side chain residues, predicted to disrupt bundle association.
Excised inside-out patches were subjected to either ambient conditions (atmospheric oxygen) or perfused with oxidizing or reducing agents on the cytoplasmic side. Although WT MscL has no cysteines, control experiments with spheroplasts carrying p5–2-2b (WT MscL) plasmid revealed that perfusion of 0.1–1% H2O2 irreversibly inactivates 30–80% of active channels in the course of 10–60 s. 25 mM DTT applied to a WT MscL patch under constant pressure, typically increased the channel activity (unpublished data). To characterize the effect of reducing versus ambient environment on WT MscL activity, we statistically treated activation curves and extracted gating parameters with and without DTT. The data from several independent patches are presented as log(Po/Pc) versus membrane tension in Fig. 6 A. The two-state equilibrium model fitted to the data yielded the energy of the transition, ΔE, and effective protein expansion, ΔA presented in Table III . The slope of dose–response curves for WT MscL remains the same after DTT addition, but the midpoint (γ0.5, tension at which Po/Pc = 1) shifts from 10.5 to 8.9 dyne/cm. An apparently nonspecific effect of reduction, also reported for cysteineless MscS (Koprowski and Kubalski, 1999), is most likely due to the cleavage of disulfide bonds in the surrounding proteins, which may change the mechanical properties of the membrane as a whole.
Figure 6.
Dose–response curves for WT MscL, S3 mutants with substituted leucines, and the Δ110–136 truncation mutant. The data is presented as Po/Pc values versus membrane tension. Four to seven curves measured on independent patches were plotted and fitted according to the two-state model (see materials and methods). The lines represent curves with average parameters ΔE and ΔA (see Table III) for each dataset. (A) WT MscL measured under ambient conditions (filled circles, black solid line) or in the presence of 25 mM DTT (blue open circles, dotted line). (B) L121C/L122C (squares, solid line) and L128C/L129C (triangles, dotted line) measured under ambient (red) or reducing (blue) conditions. (set) Single-channel traces of L128C/L129C MscL measured under ambient conditions (red) or in the presence of 25 mM DTT (blue) in the same patch, in response to a similar sequence of pressure steps. (C) L121A/L122A (red) and L121T/L122T (blue) measured under ambient conditions. (D) The data for the quadruple L121A/L122A/L128A/L129A (red circles and solid line) and L121T/L122T/L128T/L129T (blue diamonds and dotted line) mutants and for the truncated Δ110–136 MscL mutant (triangles, black dashed line). The black solid line representing WT MscL is given as reference in each panel.
TABLE III.
Energy, Expansion, and Midpoint Tension for Gating Transitions Extracted from Dose–response Curves for WT MscL and Eight Mutants
| Mutant | ΔE | ΔA | γ0.5 |
|---|---|---|---|
| kT | nm2 | dyne/cm | |
| WT | 29.8 ± 5.1 (n = 10) | 11.5 ± 2.1 | 10.5 ± 0.6 |
| WT + DTT | 28.3 ± 4.0 (n = 5) | 12.9 ± 0.9 | 8.9 ± 0.6a |
| L121C/L122C | 26.0 ± 2.3 (n = 5) | 10.1 ± 0.7 | 10.5 ± 0.5 |
| L121C/L122C + DTT | 30.3 ± 1.0 (n = 3) | 13.9 ± 0.1 | 8.8 ± 0.2 |
| L128C/L129C | 26.0 ± 3.4 (n = 8) | 9.9 ± 1.3 | 10.6 ± 0.2 |
| L128C/L129C + DTT | 30.6 ± 5.7 (n = 4) | 14.0 ± 2.8 | 8.9 ± 0.9 |
| L121A/L122A | 36.1 ± 3.9a (n = 6) | 14.0 ± 1.1a | 10.4 ± 0.5 |
| L128A/L129A | 34.6 ± 5.4 (n = 4) | 14.0 ± 2.2 | 10.1 ± 1.3 |
| L121T/L122T | 27.2 ± 4.1 (n = 7) | 10.8 ± 1.7 | 10.2 ± 0.7 |
| L121A/L122A/L128A/L129A | 33.3 ± 2.9 (n = 7) | 14.0 ± 1.3a | 9.7 ± 0.7a |
| L121T/L122T/L128T/L129T | 34.1 ± 3.7a (n = 4) | 15.0 ± 2.2a | 9.2 ± 0.5a |
| Δ110-136 | 36.1 ± 5.9a (n = 12) | 15.3 ± 2.2a | 9.5 ± 1.0a |
The double cysteine mutants were characterized under ambient oxidizing conditions as well as in the presence of 25 mM DTT (+DTT). The data obtained from L121C/L122C + DTT and L128C/L129C + DTT traces were t tested against WT + DTT. The data from WT + DTT and from all preparations without DTT were t tested against WT.
These numbers marked are significantly different from controls, with t test values < 0.05.
Based on the cross-linking patterns, we concluded that ambient oxygen is almost as effective in bridging cysteine pairs in L121C/L122C and L128C/L129C mutants as a strongly oxidizing environment. Therefore, ambient conditions were considered oxidative and compared with reducing environment. The mutants L121C/L122C and L128C/L129C exhibited a similar degree of activation as WT MscL upon perfusion with buffer containing DTT. Two current traces recorded in the same L128C/L129C patch under ambient oxygen and after 60 min of bathing in 25 mM DTT are shown as inset in Fig. 6 B. Under reducing conditions the current grows steeper in response to a similar increase of pipette pressure. The patch ruptured after reaching saturation, but, as evident from the trace, the maximal current and the total number of active channels did not increase. The data from two independent preparations of spheroplasts (8 patches total) indicated no significant difference in maximal number of active channels per patch observed under ambient or reducing conditions. A shift of activation curves observed in the double cysteine mutants upon transferring to the reducing medium was similar to one recorded for WT MscL (Fig. 6, A and B). Positions of activation curves for L121C/L122C and L128C/L129C MscL coincided under ambient conditions and changed consistently upon DTT perfusion. There was not only a shift in the midpoint, but also a statistically significant increase in the slope of the curve upon reduction. This translated into the increase in effective transition-related protein expansion from ∼10 to 14 nm2 for both mutants (see Table III).
