Abstract
Electrical activity in nerve, skeletal muscle, and heart requires finely tuned activity of voltage-gated Na+ channels that open and then enter a nonconducting inactivated state upon depolarization. Inactivation occurs when the gate, the cytoplasmic loop linking domains III and IV of the α subunit, occludes the open pore. Subtle destabilization of inactivation by mutation is causally associated with diverse human disease. Here we show for the first time that the inactivation gate is a molecular complex consisting of the III-IV loop and the COOH terminus (C-T), which is necessary to stabilize the closed gate and minimize channel reopening. When this interaction is disrupted by mutation, inactivation is destabilized allowing a small, but important, fraction of channels to reopen, conduct inward current, and delay cellular repolarization. Thus, our results demonstrate for the first time that physiologically crucial stabilization of inactivation of the Na+ channel requires complex interactions of intracellular structures and indicate a novel structural role of the C-T domain in this process.
Keywords: inactivation, long QT syndrome, sodium channel, structure, heart
INTRODUCTION
Na+ channels open in response to membrane depolarization, allowing a rapid selective influx of Na+ that serves to further depolarize excitable cells and initiate multiple cellular signals (Catterall, 2000). Within milliseconds of opening, Na+ channels enter a nonconducting inactivated state as the inactivation gate, the cytoplasmic loop linking domains III and IV of the a subunit, occludes the open pore (Stuhmer et al., 1989; Patton et al., 1992; West et al., 1992b; McPhee et al., 1994, 1995, 1998; Kellenberger et al., 1996; Catterall, 2000). Channel inactivation is necessary to limit the duration of excitable cell depolarization, and disruption of inactivation by inherited mutations, which delays cellular repolarization, is associated with a diverse range of human diseases including myotonias (Yang et al., 1994), epilepsy and seizure disorders (Lossin et al., 2002; Kearney et al., 2001), autism (Weiss et al., 2003), and sudden cardiac death (Keating and Sanguinetti, 2001).
Extensive mutational, functional, and biochemical analysis of the III-IV loop has provided evidence for a key role of this cytoplasmic linker in inactivation and identified the hydrophobic cluster (IFM) in the linker as a key motif that coordinates hydrophobic interactions between the inactivation gate and its docking sites both on transmembrane segment 6 (S6) of domain IV as well as the intracellular loop linking segments S4 and S5 of domain IV (Stuhmer et al., 1989; Patton et al., 1992; West et al., 1992b; Eaholtz et al., 1998, 1999). (McPhee et al., 1994, 1995, 1998; Kellenberger et al., 1996.)
Disease-associated mutations that disrupt inactivation may result in an increase in experimentally detectable sustained tetrodotoxin (TTX)-sensitive current (Isus) (Clancy and Kass, 2002; Clancy et al., 2002), which, though only a fraction (<1%) of the peak current elicited in response to depolarization, is sufficient to account for disease phenotype: delay in repolarization (Clancy and Rudy, 1999; Nuyens et al., 2001; Clancy et al., 2002). The disruption of inactivation by mutation of the IFM motif resembles, to a certain extent, the effects of some disease-associated mutations on Na+ channel inactivation. Some mutations, such as the ΔKPQ deletion mutation (Bennett et al., 1995) occur in the III-IV linker region and promote inactivation-free gating modes (Bennett et al., 1995). Unexpectedly, however, mutations in other regions of the channel, notably the C-T domain, can also destabilize inactivation and promote an inactivation-deficient gating mode (Wei et al., 1999a; Baroudi and Chahine, 2000; Veldkamp et al., 2000; Clancy et al., 2002; Rivolta et al., 2002).
The fact that a large number of disease-associated mutations that alter inactivation have been found in the Na+ channel COOH-terminal domain has led us to question whether or not the COOH terminus may have a more direct structural role in the control of channel inactivation, and to ask how the C-T domain affects stabilization of the inactivated Na+ channel. In a previous theoretical and experimental study, we investigated the secondary structure of the C-T domain and found evidence that it is highly structured (Cormier et al., 2002). The NH2-terminal (proximal) half (∼100 residues) of the C-T domain was predicted to form six helices, whereas the distal half (∼100 residues) is unstructured, and circular dichroism measurements were consistent with this prediction. Truncation of the unstructured distal half did not affect channel gating, but truncation of one of the predicted six helices in addition to the unstructured region greatly destabilized inactivation (Cormier et al., 2002). Here we provide evidence for direct physical interaction between the C-T domain and the III-IV linker inactivation gate. We find that the full length C-T domain, as well as a truncated construct in which the distal unstructured region is deleted, bind to the III-IV linker, but that truncation of the predicted sixth helix uncouples the COOH terminus from the III-IV linker. These biochemical data are remarkably consistent with a role of the COOH-terminal/III-IV linker in stabilization of the inactivated state. Our data show for the first time that the inactivation gate of the voltage-dependent Na+ channel is a molecular complex that consists of the III-IV linker and the COOH-terminal domain of the channel. Uncoupling of this complex destabilizes inactivation and increases the likelihood of channel reopening during prolonged depolarization.
MATERIALS AND METHODS
Expression of Recombinant Na+ Channels and Electrophysiology
Na+ channels were expressed in human embryonic kidney (HEK) 293 cells at 22°C as previously described (Abriel et al., 2001). CD8-positive cells were identified using Dynabeads (Dynal, M-450) and were patch clamped 48 h after transfection. Membrane currents were measured using whole-cell patch-clamp procedures, with Axopatch 200B amplifiers (Axon Instruments, Inc.). Protocols and solutions for measurement of Na+ channel currents TTX-sensitive Isus are described in detail in a previous publication (Clancy et al., 2003).