If the COMP-like structure is correct, an introduction of small side chain substitutions for the highly conserved leucines in S3 is expected to weaken the interactions between S3 domains. The L121A/L122A, L128A/L129A, and L121T/L122T double mutants as well as the quadruple L121A/L122A/L128A/L129A and L121T/L122T/L128T/L129T mutants well expressed and formed functional channels. The dose–response curves for the double mutants were similar to the curve for WT control, with a slightly higher slope for the alanine mutants (Fig. 6 C). The curves for the quadruple mutants (Fig. 5 C) are similar to that of Δ110–136 mutant (see below), with a slightly lower γ0.5 and higher slope relative to WT. The thermodynamic parameters derived from these curves are presented in Table III.
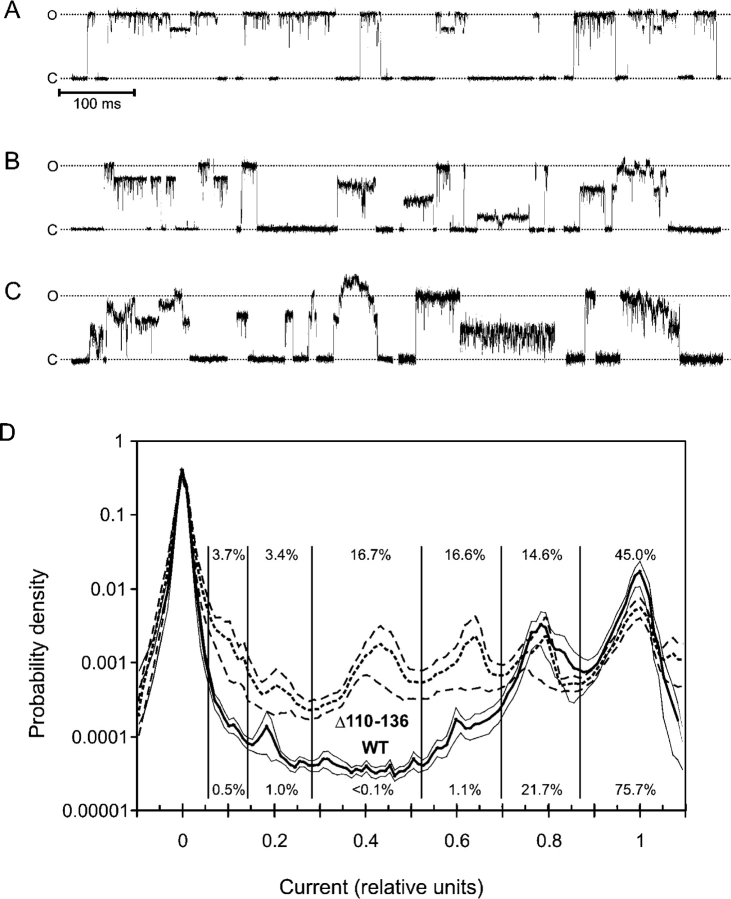
Although dose–response curves calculated from integral currents and gating parameters were only slightly altered with either cysteine cross-linking or leucine replacement mutations, the patterns of channel gating changed remarkably. Double alanine substitutions for L121/L122 and L128/L129 significantly increased the occupancy of short-lived low-conductance substates, as illustrated by the trace and all-points histograms (Fig. 7) . More polar threonines in positions 121 and 122 caused further reduction of the occupancy of the fully open state, favoring multiple short-lived substates, also long-lived states of ∼0.65, 0.45, and 0.2 of full conductance (Fig. 7, middle). The L121C/L122C mutant gated similarly to WT under ambient oxygen, but changed the pattern upon reduction, occupying more subconducting states. Indeed, reduced cysteines are smaller and more polar than leucines. Therefore, the change of gating pattern upon disulfide reduction is expected to be similar to the effect of alanine substitution at the same positions. Peculiarly, the quadruple mutants with all four leucines substituted exhibit less pronounced short substates than the double mutants, but considerably more pronounced long-lived substates with amplitudes of 0.8, 0.65, 0.45, and 0.25 relative to the fully open state (Fig. 7). The conductance of the fully open state was 3.48 ± 0.08 nS for L121A/L122A/L128A/L129A and 3.49 ± 0.06 nS for L121T/L122T/L128T/L129T, which is ∼14% higher than that for WT and cysteine mutants (3.07 ± 0.08 nS) recorded in the same solution.
Figure 7.
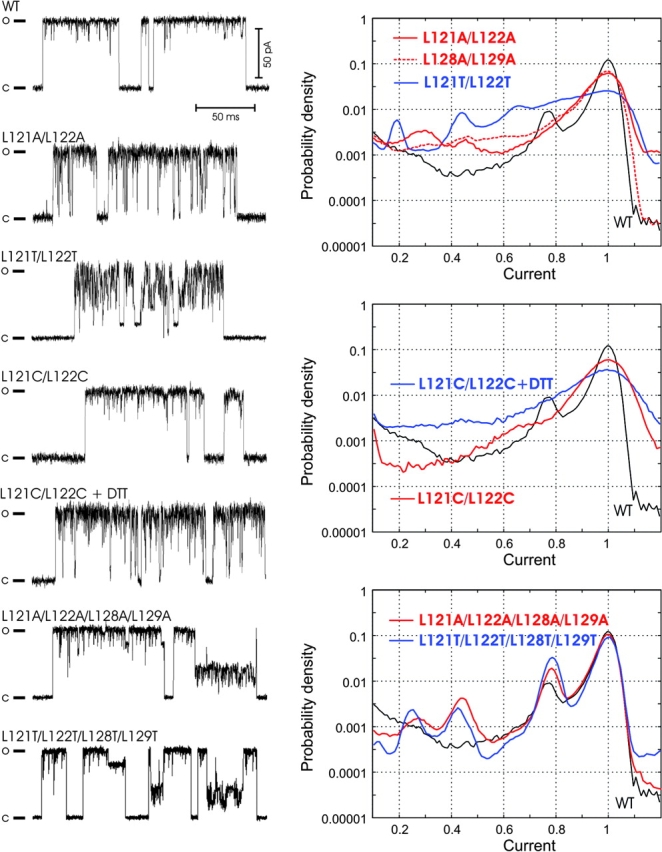
Representative single-channel traces (left) and probability density amplitude histograms (right) for WT MscL and mutants with alanines, threonines, or cysteines substituting for conserved leucines in S3. Cysteine mutants were characterized under ambient or reducing conditions. The histograms for WT are shown in all panels as reference (black solid line).