Engineering of Fusion Proteins
The GST- and His-tagged SCN5A constructs were engineered using conventional PCR protocols. The PCR products were subcloned into the EcoRV site of the vector pBKS (Invitrogen) and after restriction digestion subcloned into either the pGEX (Amersham Biosciences) or pET 28 expression vectors (Novagen). The His-tagged COOH-terminal constructs of SCN5A were made as previously described (Cormier et al., 2002). The full-length C-T extends from residues 1773 through 2016. The mutants 1885Stop and 1921Stop refer to the site where a stop codon was placed in order to express a truncated C-T. All fusion constructs were verified by sequence analysis.
Mutations were introduced using the Quik Change Mutagenesis kit (Stratagene). Mutant clones for fusion proteins were selected by expression screening of small (2 ml) cultures inoculated with 0.1 ml of over grown culture. Proteins were visualized by Coomassie-stained gels using PAGE. Plasmid DNA of those clones that expressed protein of correct mass was purified (Promega), and the mutations verified by sequence analysis. Final constructs were transformed into DE3 cells (Stratagene).
Large-scale Fusion Protein Cultures
Fusion proteins were grown to large scale by first inoculating two 10-ml cultures of 2xyt media overnight for each 500-ml culture. These cultures were used to spike the large scale cultures and grown for 3–4 h at 37°C. IPTG was added (1-mM final concentration) once the culture reached an OD of 0.6 at 600 λ. The temperature was decreased to 32°C and the induction continued for 3–4 h. Some of the larger fusion proteins were similarly grown but induced at 16°C for 70 h. After induction the cultures were pelleted by centrifugation for 10 min at 3,000 rpm in a JA-10 (Beckman Coulter) rotor. The resultant pellets were resuspended in cold PBS (10 ml per 500 ml culture) supplemented with EDTA-free protease inhibitor tablets (Roche). The culture was divided into two 15-ml tubes and solubilized with Triton X-100 (1% final concentration). The samples were sonicated sequentially three times for 20 s and rocked overnight at 4°C. The bacterial lysate was obtained by centrifugation for 20 min at 10,000 RPM in a JA-13.1 swinging bucker rotor (Beckman Coulter). The supernatant was aliquoted and stored at −80°C.
GST Pull-down Assays
GST fusion protein lysates (500 μl) were incubated for 2 h with 50 μl of PBS-washed glutathione sepharose beads (Amersham Biosciences). Bound GST fusion proteins were washed with PBS and quantified by PAGE and titered by dilution with unbound glutathione sepharose beads accordingly. For each pull-down assay 5–25 μl of bound GST fusion protein was balanced with unbound glutathione sepharose to a final volume of 50 μl. His-tagged constructs were diluted fourfold to bring the Triton X-100 to 0.25% final concentration with a modified RIPA buffer (in mM: Tris-HCl, 50; NaCl, 150; EDTA, 1) made in the absence of Triton X-100. 250–500 μl of the diluted His-tagged construct was incubated with each GST fusion protein pull down. The reaction mix was balanced to 500 μl with RIPA buffer containing 0.25% Triton X-100 and incubated overnight at 4°C.
GST fusion proteins were washed 4–5 times with cold RIPA buffer, solubilized with 15 μl 5× sample buffer and electrophoresed on 4–20% gradient gels (Biorad). Proteins were transferred to nitrocellulose and detected using Penta-His monoclonal antibody (QIAGEN) and developed with the reagents supplied with the Immuno-Star HRP (Bio-Rad Laboratories) kit.
Pull-down assays were quantified by scanning the developed bands from the Western blots as an estimate of III-IV–C-T interaction and the Ponceau-stained bands as a measure for total loaded GST protein using the densitometric functions of the Gel Doc 2000 (Bio-Rad Laboratories). Pull-down assays were corrected for total loaded protein, normalized to the wild-type construct, and background subtracted (GST values) to determine the relative affinity of the constructs.
Theoretical Analysis of III-IV Loop Structure
We used local structure prediction methods that are useful in predicting local structure conformations from local sequence segments (Yang and Wang, 2002, 2003), and analyzed the sequence profile for SCN5A homologues that is found in the NCBI nr sequence database (Fig. 3 A). The sequences were selected with five PSI-BLAST iterations (default parameters) (Altschul et al., 1997) with e-value cutoff <10−2 using the SCN5A 1471–1523 as the seed (first row of the sequence profile in Fig. 3 A). Only representative sequences are shown in the figure; no sequence pair in the profile has >90% sequence identity. The multiple alignment was generated with the CLUSTALW program with default parameters (Thompson et al., 1994).
Figure 3.

Sequence profile and molecular model of the Na+ channel III-IV loop. (A) Sequence profile for the SCN5A homologues found in the NCBI nt sequence database. The sequences are selected with five PSI-BLAST iterations (default parameters) with e-value cutoff equal to 10−2 using the SCN5A 1471–1523 as the seed (first row of the sequence profile). Only representative sequences are shown in the figure; no sequence pair in the profile has >90% sequence identity. (B) Hypothetical molecular model of the Na+ channel III-IV loop (residues 1471–1523) generated by local structural predictions. Shown in the figure are structures predicted by modeling based on the III-IV loop linear sequence. Shown is the predicted helix-turn-helix structure illustrating the predicted locations of the IFM motif (highlighted in red), KPQ (highlighted in green), and a P-rich region (highlighted in orange) discussed in the text and linked to functional and structural integrity of the channel. Underlined residues in the linear sequence correspond to predicted α helices in the model.
Analysis of Experimental Data
Analysis for all experiments was in Excel (Microsoft) and Origin 6.1 (Microcal Software). Data are represented as mean ± SEM. Statistical significance was determined using unpaired Student's t test; P < 0.05 was considered statistically significant.