Gating Properties of the Δ110–136 Truncation Mutant
Previous studies reported that the removal of the COOH-terminal domain was tolerated by MscL (Blount et al., 1996b), producing only subtle effect on gating (Hase et al., 1997). Fig. 8 A shows representative fragments of Δ110–136 MscL traces recorded within first seconds of pressure application. The traces appear similar to WT; however, the average single-channel conductance measured relative to MscSs present in the same patch was 3.38 ± 0.9 nS, i.e., 10% higher than WT. Several types of unusual behavior were recorded in ∼60% of all Δ110–136 traces (Fig. 8, B and C), typically after a period of “normal” gating. Like WT, Δ110–136 MscL occupied a number of subconducting states. Note, however, that WT MscL usually undergoes transitions to the fully open state, from which it visits subconducting states and returns back to the closed state predominantly from the fully open level. In contrast, Δ110–136 tends to open from the closed state directly to a long-lived substate and frequently close from the substate without visiting the fully open state. A similar behavior was observed in quadruple alanine and threonine mutants. A specific feature frequently observed in the truncated mutant, but practically never in the wild-type MscL, were irregular amplitudes of fully open and subconducting states (Fig. 8 C), as if the channel occupies a continuum of intermediate states rather than discrete states. Another pattern shows multistep openings with stair-like transitions between adjacent subconducting states. About a quarter of Δ110–136 traces demonstrated currents with amplitudes exceeding that of the fully open state. Such “extra” conductances were often characterized by “floating” amplitudes and irregular oscillations. In about a third of the recorded traces Δ110–136 MscL showed extended periods of fast flickering between the baseline and an intermediate conductance level (Fig. 8 C). We also found that the patches containing Δ110–136 MscL were in average mechanically less stable as they disrupted at pipette pressures 30% lower than patches with WT MscL (unpublished data).
Figure 8.
Fragments of typical patch-clamp traces recorded from giant spheroplasts expressing Δ110–136 MscL. The mutant shows WT-like gating patterns (A), but greater variability of single-channel amplitudes and kinetic patterns (B and C). (D) Probability density distributions of current amplitudes for Δ110–136 and WT calculated from all-point histograms. To emphasize only long-lived subconducting states, fragments of the traces were low-pass filtered at a 500 Hz cut-off. The fragments of traces characterized with Po between 0.4 and 0.5 were selected. The current was normalized to the amplitude of the fully open state and the range from −0.1 to 1.1 was divided into 120 bins. Thick lines represent the mean of 11 traces for WT and 10 traces for Δ110–136. Thin lines delineate standard error intervals calculated for each bin. The relative occupancy for each substate indicated by numbers was calculated as the fraction of data points between the two minimums separating the adjacent peaks.
To characterize the amplitudes and occupancies of stable subconducting states in Δ110–136 MscL, we generated combined all-point histograms for large datasets, which included fragments of 11 and 10 independently recorded traces for WT and Δ110–136, representing 180 s of channel activity in total. The fragments were recorded near P0.5, i.e., at pressures that provide 40–50% activation of channels. Averaged probability density distributions of current amplitudes calculated for these traces are presented in Fig. 8 D. The positions of peaks on both histograms indicate that the amplitudes of most substates for Δ110–136 and WT essentially coincide. Discernible peaks with maximums positioned at 0.18, 0.6, and 0.78 for WT indicate the same current amplitudes as for Δ110–136; however, the mutant shows an additional prominent substate at 0.43. Δ110–136 is characterized by considerably higher occupancies of substates and by somewhat decreased relative probability of being in the fully open state, as indicated by numbers by the histograms. The standard error of the probability of occupying a substate is also larger for Δ110–136 than that for WT due to the variability of the gating pattern described above.
A dose–response curve for Δ110–136 MscL calculated from amplitude histograms are shown in Fig. 6 D along with the data for the quadruple alanine and threonine mutants. The line representing the average for seven patches is shifted to the left from that for WT and is slightly steeper. The effective energy and expansion parameters of the Δ110–136 transition (Table III) are similar to that of the quadruple mutants and comparable to those of WT. Note that the quadruple mutants exhibited similar propensity to long-lived subconducting states (Fig. 7, bottom) and their full conductance was also higher than that of WT. The data suggest that a disassembled state of S3 bundle or complete absence of S3 both increase the multiplicity of conducting states, slightly reduce γ0.5, and have similar but rather subtle effects on the energetics of MscL-gating transition.