RESULTS
We previously provided evidence that the cardiac Na+ channel (Fig. 1, A and B) C-T domain consists of helical (proximal half) and nonstructured (distal half) regions (Fig. 1 B) (Cormier et al., 2002) and that the structured region plays a role in stabilizing inactivation. Fig. 1 C illustrates the functional effects of a three amino acid deletion mutation of the III-IV loop (ΔKPQ, left trace) associated with cardiac arrhythmia (Bennett et al., 1995), a C-T truncation that results in loss of the sixth helix and the unstructured region (1885stop, middle trace), and the combination of both mutations (ΔKPQ + 1885stop (K1885), right trace) on INa in response to depolarization (−10 mV) at low and high gain. Wild-type channel activity is shown in the figure for comparison. At low resolution (gain), each trace shows a large inward current (as Na+ channels activate) followed by rapid decay as channels inactivate. Closer examination (high gain, insets) reveals sustained current (Isus, arrow) that is affected both by mutation of the III-IV loop (ΔKPQ) and by 1885stop. The 1885stop C-T truncation (Cormier et al., 2002) results in a similar Isus amplitude as the ΔKPQ (Bennett et al., 1995) mutation, whereas truncation of the C-T distal to the structured region (1921stop) does not destabilize inactivation nor increase Isus (Cormier et al., 2002) (Fig. 1 D, bar graph). In addition, unlike the ΔKPQ + 1885stop combined mutation, which results in a 15–20-fold increase of Isus compared with wild type channels (K1885, Fig. 1 C, right, and D, bar graph), the 1921stop truncation mutation in combination with ΔKPQ does not further increase Isus (K1921, bar graph). These results demonstrate a synergistic effect on Isus, of the combined disruption of the sixth helix of the C-T and the deletion mutation in the III-IV linker. These data thus raise the possibility of a direct interaction between the III-IV linker and the C-T domain, which we then investigated.
Figure 1.
Mutation of the Na+ channel C-T domain and III-IV loop synergistically disrupts inactivation and increases Isus. (A) Membrane-spanning model of the SCN5A sodium channel indicating the COOH domain and IFM motif in the III-IV loop. (B) Expanded view indicates the six predicted α helices within the C-T and sites of the deletion mutants 1885stop and 1921stop. (C) Whole-cell currents are shown at low and high gain (insets, peak currents are off-scale) recorded in HEK 293 cells expressing SCN5A constructs. Shown are normalized and averaged TTX-sensitive current traces (materials and methods) in response to voltage steps (−10 mV, 150 ms) (left to right, wild-type (WT), (n = 4); ΔKPQ (n = 4); 1885stop (n = 6); and K1885 (n = 5); arrow indicates ISUS). Bars: 20 ms high and 50 ms low gain traces; 1% peak current, high gain traces. (D) Mean ± SEM Isus (calculated as the percentage of peak current) measured at 150 ms during depolarization. ** and ##, P < 0.001 vs. WT; *, NS vs. WT. Summarized are data for the following channels: wild-type (WT); 1921stop; ΔKPQ; 1885stop; K1921; K1885.
We tested for direct interactions between the full length, 1921stop (structural domain preserved) and 1885stop (truncation of the sixth helix of the structured domain) C-T domains (Fig. 2 A, schematics on left) and the III-IV linker using GST–pull-down assays (materials and methods) and found that the full-length C-T domain interacts with the wild-type III-IV loop (Fig. 2 A, top row, first lane). This interaction is not disrupted by truncation of the distal unstructured C-T domain (1921stop, Fig. 2 A, middle row, first lane), but is abolished if the last helical region is deleted (1885stop, Fig. 2 A, bottom row, first lane). Importantly, neither mutation of the inactivation latch (IFM/QQQ, Fig. 2 A, second lane from left), which has been shown by others to interrupt the interaction between the III-IV linker and its docking sites on DIV-S6 (McPhee et al., 1995, 1998), nor the ΔKPQ deletion mutation of the III-IV loop (Fig. 2 A, third lane from left) affects the interaction with the full-length or 1921stop C-T domain (Fig. 2 A, top and middle rows, respectively). The 1885stop truncation, however, completely abolished the interaction with any of the IIII-V linker constructs (Fig. 2 A, bottom row). Consistent with these results is the observation that the smallest pretermination translation products for the full-length and 1921 His–tagged C-T that are smaller in mass than 1885 are also not pulled down by the III-IV loop. These data (summarized in Fig. 2 B) confirm the electrophysiological results that suggest independence between previously determined docking sites for the inactivation gate (IFM motif) and its interaction with the C-T domain.
Figure 2.
The Na+ channel III-IV loop and C-T domain interact. Bacteria expressing His-tagged full-length (WT C-T) and truncated (1921stop, 1885stop) C-T variants (schematics in A, left) and GST-tagged III-IV loop variants (WT, ΔKPQ, IFM/QQQ) were lysed and used in pull-down assays to test for III-IV loop/C-T interactions. Pretermination translation products for the WT C-T and 1921stop construct were observed as smaller bands and detected by the His Western blot. (A) Shown is a representative experiment in which His-tagged C-T constructs (WT, 1921stop, and 1885stop, respectively; see materials and methods) are illustrated in the top, middle, and bottom panels were immobilized using GST-tagged III-IV loop variants (WT, IFM/QQQ, and ΔKPQ in the first second and third lanes, respectively) and detected using an anti-His monoclonal antibody. The extent of the III-IV linker used in this study ranged from D1471 through D1523. The mutations IFM and ΔKPQ refer to changes at residues 1485–1487 and 1505–1507, respectively. The control lane (GST alone, fourth lane) indicates specificity of the reactions. The last lane is a Western blot detecting the His-tagged C-T in the bacterial lysates. Note that the pretermination translation products for the WT C-T and 1921 smaller than 1885stop were also not pulled down by the III-IV linker. (B) Histogram of scanned pull downs (±SEM) normalized to total protein and corrected for background and total GST loaded (materials and methods). The number of experiments is indicated in the figure. Open bars correspond to interactions between His-tagged full length C-T construct and labeled III-IV linker variants. Filled bars correspond to wild-type III-IV loop interacting with 1921stop and 1885stop C-T constructs. **, NS vs. WT; *, P < 0.007 vs. WT.