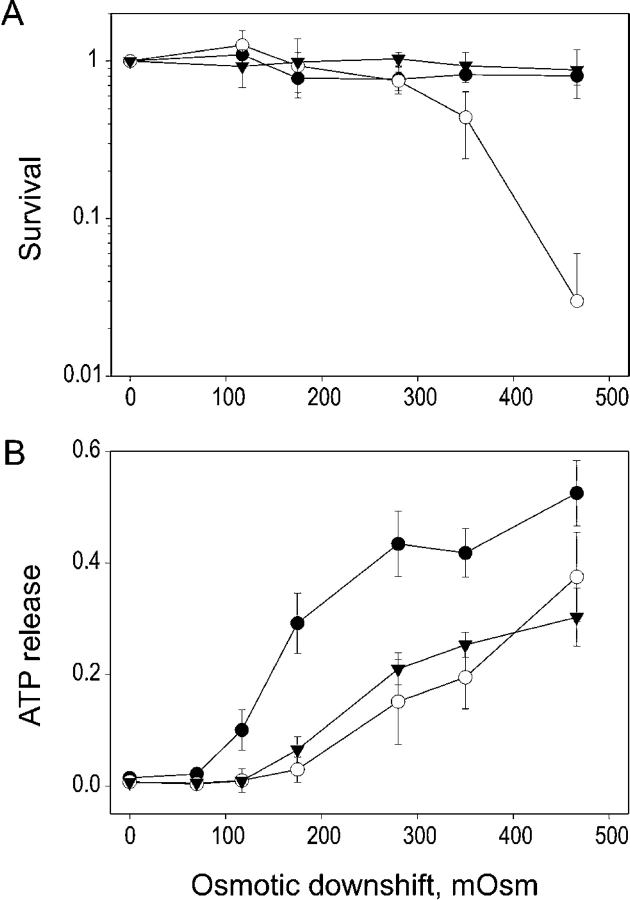
Osmotic Shock Survival and Release of a Cytoplasmic Marker in the Δ110–136 Truncated Mutant
The physiological effect of truncation was assessed by measuring the extent to which the Δ110–136 mutant could functionally substitute for WT MscL in terms of rescuing the osmotically fragile MJ455 cells devoid of both mechanosensitive channels MscL and MscS (Levina et al., 1999) under varied osmotic shock conditions. Fig. 9 A shows that control cells carrying the empty vector start dying at osmotic downshifts beyond 350 mOsm, and 2–6% remain alive upon a 470 mOsm downshift. Cells expressing WT or Δ110–136 MscL are essentially insensitive to shocks of this magnitude. The amount of cytoplasmic marker ATP released in the medium after the shock was substantially different between the two MscL-expressing cultures. Fig. 9 B shows that at around 300 mOsm shock, the Δ110–136 cells release about half of their total ATP, whereas control cells and WT release only ∼20%. The small difference in ATP release between cells expressing WT MscL and empty vector control suggests that WT MscL hardly passes ATP. This is consistent with the data reported by Ajouz et al. (1998) who found no difference in osmotic release of glutamate, trehalose, or glycine betaine from WT and mscL knock-out bacteria. Apparently those organic osmolytes may exit cells through alternative pathways. Expression of Δ110–136 MscL, however, significantly increases the osmotic ATP release, suggesting that the truncated channel is more permeable to ATP than the WT.
Figure 9.
The survival of MJF 455 E. coli cells (A) and ATP release (B) under conditions of varied osmotic shock. The cells were grown in a rich medium of 850 mOsm and subjected to osmotic downshifts of indicated magnitudes. The ATP in the shock medium was assayed, and the same samples were plated for colony count. The empty vector control is shown by open circles, WT-expresser cells by triangles, and Δ110–136 cells by filled circles. The data represent means of three independent experiments (± SEM).
DISCUSSION
The data presented above strongly support the COMP-like conformation of the S3 bundle of EcoMscL depicted in Fig. 2 B. The LLxEIRDLLK motif appears to be critical for pentamerization of S3, based on the role of a similar sequence in the oligomerization domain of the cartilage protein COMP (Malashkevich et al., 1996). The high conservation of this sequence suggests that the arrangement of COOH-terminal domains across the family of MscL channels is likely to be the same. Although the primary structural information was obtained on TbMscL, our choice of EcoMscL for modeling, simulation, and experimentation was dictated by the fact that most of the single-channel data has been obtained on the latter version of the channel and it remains the focus for many research groups.
Previously, the assembly of COOH-terminal fragments from the crystal structure of TbMscL has been MD simulated and shown to be unstable. The backbone of each chain moved away from the near-axial position by ∼5 Å (RMSD) within 1,500 ps and had a tendency to separate further (Elmore and Dougherty, 2001), suggesting that the crystal conformation may have been stabilized by low pH and detergent (Chang et al., 1998). An additional factor that could have stabilized the crystal conformation is the presence of two valines preceding the first pair of conserved leucines, which disrupt the strictly amphipathic character of this sequence in the mycobacterial MscL. The MD simulation of the EcoMscL S3 assembly modeled after TbMscL presented in this study (Fig. 3 A) also revealed an outward movement of helices, higher amplitudes of disconcerted fluctuations (Table I), and increasing volume of water trapped inside, all suggesting loose association in the bundle.
The arrangement of EcoMscL with leucines inside (Fig. 2 B) better satisfies the hydrophobicity criteria and was found stable during 1-ns molecular dynamic simulations at 310 K, as well as at 360 K (85°C). The helices did not uncoil, which is consistent with high temperature stability of the secondary structure of MscL studied previously by CD spectroscopy (Arkin et al., 1998). Pentamerization itself may increase helical stability, as has been demonstrated recently for chemically synthesized COOH-terminal domains of TbMscL preassembled on a template (Kochendoerfer et al., 2002). The EcoMscL bundle also remained stable under stress during the virtual “pull” experiments (Fig. 4). The extremely low RMSDd derived from relative distances for residues 121–129 (Table I) indicated that the bundle moves as one unit, without a tendency to spread apart.
Several types of intermolecular interactions were found to stabilize the oligomerization domains of COMP (Malashkevich et al., 1996). In the COMP heptad that resembles the S3 domain of MscL, ∼48% of its surface area, largely hydrophobic, is buried, and there are two potential “layers” of intersubunit ionic pairs on the periphery, which were also predicted by modeling (Kajava, 1996). Similar interactions apparently hold together the S3 bundle of MscL. With leucines positioned inside the bundle, the MD-equilibrated COMP-like model is characterized by the total buried area of aliphatic sidechains of ∼2,500 Å2 versus 700 Å2 calculated for the simulated Tb-like model. The MD simulation revealed the latter in a highly hydrated state. Use of the standard atomic solvation parameters (Wesson and Eisenberg, 1992), which take into account only interactions of solvent-accessible surfaces with water, predict the COMP-like orientation to be more stable by ∼5.0 kcal/mole (8.5 kT). This estimation ignores extensive interchain Van-der-Walls interactions inside the protein. Using the empirical energy of transfer for aliphatic carbons from water to protein interior of 19 cal/mole Å2 (Lomize et al., 2002) one may find a much stronger stabilizing effect of combined apolar interactions of ∼34 kcal/mole (57 kT). The statistics of intersubunit salt bridge formation in the second half of each simulation indicated an average of 8.5 ionic pairs per bundle for the COMP-like model and 10.5 pairs for the Tb-like structure. A rough estimation based on 2.2 kcal/mole (3.8 kT) per ionic pair (Kumar and Nussinov, 1999), makes electrostatic interactions more favorable in the Tb-like model; however, the domination of apolar interactions predicts the COMP-like structure to be decisively more stable, by ∼29 kcal/mole.
Molecular modeling sometimes predicts distances between critical residues sufficiently well to design cysteine mutants for disulfide-trapping experiments (Sukharev et al., 2001a). The MD simulation, although covering a relatively short time scale, provides a statistical assessment of proximities based on the dynamics of the backbone and sidechains. The cross-linking data in Fig. 5 are consistent with the distances between Cβ of the corresponding residues and the ranges of thermal fluctuations presented in Table I. Although the distances between adjacent Leu121 and adjacent I125 are similar, the distribution for L121 was skewed to the shorter range. Probably for this reason L121C shows a slightly higher degree of dimer formation under ambient conditions than does I125C. One should remember that due to the odd number of subunits in the complex, it is impossible to achieve a 100% dimerization with one cysteine per chain. With double cysteine substitutions, a complete interlinking of the bundle is possible, as indicated by the presence of pentamer bands in L121C/L122C and L128C/L129C mutants. Such cross-links appear to be sterically impossible in the Tb-like arranged bundle, especially under ambient oxidation conditions. Some proteins are sufficiently dynamic that when cysteines are introduced in two positions far apart in the native conformations, disulfide bridges still form under strongly oxidizing conditions (Falke and Koshland, 1987). Such large fluctuations are typically too rare to be simulated using MD. Thus, careful controls must be used in order to argue that formation of disulfide bridges between introduced cysteines indicates proximity of the mutated residues in the native protein. We chose E119 and T123 as control residues because the distance between each of these residues on adjacent subunits is large when the EcoMscL S3 bundle is modeled after COMP, but is short in the Tb-like structure. The very low cross-linking in E119C or T123C mutants (Fig. 5 D) corroborates with the mean distances and deviations for these residues derived from MD simulations (Tables 1 and 2). The conclusion that the bundle has COMP-like structure and is rather stable is supported by the fact that cysteines 119 or 123 do not cross-link substantially even under strongly oxidizing conditions, whereas cysteines substituting for hydrophobic residues in the core of the COMP-like model cross-link under ambient conditions in the absence of oxidizing reagents. Having all cross-linking corollaries and controls, however, we admit that a high-resolution study of S3 bundle structure would still be required to show that the LLxEIRDLLK motif is indeed a pentamerization signature.
Here we attempted to answer the long-standing question of whether or not the S3 bundle separates upon channel opening. In the original model of MscL gating transition (Sukharev et al., 2001b), the S3 bundle was separated in the open state, making the pore wide-open, as initially suggested by pore size estimations (Cruickshank et al., 1997) and thioredoxin permeation experiments (Ajouz et al., 1998), which were later contested (Vazquez-Laslop et al., 2001). The rationale for placing S3 domains parallel to the membrane surface was to compensate for dramatic flattening of the transmembrane barrel in the open state (Sukharev et al., 2001b); however, no experimental evidence for such conformation existed. Contrary to the initial models, the behavior of double-cysteine mutants, mutants with the weakened association of S3 domains as well as the truncated mutant presented here strongly suggest that the bundle does not separate during MscL gating. First of all, the channel gated normally with S3 domains interlinked by disulfide bonds and there was no increase in the number of active channels upon reduction of bonds. There was no substantial difference in the position of dose–response curves and thermodynamic parameters of gating upon addition of DTT beyond the nonspecific curve shift, which was observed in controls with cysteineless WT MscL (Fig. 6). Nor was a significant change in gating parameters observed in mutants with diminished hydrophobic interactions between S3 domains and apparently loose bundle structure. The L121A /L122A substitutions should cause a decrease in the domain association energy by at least 7–9 kcal/mole (11–14 kT) for the entire bundle, as estimated from solvent-accessible area of these residues and standard hydration parameters (Guy, 1985). If the work of bundle dissociation had to be produced by membrane tension during the opening, then the effect of these mutations or COOH-terminal truncation would be readily seen as a decrease in ΔE. In reality, the apparent energy of the transition does not decrease (see Table III). This indicates that the state of the gate is not rigidly coupled to the conformation of the S3 bundle. The rounds of MD simulations with a radial “pull” of residues 104 or 114 illustrate that indeed the linkers are long enough to accommodate the opening without breakdown of the bundle. Finally, the channels cross-linked via S3 exhibited the same unitary conductance as WT, whereas the bundle-destabilizing quadruple alanine or threonine mutations or complete Δ110–136 deletion slightly increased the conductance.