We next used a theoretical analysis of the putative structure of the III-IV loop region to help identify possible sites that might interact with the C-T domain. Fig. 3 A shows the sequence profile for the SCN5A III-IV loop homologues found in the NCBI nr sequence database. Local structure predictive analysis (materials and methods) of the III-IV loop showed that two regions flanking the IFM motif (residues 1471–1480 and 1487–1501) are likely to form helical structures (Fig. 3 B). The predicted helix downstream of the IFM motif is consistent with the experimental observation that the residues between T1488 and K1499 form a stable helical structure in the isolated peptide chain (Rohl et al., 1999). The sequence region up-stream from the IFM motif has been predicted to form helical structures (Sirota et al., 2002), but the relative orientation between the two predicted helices has not been elucidated. Local structure predictions indicate that the two predicted helices are likely to pack to form a helix-turn-helix structure with the IFM motif situated at the turn region (Fig. 3 B, residues marked in red) (Yang and Wang, 2002).
Analysis of the sequence profile for SCN5A homologues found in the NCBI nr sequence database reveals two invariant residues in the sequence profile: T1488 and P1513 (materials and methods, and boxed areas in the sequences). T1488 is part of the conserved IFMT motif (Rohl et al., 1999), but the role of the invariant P1513 has not been identified previously. The local structure predictions suggest that the sequence near this proline-rich (P-rich) region (residues 1502–1523) is not likely to form regular secondary structure, suggesting that the prolines in this region are not likely to have roles as helix breakers. Nevertheless, the sequence profile analysis indicates that the known III-IV loop homologues have no tolerance for insertions or deletion in this region. These sequence-structure features and the propensity for a P-rich region to form a potential protein–protein interaction site (Kini and Evans, 1995) suggest that this region near the P-rich motif (KPQKPIPRP) might prove important in mediating interactions with the C-T domain.
To investigate this possibility, residues in the III-IV loop were mutated in groups of four to glutamates in order to map a putative interacting interface between the III-IV linker and the C-T domain (Fig. 4 A). This approach disrupts hydrophobic interactions while maintaining helical integrity because four consecutive charged residues on a helical structure will abolish hydrophobic interactions in close contact from any direction to the helix surface. These constructs eliminate the possibility that a helix could “roll” to bind to its partner to avoid burying a charged residue. Glutamic acid, instead of aspartate, is used to introduce real charges on the structure because the aspartic acid side chain is known to be a helix breaker, whereas the glutamic acid side chain has little effect on the intrinsic stability of an α-helix. Arginine and lysine have long aliphatic side chains and are thus less precise in placing real charges on the helix surface. Each mutant III-IV linker construct was expressed as a GST fusion protein, and assayed by the ability to interact with the His tagged full length C-T (materials and methods).
Figure 4.

Mutation of a P-rich region of the Na+ channel III-IV loop disrupts interactions with the C-T domain and destabilizes inactivation. (A) III-IV loop linear sequence. (B) Pull down of His-tagged full-length C-T fusion proteins and glutamic acid scan of the GST-tagged III-IV linker constructs in which groups of four residues were systematically mutated to glutamic acid and detected using anti-His. (C) Histogram (±SEM) of C-T pull downs by the mutant III-IV GST fusion proteins normalized as described in materials and methods. The number of experiments for each construct is indicated in parentheses. *, P < .001 vs. PIPR; #, NS vs. PIPR.
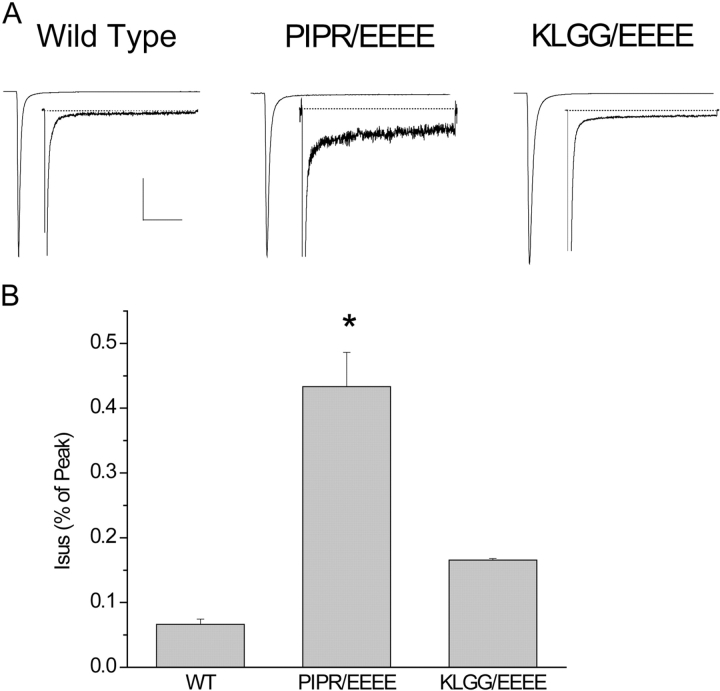
Most of the mutant linker constructs pulled down the C-T, confirming the physical interaction between the C-T and III-IV loop (Fig. 4 B). However, as the mutations approach the putatively unstructured and P-rich region, the intensities of the pull-down interactions decrease, suggesting that mutation of this region might be important in disruption of the interaction of the III-IV–C-T interaction. The pull-down data suggest that the P-rich region, identified by theoretical analysis as a region likely to have structural and/or functional importance (see above), may be particularly sensitive to disruption by mutation. To test if the biochemical results of the quadruple glutamate mutations in the pull-down assays also translated to functional sodium channels, we expressed similar mutations in the full length Na+ channel. Fig. 5 compares effects of the quadruple glutamate mutation of the PIPR residues with that of residues KLGG, which in pull-down assays were less disruptive than mutation of PIPR. Here TTX-sensitive sustained whole-cell current was measured in cells expressing wild-type (WT), PIPR/EEEE mutated, and KLGG/EEEE mutated channels. We find, in fact, that the PIPR/EEEE mutation, which ablates the physical interaction between the III-IV loop and the C-T domain, destabilizes inactivation and causes a significant increase in Isus (0.43 ± 0.05, n = 6 vs. 0.07 ± 0.001, P < 0.001, units in percentage of peak current) (Fig. 5 A). Replacement of KLGG residues (which are upstream of PIPR) by EEEE causes a significantly smaller change in sustained channel activity (0.166 ± 0.002, P < 0.001 vs. PIPR/EEEE channels), providing further evidence for a role of III-IV linker–CT domain interactions in stabilizing inactivation and suggesting that the region near the PIPR motif may be important to this interaction. Although the PIPR/EEEE mutation enhances sustained channel activity (Fig. 5), it affects neither the voltage dependence (Fig. 6 A) nor onset kinetics (Fig. 6 A) of channel inactivation; and flecainide block, which depends both on channels opening and inactivating (Liu et al., 2002, 2003), of PIPR/EEEE channels is use and frequency dependent (Fig. 6 B). In fact, flecainide use–dependent block of PIPR/EEEE channels is slightly greater than for wild-type channels, which may be due to mutation-induced repetitive channel openings during the test pulses.