How could we be sure that the small sidechain substitutions at positions 121, 122, 128, and 129 indeed loosen the S3 association? The most obvious effect of the double mutations is the increase in the occupancy of short-lived low-conducting substates, no matter which pair of leucines was substituted. This can be ascribed to partial bundle dissociation and increased dynamics of S3 domains or M2-S3 linkers, which now acquire enough conformational freedom to interact with the conducting pathway of the pore. The observation that the L121C/L122C mutant behaves more like WT under ambient oxidizing conditions, but switches to a regime resembling L121A/L122A gating upon perfusion of DTT (Fig. 7), corroborates with this hypothesis. Because the increase in the substate occupancy is not accompanied with changes in the thermodynamic parameters of gating, one may conclude that the substates arise form flapping of COOH termini or linkers into the pore rather than from their influence on the main gate. The substates are less prominent in quadruple mutants, probably because their bundles are completely disrupted and there is no interaction that would hold disordered COOH termini near the pore entrance.
The long-lived substates, however, may be of different nature. Note that the long-lived substates of low conductance frequently observed in L121T/L122T are characterized by low noise. These could be due to channels that are prevented from closing completely by the “foot-in-the-door” action of loose S3 domains or linkers. On the other hand, the long-lived substates have amplitudes similar to the long-lived substates of the quadruple alanine and threonine mutants and of truncated Δ110–136 MscL and may signify a destabilization of the fully open state of the barrel when the COOH-terminal ends are not secured together. Such slow transitions may be attributed to asymmetric barrel conformations (Shapovalov et al., 2003) more populated in the mutants in which COOH termini are disordered or absent.
In summary, immobilization of S3 with covalent cross-links does not evoke visible changes in gating compared with WT, whereas destabilizing mutations do. It is evident, therefore, that in WT MscL S3 domains remain stably associated in both closed and open states. The revised model of the Eco MscL open state with S3 domains bundled together is presented in Fig. 10 . The M2-S3 linkers are depicted in an arbitrary extended-relaxed conformation with five windows allowing enough space for conduction and permeation.
Figure 10.
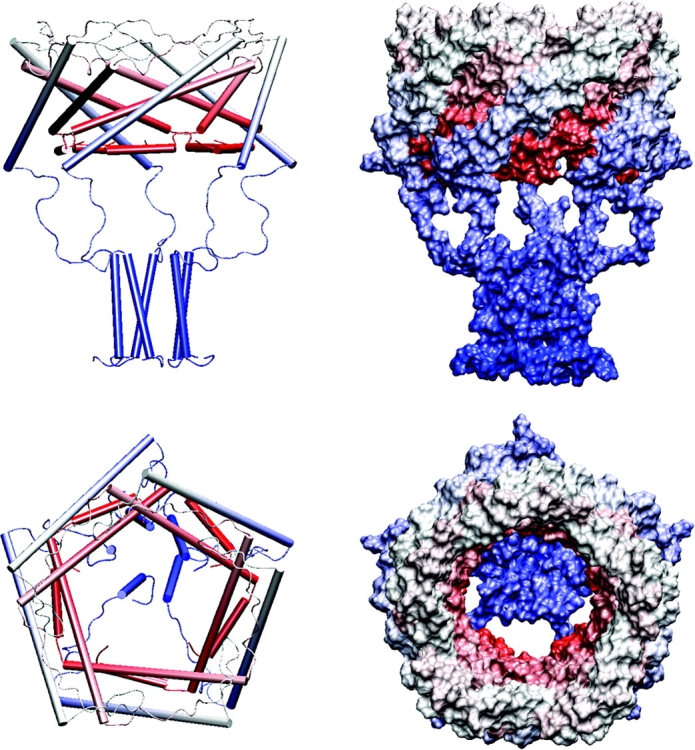
Structural models of EcoMscL in the open conformation. The right side shows α helices as cylinders and coiled segments as strings, the left side is a space-filled model. The structure begins with red at the NH2 terminus and ends with blue at the COOH terminus. The top figures show a side view and the bottom shows a view from the outside through the pore. The five windows of the sieve are formed by the linkers that connect the M2 transmembrane segments to the S3 helices.
Stable association of COOH-terminal segments in all channel conformations suggests several possible roles for such an assembly. The first hypothesis is that in the resting conformation M2-S3 linkers and the S3 bundle itself serve as a “sheath” covering the S1 bundle. The latter is a sensitive element in the channel that functions as a cytoplasmic gate (Sukharev et al., 2001a). The S3 assembly, nicknamed as “hanging basket” or “gondola” as have been cytoplasmic domains of voltage-gated K+ channels (Sokolova et al., 2001), apparently remains stable when the channel opens. The 20–amino acid M2-S3 linkers are sufficiently long to allow for barrel opening without a separation of S3 domains. The “basket” then covers the inner entrance to the pore, forming a “sieve” that permits ion passage but prevents efflux and loss of essential metabolites under severe osmotic shock. The behavior of MscL without S3 domains is highly consistent with this representation. Indeed, the osmotic efflux of ATP from cells expressing the truncated channel is considerably higher than efflux from control WT and mscL knock-out cells (Fig. 9). It seems important that the “basket” rejects cytoplasmic molecules from entering the pore as they may get stuck in the channel when it attempts to close. Incomplete closure would mean permanent leakage and energy decoupling for the cell. Therefore, in the event of strong osmotic shock, MscL would ensure a fast exchange of only ions and small molecules. Besides the “prefilter” function, the stable association of COOH-terminal ends of all subunits may partially restrain the mobility of transmembrane helices (Betanzos et al., 2002), maintain the fivefold symmetry of the entire complex in the intermediate, and open states, thus making the gating transition more orderly.
In conclusion, the data support the results of molecular modeling and MD simulations of S3 bundle of EcoMscL that can be represented as a fivefold coiled-coil structure similar to COMP. It is stabilized by hydrophobic interactions between highly conserved pairs of aliphatic sidechains inside and by ionic pairs on the periphery of the bundle. The bundle remains stable in the open, closed, and likely intermediate conformations. The association of S3 domains brings the COOH termini of all five subunits together; this forms a “hanging basket” that may protect the NH2-terminal channel gate in the closed state, assist a more ordered transition, and apparently sets the size cut-off for the molecular permeation through the open channel.
Acknowledgments
The authors thank Mrs. Monica Betanzos for assistance in site-directed mutagenesis.
The major part of this work was supported by NASA (NAG2-1352) and the National Institutes of Health (NS39314-01) grants to S. Sukharev.
Olaf S. Andersen served as editor.
Footnotes
Abbreviations used in this paper: EcoMscL, E. coli MscL; MD, molecular dynamic.
References
- Ajouz, B., C. Berrier, A. Garrigues, M. Besnard, and A. Ghazi. 1998. Release of thioredoxin via the mechanosensitive channel MscL during osmotic downshock of Escherichia coli cells. J. Biol. Chem. 273:26670–26674. [DOI] [PubMed] [Google Scholar]
- Arkin, I.T., S.I. Sukharev, P. Blount, C. Kung, and A.T. Brunger. 1998. Helicity, membrane incorporation, orientation and thermal stability of the large conductance mechanosensitive ion channel from E. coli. Biochim. Biophys. Acta. 1369:131–140. [DOI] [PubMed] [Google Scholar]
- Betanzos, M., C.-S. Chiang, H.R. Guy, and S. Sukharev. 2002. A large iris-like expansion of a mechanosensitive channel protein induced by membrane tension. Nat. Struct. Biol. 9:704. [DOI] [PubMed] [Google Scholar]
- Bian, S., I. Favre, and E. Moczydlowski. 2001. Ca2+-binding activity of a COOH-terminal fragment of the Drosophila BK channel involved in Ca2+-dependent activation. Proc. Natl. Acad. Sci. USA. 98:4776–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount, P., S.I. Sukharev, P.C. Moe, M.J. Schroeder, H.R. Guy, and C. Kung. 1996. a. Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J. 15:4798–4805. [PMC free article] [PubMed] [Google Scholar]
- Blount, P., S.I. Sukharev, M.J. Schroeder, S.K. Nagle, and C. Kung. 1996. b. Single residue substitutions that change the gating properties of a mechanosensitive channel in Escherichia coli. Proc. Natl. Acad. Sci. USA. 93:11652–11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, G., R.H. Spencer, A.T. Lee, M.T. Barclay, and D.C. Rees. 1998. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science. 282:2220–2226. [DOI] [PubMed] [Google Scholar]
- Cortes, D.M., L.G. Cuello, and E. Perozo. 2001. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 117:165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank, C.C., R.F. Minchin, A.C. Le Dain, and B. Martinac. 1997. Estimation of the pore size of the large-conductance mechanosensitive ion channel of Escherichia coli. Biophys. J. 73:1925–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden, T., D. York, and L. Pedersen. 1993. Particle Mesh Ewald - an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- Elmore, D.E., and D.A. Dougherty. 2001. Molecular dynamics simulations of wild-type and mutant forms of the mycobacterium tuberculosis MscL channel. Biophys. J. 81:1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke, J.J., and D.E. Koshland, Jr. 1987. Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science. 237:1596–1600. [DOI] [PubMed] [Google Scholar]
- Feller, S.E., Y.H. Zhang, R.W. Pastor, and B.R. Brooks. 1995. Constant-pressure molecular-dynamics simulation - the Langevin piston method. J. Chem. Phys. 103:4613–4621. [Google Scholar]
- Fraczkiewicz, R., and W. Braun. 2002. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comp. Chem. 19:319–333. [Google Scholar]
- Gullingsrud, J., D. Kosztin, and K. Schulten. 2001. Structural determinants of MscL gating studied by molecular dynamics simulations. Biophys. J. 80:2074–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, H.R. 1985. Amino acid side-chain partition energies and distribution of residues in soluble proteins. Biophys. J. 47:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase, C.C., A.C. Le Dain, and B. Martinac. 1997. Molecular dissection of the large mechanosensitive ion channel (MscL) of E. coli: mutants with altered channel gating and pressure sensitivity. J. Membr. Biol. 157:17–25. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202. [DOI] [PubMed] [Google Scholar]
- Kajava, A.V. 1996. Modeling of a five-stranded coiled coil structure for the assembly domain of the cartilage oligomeric matrix protein. Proteins. 24:218–226. [DOI] [PubMed] [Google Scholar]
- Kale, L., R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan, and K. Schulten. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comp. Physics. 151:283–312. [Google Scholar]
- Kneller, D.G., F.E. Cohen, and R. Langridge. 1990. Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 214:171–182. [DOI] [PubMed] [Google Scholar]
- Kochendoerfer, G.G., J.M. Tack, and S. Cressman. 2002. Total chemical synthesis of a 27 kDa TASP protein derived from the MscL ion channel of M. tuberculosis by ketoxime-forming ligation. Bioconjug. Chem. 13:474–480. [DOI] [PubMed] [Google Scholar]
- Koprowski, P., and A. Kubalski. 1999. Glutathione (GSH) reduces the open probability of mechanosensitive channels in Escherichia coli protoplasts. Pflugers Arch. 438:361–364. [DOI] [PubMed] [Google Scholar]
- Kreusch, A., P.J. Pfaffinger, C.F. Stevens, and S. Choe. 1998. Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature. 392:945–948. [DOI] [PubMed] [Google Scholar]
- Kumar, S., and R. Nussinov. 1999. Salt bridge stability in monomeric proteins. J. Mol. Biol. 293:1241–1255. [DOI] [PubMed] [Google Scholar]