Figure 5.
Preferential destabilization of inactivation by mutation of a P-rich region of the Na+ channel III-IV loop. (A) Whole-cell recordings of TTX-sensitive current in HEK 293 cells expressing wild-type (WT)-, PIPR/EEEE-, and KLGG/EEEE-mutated channels shown at low and high (insets, peak current off scale) gain. Shown are normalized (to peak current) and averaged (WT, n = 7; PIPR/EEEE, n = 6; KLGG/EEEE, n = 4) traces for each construct. Bars: 20 ms high and 50 ms low gain traces; 1% peak current, high gain traces. (B) The bar graph summarizes normalized mean (±SEM) Isus measured at 150 ms during voltage pulses to −10 mV and calculated as a percentage of peak current. *, P < 0.01 vs. WT and KLGG/EEEE.
Figure 6.
Kinetics, voltage dependence, and flecainide block of PIPR/EEEE channels. (A) The PIPR/EEEE mutation does not affect the kinetics of the onset of inactivation (left) or the voltage dependence of inactivation. Plotted in the left-hand graph are onset kinetics measured as the time to decay to half peak current (T1/2) vs. test pulse voltage for wild-type (WT) channels (n = 5) and PIPR/EEEE mutant channels (n = 4). There is no significant difference between these parameters at any voltage tested. Plotted in the right-hand graph are steady-state inactivation curves measured by preceding test pulses to −10 mV by a series of 500-ms conditioning pulses and normalizing measured test pulse current to that measured after the most negative (−140 mV) conditioning pulse. Normalized current is plotted against conditioning pulse voltage for WT (n = 6) and PIPR/EEEE channels (n = 8). Traces shown are normalized and averaged recordings of currents evoked by test pulses to −10 mV for WT (n = 5) and PIPR/EEEE (n = 4) channels. (B) Use-dependent block (UDB) of WT and PIPR/EEEE channels by flecainide (10 mM). Cells were held at −100 mV and trains of 50-test pulses (20 ms; −10 mV) were applied at pulse frequencies of 1 and 10 Hz. In each panel current traces are shown after the first (P1) and fiftieth (P50) pulse of the conditioning train in the presence of 10 mM flecainide. The bar graphs in the lower panel plot the percentage block ((P1 − P50)/P1)*100) recorded after applying trains at 1 Hz and 10 Hz for WT (n = 4) and PIPR/EEEE (n = 3) channels. *, P < 0.01.
The inactivation properties of PIPR/EEEE channels compared with wild-type channels suggest that the mutation does not cause marked structural changes in the III-IV linker that underlies voltage-dependent inactivation. Thus, we next focused more on the PIPR region in control of sustained current. As illustrated in Fig. 7, mutation of the P-rich region of the III-IV loop by glutamate substitution increases Isus, much as the C-T truncation (1885stop) or ΔKPQ mutation (compare data in Figs. 1 and 5). However, if we replace the PIPR residues by alanine or glutamine residues, the III-IV loop–C-T physical interactions are not disrupted (Fig. 7 A), and there is no significant effect on Isus, a marker of stabilization of inactivation (Fig. 7 B). These data suggest that the characteristics of the PIPR region, somewhat basic and predicted to be lacking in regular secondary structural elements, might be important in coordinating III-IV linker–C-T domain interactions and the stabilization of inactivation, but also raise the possibility that these effects are due to indirect effects of the quadruple glutamate mutation. To test for this, we compared the effects of single and double glutamate mutations of the PIPR region on both III-IV linker–C-T domain interactions (Fig. 8 A) and sustained channel activity (Fig. 8, B and C). We again found correlation between disruption of the protein–protein interaction as assayed by the pull-down experiments and disruption of inactivation as measured by sustained channel activity.
Figure 7.