- Levina, N., S. Totemeyer, N.R. Stokes, P. Louis, M.A. Jones, and I.R. Booth. 1999. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 18:1730–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize, A.L., M.Y. Reibarkh, and I.D. Pogozheva. 2002. Interatomic potentials and solvation parameters from protein engineering data for buried residues. Protein Sci. 11:1984–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell, A.D., D. Bashford, M. Bellott, R.L. Dunbrack, J.D. Evanseck, M.J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- Malashkevich, V.N., R.A. Kammerer, V.P. Efimov, T. Schulthess, and J. Engel. 1996. The crystal structure of a five-stranded coiled coil in COMP: a prototype ion channel? Science. 274:761–765. [DOI] [PubMed] [Google Scholar]
- Martinac, B. 2001. Mechanosensitive channels in prokaryotes. Cell. Physiol. Biochem. 11:61–76. [DOI] [PubMed] [Google Scholar]
- Martinac, B., M. Buechner, A.H. Delcour, J. Adler, and C. Kung. 1987. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. USA. 84:2297–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulef, K., G.E. Flynn, and W.N. Zagotta. 1999. Molecular rearrangements in the ligand-binding domain of cyclic nucleotide-gated channels. Neuron. 24:443–452. [DOI] [PubMed] [Google Scholar]
- Maurer, J.A., D.E. Elmore, H.A. Lester, and D.A. Dougherty. 2000. Comparing and contrasting Escherichia coli and mycobacterium tuberculosis mechanosensitive channels (MscL). New gain of function mutations in the loop region. J. Biol. Chem. 275:22238–22244. [DOI] [PubMed] [Google Scholar]
- Moe, P.C., P. Blount, and C. Kung. 1998. Functional and structural conservation in the mechanosensitive channel MscL implicates elements crucial for mechanosensation. Mol. Microbiol. 28:583–592. [DOI] [PubMed] [Google Scholar]
- Moe, P.C., G. Levin, and P. Blount. 2000. Correlating a protein structure with function of a bacterial mechanosensitive channel. J. Biol. Chem. 275:31121–31127. [DOI] [PubMed] [Google Scholar]
- Nakahira, K., M.F. Matos, and J.S. Trimmer. 1998. Differential interaction of voltage-gated K+ channel beta-subunits with cytoskeleton is mediated by unique amino terminal domains. J. Mol. Neurosci. 11:199–208. [DOI] [PubMed] [Google Scholar]
- Ouali, M., and R.D. King. 2000. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 9:1162–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakula, A.A., and M.I. Simon. 1992. Determination of transmembrane protein structure by disulfide cross- linking: the Escherichia coli Tar receptor. Proc. Natl. Acad. Sci. USA. 89:4144–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, M., and L. Salkoff. 1997. A novel calcium-sensing domain in the BK channel. Biophys. J. 73:1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewing, S., J. Roeper, and O. Pongs. 1996. Kv beta 1 subunit binding specific for shaker-related potassium channel alpha subunits. Neuron. 16:455–463. [DOI] [PubMed] [Google Scholar]
- Shapovalov, G., R. Bass, D.C. Rees, and H.A. Lester. 2003. Open-state disulfide cross-linking between M. tuberculosis mechanosensitive channel subunits. Biophys. J. 84:1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng, S.L., C.A. Cukras, J. Harwood, and C.G. Nichols. 2000. Structural determinants of PIP(2) regulation of inward rectifier K(ATP) channels. J. Gen. Physiol. 116:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova, O., L. Kolmakova-Partensky, and N. Grigorieff. 2001. Three-dimensional structure of a voltage-gated potassium channel at 2.5 nm resolution. Structure. 9:215–220. [DOI] [PubMed] [Google Scholar]
- Spencer, R.H., G. Chang, and D.C. Rees. 1999. ‘Feeling the pressure’: structural insights into a gated mechanosensitive channel. Curr. Opin. Struct. Biol. 9:448–454. [DOI] [PubMed] [Google Scholar]
- Sukharev, S. 2002. Purification of the small mechanosensitive channel of E. coli (MscS): the subunit structure, conduction and gating characteristics in liposomes. Biophys. J. 83:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev, S., M. Betanzos, C.S. Chiang, and H.R. Guy. 2001. a. The gating mechanism of the large mechanosensitive channel MscL. Nature. 409:720–724. [DOI] [PubMed] [Google Scholar]
- Sukharev, S., S.R. Durell, and H.R. Guy. 2001. b. Structural models of the MscL gating mechanism. Biophys. J. 81:917–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev, S.I., P. Blount, B. Martinac, F.R. Blattner, and C. Kung. 1994. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature. 368:265–268. [DOI] [PubMed] [Google Scholar]
- Sukharev, S.I., B. Martinac, V.Y. Arshavsky, and C. Kung. 1993. Two types of mechanosensitive channels in the Escherichia coli cell envelope: solubilization and functional reconstitution. Biophys. J. 65:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev, S.I., M.J. Schroeder, and D.R. McCaslin. 1999. a. Stoichiometry of the large conductance bacterial mechanosensitive channel of E. coli. A biochemical study. J. Membr. Biol. 171:183–193. [DOI] [PubMed] [Google Scholar]
- Sukharev, S.I., W.J. Sigurdson, C. Kung, and F. Sachs. 1999. b. Energetic and spatial parameters for gating of the bacterial large conductance mechanosensitive channel, MscL. J. Gen. Physiol. 113:525–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop, N., H. Lee, R. Hu, and A.A. Neyfakh. 2001. Molecular sieve mechanism of selective release of cytoplasmic proteins by osmotically shocked Escherichia coli. J. Bacteriol. 183:2399–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesson, L., and D. Eisenberg. 1992. Atomic solvation parameters applied to molecular dynamics of proteins in solution. Protein Sci. 1:227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, K., A. Batiza, M. Schroeder, P. Blount, and C. Kung. 1999. Hydrophilicity of a single residue within MscL correlates with increased channel mechanosensitivity. Biophys. J. 77:1960–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]