Disruption of interactions in a P-rich region destabilizes inactivation. (A, top) Pull down of His-tagged full-length C-T fusion by GST-tagged III-IV constructs in which the PIPR region was mutated either to alanine, glutamic acid, or glutamine and detected using anti-His. The Western blot lane demonstrates the migration of the His tagged C-T. The Ponceau red stain of the identical membrane used in the pull-down assay is shown in the lower row. (B) Mutation of PIPR motif (PIPR/EEEE) increases Isus. Whole-cell recordings of TTX-sensitive current in HEK 293 cells expressing wild-type (WT)-, PIPR/EEEE-, PIPR/AAAA-, and PIPR/QQQQ-mutated channels shown at low and high (insets, peak current off scale) gain. Shown are normalized (to peak current) and averaged (WT, n = 7; PIPR/EEEE, n = 6; PIPR/AAAA, n = 4; PIPR/QQQQ, n = 4) traces for each construct. Bars: 20 ms high and 50 ms low gain traces; 1% peak current, high gain traces. The bar graph summarizes normalized mean (±SEM) Isus measured at 150 ms during voltage pulses to −10 mV and calculated as a percentage of peak current. *, P <0.01, vs. WT.
Figure 8.
Disruption of III-IV loop–C-T interactions and sustained current is sensitive to the number of glutamate mutations in the PIPR region. (A) Pull down of His-C-T by WT, PIPR/EEEE, PIPR/PEPE, and PIPR/PIPE by III-IV GST fusion proteins. The His–C-T lane is a Western blot reflecting the C-T detected with a monoclonal His antibody. The Ponceau red stain of the identical membrane used in the pull-down assay is shown in the lower row. (B) Mutation of PIPR motif (PIPR/EEEE) increases Isus. Whole-cell recordings of TTX-sensitive current in cells expressing wild-type (WT)-, PIPR/EEEE-, PIPR/PEPE-, and PIPR/PIPE-mutated channels shown at low and high (insets, peak current off scale) gain. Shown are normalized (to peak current) and averaged (WT, n = 7; PIPR/EEEE, n = 6; PIPR/PEPE, n = 4; PIPR/PIPE, n = 7) traces for each construct. Bars: 20 ms high and 50 ms low gain traces; 1% peak current, high gain traces. (C) The bar graph summarizes normalized mean (±SEM) Isus measured at 150 ms during voltage pulses to −10 mV and calculated as percentage peak current. *, P < 0.01 vs. WT.
DISCUSSION
A Novel Structural Role of the Na+ Channel C-T Domain
Our results indicate that the inactivation gate of the cardiac voltage-gated Na+ channel is a molecular complex consisting of the III-IV linker and the C-T domain of the channel. Using computational and experimental analysis, we have identified a novel P-rich motif of the III-IV linker, distinct from the previously defined IFM motif (West et al., 1992a) that appears to be in a region important in coordinating this interaction. Local structure analysis suggests that the P-rich motif is in an unstructured region of the III-IV loop (Yang and Wang, 2003); and replacement of the residues PIPR by glutamates disrupts the interaction between the C-T domain and the III-IV loop, increases sustained Na+ channel current, but does not affect the voltage dependence or kinetics of channel inactivation. These data suggest that the PIPR/EEEE mutation does not significantly alter the structure of the III-IV loop in a manner that would be reflected in altered inactivation properties, a suggestion that must be confirmed by direct structural measurements. Instead, because the PIPR/AAAA and PIPR/QQQQ mutations neither ablated physical interactions between the C-T domain and the III-IV loop nor increased Isus in functional recordings, our experimental data suggest that it is not the presence of these prolines that is crucial to the protein–protein interactions of these two intracellular segments of the channel, but perhaps the rather generally basic nature of this region, and the prediction that there are no predicted secondary structural elements. Consistent with this is the fact that the PIPR/AAAA mutation does not ablate the interaction between the III-IV loop and the C-T domain or destabilize inactivation because alanine residues, while stable in α helices and β sheets, would not likely confer structure on this region due to prolines both upstream and downstream of their location. Introduction of alanines would also not substantially change the generally basic nature of this region. The mutation of PIPR to glutamine residues also does not disrupt the III-IV–C-T interaction or inactivation. This result suggests that the effectiveness of the disruption of the putative interaction depends in part on the (chemical) nature of the residues substituted, as the major difference between glutamate and glutamine, which are both hydrophilic and polar, is the very acidic nature of glutamate. The effects of the single and double glutamate mutations of this region illustrated in Fig. 8 suggest that this is, in fact, the case. However, confirmation of these suggestions must await structural measurements of the complete III-IV loop. Finally, our data also indicate that the last predicted helical region of the C-T domain is critical to this interaction.
Interactions between the III-IV Linker and the C-T Domain Stabilize Inactivation
Because disruption of the III-IV linker–C-T interaction does not abolish inactivation, but instead increases the fraction of channels that can reopen during sustained depolarization, our results suggest that the role of the C-T in this complex is to stabilize the inactivation gate–occluded channel. The role of the C-T domain thus resembles a latch that is necessary to ensure that the inactivation gate, once docked via the IFM motif, remains closed. Disruption of the complex by mutation thus removes this physiologically key structural component of the channel and allows for “leakage” of Na+ into cells under conditions that normally exclusively prevent Na+ entry. This Na+ leak, though only a fraction of total Na+ entry during an action potential, can have severe physiological consequences.
The results of this study thus provide a new structural framework to begin asking how multiple disease-associated mutations of the heart Na+ channel C-T domain may act to destabilize inactivation and promote physiological dysfunction and how sequence differences in C-T regions of Na+ channel variants may contribute to differences in gating differences of the variants (Mantegazza et al., 2001). A review of the literature reveals multiple mutations reported in the cardiac C-T domain that destabilize inactivation. Disease associated mutations that have been reported to destabilize the inactivated state include those in the acid-rich region of the proximal half of the C-T (Bezzina et al., 1999; Wei et al., 1999b; Baroudi and Chahine, 2000; Veldkamp et al., 2000), as well as unstructured loops that are proposed to link putative helical structures in this region of the C-T domain (R1826H, Ackerman et al., 2001; L1825P, Makita et al., 2002). Our previous theoretical analysis suggested that the proximal half of the SCN5A C-T domain consists of four helical segments joined by unstructured regions that are likely to fold into two E-F hand pairs (Cormier et al., 2002). It is interesting to speculate whether or not these putative structures participate in and/or underlie interactions between the C-T domain and the III-IV linker either directly, or as our data suggest, via the distal half of the COOH terminus. Answers to these questions must await more detailed structural information for the C-T domain.
Acknowledgments
We thank Joseph W. Cormier and Colleen Clancy for helpful discussions; and Jenny Rao for technical assistance.
This work was supported by National Institutes of Health grants (R01 HL56810 and P01-HL67849) to R.S. Kass.
Lawrence G. Palmer served as editor.
Abbreviation used in this paper: C-T, COOH terminus.
References
- Abriel, H., C. Cabo, X.H. Wehrens, I. Rivolta, H.K. Motoike, M. Memmi, C. Napolitano, S.G. Priori, and R.S. Kass. 2001. Novel arrhythmogenic mechanism revealed by a long-qt syndrome mutation in the cardiac Na+ channel. Circ. Res. 88:740–745. [DOI] [PubMed] [Google Scholar]
- Ackerman, M.J., B.L. Siu, W.Q. Sturner, D.J. Tester, C.R. Valdivia, J.C. Makielski, and J.A. Towbin. 2001. Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. JAMA. 286:2264–2269. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., T.L. Madden, A.A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D.J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudi, G., and M. Chahine. 2000. Biophysical phenotypes of SCN5A mutations causing long QT and Brugada syndromes. FEBS Lett. 487:224–228. [DOI] [PubMed] [Google Scholar]
- Bennett, P.B., K. Yazawa, N. Makita, and A.L. George. 1995. Molecular mechanism for an iniherited cardiac arrhythmia. Nature. 376:683–685. [DOI] [PubMed] [Google Scholar]
- Bezzina, C., M.W. Veldkamp, M. P.van den Berg, A.V. Postma, M.B. Rook, J.W. Viersma, I.M. van Langen, G. Tan-Sindhunata, M.T. Bink-Boelkens, A.H. Der Hout, et al. 1999. A single Na+ channel mutation causing both long-QT and Brugada syndromes. Circ. Res. 85:1206–1213. [DOI] [PubMed] [Google Scholar]
- Catterall, W.A. 2000. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 26:13–25. [DOI] [PubMed] [Google Scholar]
- Clancy, C.E., and R.S. Kass. 2002. Defective cardiac ion channels: from mutations to clinical syndromes. J. Clin. Invest. 110:1075–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, C.E., and Y. Rudy. 1999. Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature. 400:566–569. [DOI] [PubMed] [Google Scholar]
- Clancy, C.E., M. Tateyama, and R.S. Kass. 2002. Insights into the molecular mechanisms of bradycardia-triggered arrhythmias in long QT-3 syndrome. J. Clin. Invest. 110:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, C.E., M. Tateyama, H. Liu, X.H. Wehrens, and R.S. Kass. 2003. Non-equilibrium gating in cardiac Na+ channels: an original mechanism of arrhythmia. Circulation. 107:2233–2237. [DOI] [PubMed] [Google Scholar]
- Cormier, J.W., I. Rivolta, M. Tateyama, A.S. Yang, and R.S. Kass. 2002. Secondary structure of the human cardiac Na+ channel C terminus. Evidence for a role of helical structures in modulatin of channel inactivation. J. Biol. Chem. 277:9233–9241. [DOI] [PubMed] [Google Scholar]
- Eaholtz, G., A. Colvin, D. Leonard, C. Taylor, and W.A. Catterall. 1999. Block of brain sodium channels by peptide mimetics of the isoleucine, phenylalanine, and methionine (IFM) motif from the inactivation gate. J. Gen. Physiol. 113:279–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaholtz, G., W.N. Zagotta, and W.A. Catterall. 1998. Kinetic analysis of block of open sodium channels by a peptide containing the isoleucine, phenylalanine, and methionine (IFM) motif from the inactivation gate. J. Gen. Physiol. 111:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney, J.A., N.W. Plummer, M.R. Smith, J. Kapur, T.R. Cummins, S.G. Waxman, A.L. Goldin, and M.H. Meisler. 2001. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioral abnormalities. Neuroscience. 102:307–317. [DOI] [PubMed] [Google Scholar]
- Keating, M.T., and M.C. Sanguinetti. 2001. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 104:569–580. [DOI] [PubMed] [Google Scholar]
- Kellenberger, S., T. Scheuer, and W.A. Catterall. 1996. Movement of the Na+ channel inactivation gate during inactivation. J. Biol. Chem. 271:30971–30979. [DOI] [PubMed] [Google Scholar]
- Kini, R.M., and H.J. Evans. 1995. A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem. Biophys. Res. Commun. 212:1115–1124. [DOI] [PubMed] [Google Scholar]
- Liu, H., J. Atkins, and R.S. Kass. 2003. Common molecular determinants of flecainide and lidocaine block of heart Na+ channels: evidence from experiments with neutral and quaternary flecainide analogues. J. Gen. Physiol. 121:199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., M. Tateyama, C.E. Clancy, H. Abriel, and R.S. Kass. 2002. Channel openings are necessary but not sufficient for use-dependent block of cardiac Na+ channels by flecainide: evidence from the analysis of disease-linked mutations. J. Gen. Physiol. 120:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossin, C., D.W. Wang, T.H. Rhodes, C.G. Vanoye, and A.L. George, Jr. 2002. Molecular basis of an inherited epilepsy. Neuron. 34:877–884. [DOI] [PubMed] [Google Scholar]
- Makita, N., M. Horie, T. Nakamura, T. Ai, K. Sasaki, H. Yokoi, M. Sakurai, I. Sakuma, H. Otani, H. Sawa, and A. Kitabatake. 2002. Drug-induced long-QT syndrome associated with a subclinical SCN5A mutation. Circulation. 106:1269–1274. [DOI] [PubMed] [Google Scholar]
- Mantegazza, M., F.H. Yu, W.A. Catterall, and T. Scheuer. 2001. Role of the C-terminal domain in inactivation of brain and cardiac sodium channels. Proc. Natl. Acad. Sci. USA. 98:15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee, J.C., D.S. Ragsdale, T. Scheuer, and W.A. Catterall. 1994. A mutation in segment IVS6 disrupts fast inactivation of sodium channels. Proc. Natl. Acad. Sci. USA. 91:12346–12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee, J.C., D.S. Ragsdale, T. Scheuer, and W.A. Catterall. 1995. A critical role for transmembrane segment IVS6 of the sodium channel alpha subunit in fast inactivation. J. Biol. Chem. 270:12025–12034. [DOI] [PubMed] [Google Scholar]
- McPhee, J.C., D.S. Ragsdale, T. Scheuer, and W.A. Catterall. 1998. A critical role for the S4-S5 intracellular loop in domain IV of the sodium channel alpha-subunit in fast inactivation. J. Biol. Chem. 273:1121–1129. [DOI] [PubMed] [Google Scholar]
- Nuyens, D., M. Stengl, S. Dugarmaa, T. Rossenbacker, V. Compernolle, Y. Rudy, J.F. Smits, W. Flameng, C.E. Clancy, L. Moons, et al. 2001. Abrupt rate accelerations or premature beats cause life-threatening arrhythmias in mice with long-QT3 syndrome. Nat. Med. 7:1021–1027. [DOI] [PubMed] [Google Scholar]
- Patton, D.E., J.W. West, W.A. Catterall, and A.L. Goldin. 1992. Amino acid residues required for fast Na+-channel inactivation: charge neutralizations and deletions in the III-IV linker. Proc. Natl. Acad. Sci. USA. 89:10905–10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta, I., C.E. Clancy, M. Tateyama, H. Liu, S.G. Priori, and R.S. Kass. 2002. A novel SCN5A mutation associated with long QT-3: altered inactivation kinetics and channel dysfunction. Physiol. Genomics. 10:191–197. [DOI] [PubMed] [Google Scholar]
- Rohl, C.A., F.A. Boeckman, C. Baker, T. Scheuer, W.A. Catterall, and R.E. Klevit. 1999. Solution structure of the sodium channel inactivation gate. Biochemistry. 38:855–861. [DOI] [PubMed] [Google Scholar]
- Sirota, F.L., P.G. Pascutti, and C. Anteneodo. 2002. Molecular modeling and dynamics of the sodium channel inactivation gate. Biophys. J. 82:1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhmer, W., F. Conti, H. Suzuki, X. Wang, M. Noda, N. Yahagi, H. Kubo, and S. Numa. 1989. Structural parts involved in activation and inactivation of the sodium channel. Nature. 339:597–603. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., D.G. Higgins, and T.J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldkamp, M.W., P.C. Viswanathan, C. Bezzina, A. Baartscheer, A.A. Wilde, and J.R. Balser. 2000. Two distinct congenital arrhythmias evoked by a multidysfunctional Na+ channel. Circ. Res. 86:E91–E97. [DOI] [PubMed] [Google Scholar]
- Wei, J., D.W. Wang, M. Alings, F. Fish, M. Wathen, D.M. Roden, and A.L. George, Jr. 1999. a. Congenital long-QT syndrome caused by a novel mutation in a conserved acidic domain of the cardiac Na+ channel. Circulation. 99:3165–3171. [DOI] [PubMed] [Google Scholar]
- Wei, J., D.W. Wang, M. Alings, F. Fish, M. Wathen, D.M. Roden, and A.L. George, Jr. 1999. b. Congenital long-QT syndrome caused by a novel mutation in a conserved acidic domain of the cardiac Na+ channel. Circulation. 99:3165–3171. [DOI] [PubMed] [Google Scholar]
- Weiss, L.A., A. Escayg, J.A. Kearney, M. Trudeau, B.T. MacDonald, M. Mori, J. Reichert, J.D. Buxbaum, and M.H. Meisler. 2003. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol. Psychiatry. 8:186–194. [DOI] [PubMed] [Google Scholar]
- West, J.W., R. Numann, B.J. Murphy, T. Scheuer, and W.A. Catterall. 1992. a. Phosphorylation of a conserved protein kinase C site is required for modulation of Na+ currents in transfected Chinese hamster ovary cells. Biophys. J. 62:31–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, J.W., D.E. Patton, T. Scheuer, Y. Wang, A.L. Goldin, and W.A. Catterall. 1992. b. A cluster of hydrophobic amino acid residues required for fast Na+ channel inactivation. Proc. Natl. Acad. Sci. USA. 89:10910–10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, A.S., and L.Y. Wang. 2002. Local structure-based sequence profile database for local and global protein structure predictions. Bioinformatics. 18:1650–1657. [DOI] [PubMed] [Google Scholar]
- Yang, A.S., and L.Y. Wang. 2003. Local structure prediction with local structure-based sequence profiles. Bioinformatics. 19:1267–1274. [DOI] [PubMed] [Google Scholar]
- Yang, N., S. Ji, M. Zhou, L.J. Ptacek, R.L. Barchi, R. Horn, and A.L. George, Jr. 1994. Sodium channel mutations in paramyotonia congenita exhibit similar biophysical phenotypes in vitro. Proc. Natl. Acad. Sci. USA. 91:12785–12789. [DOI] [PMC free article] [PubMed] [Google Scholar]