Abstract
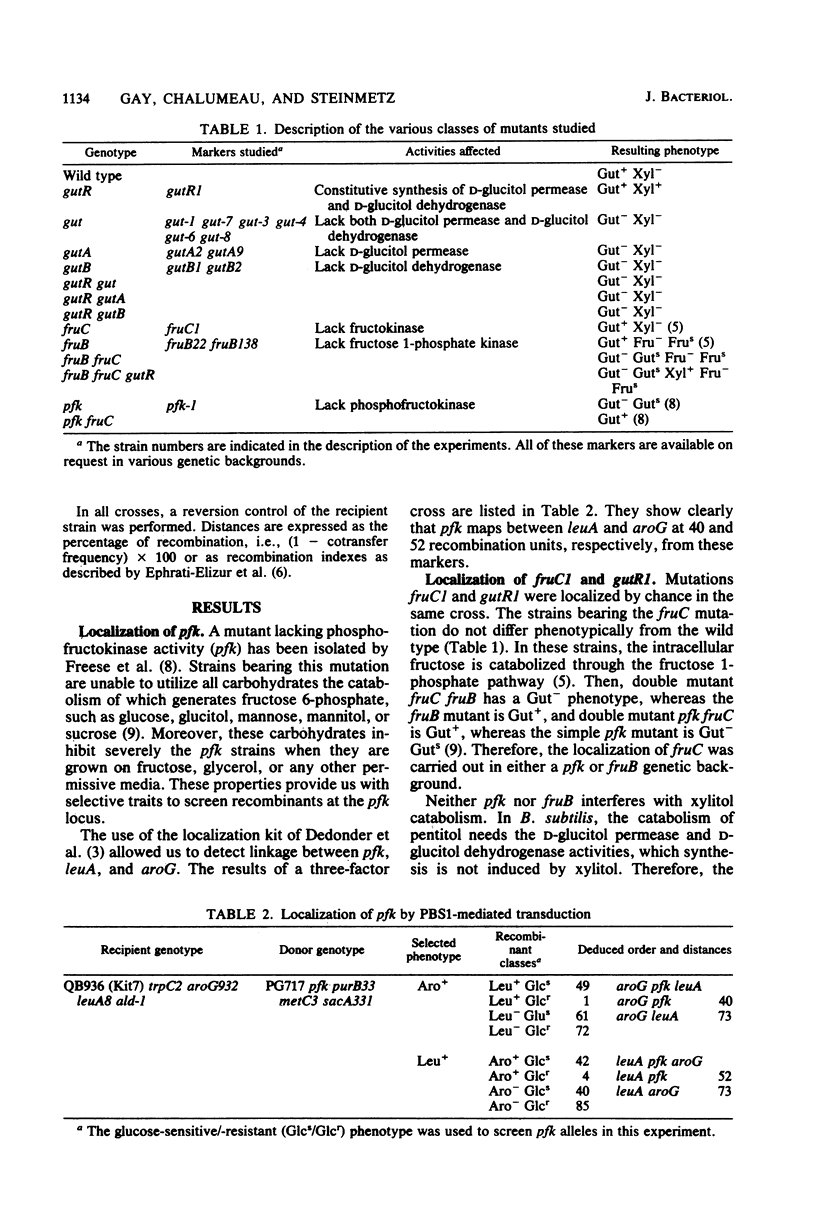
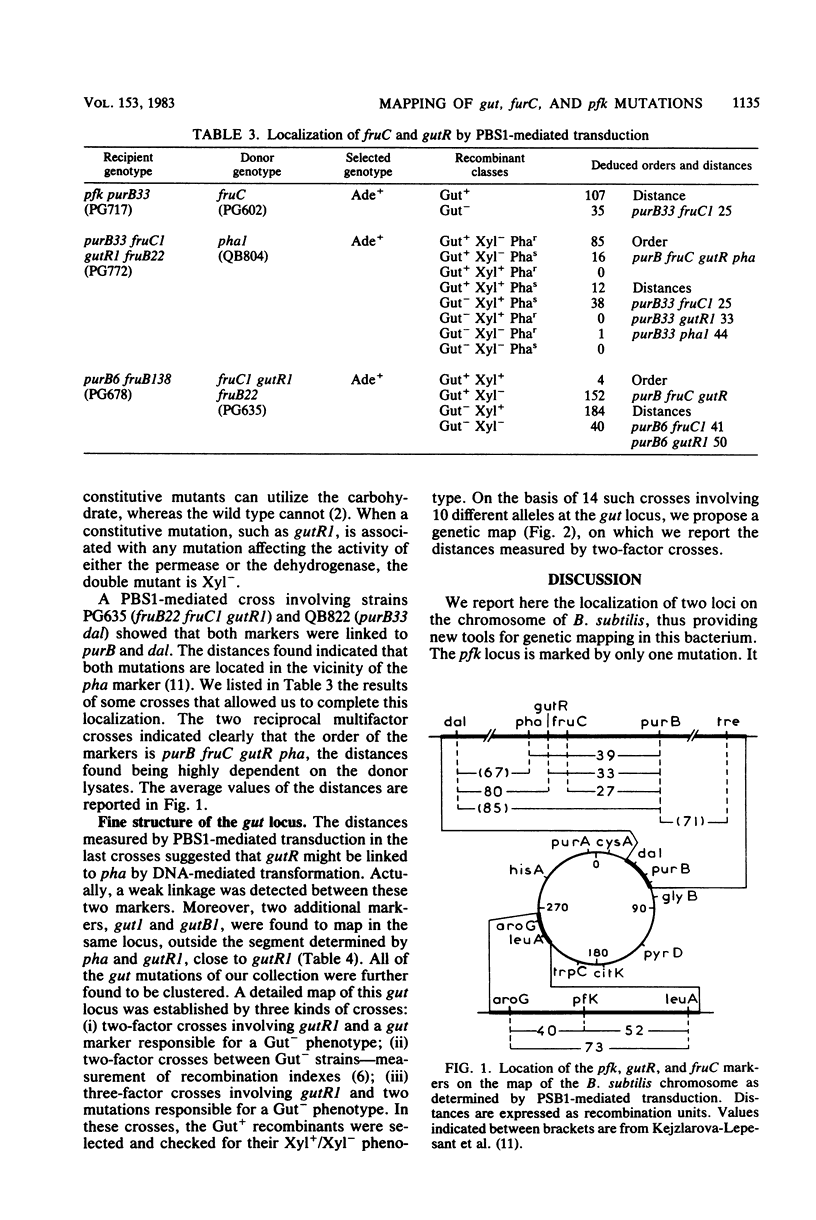
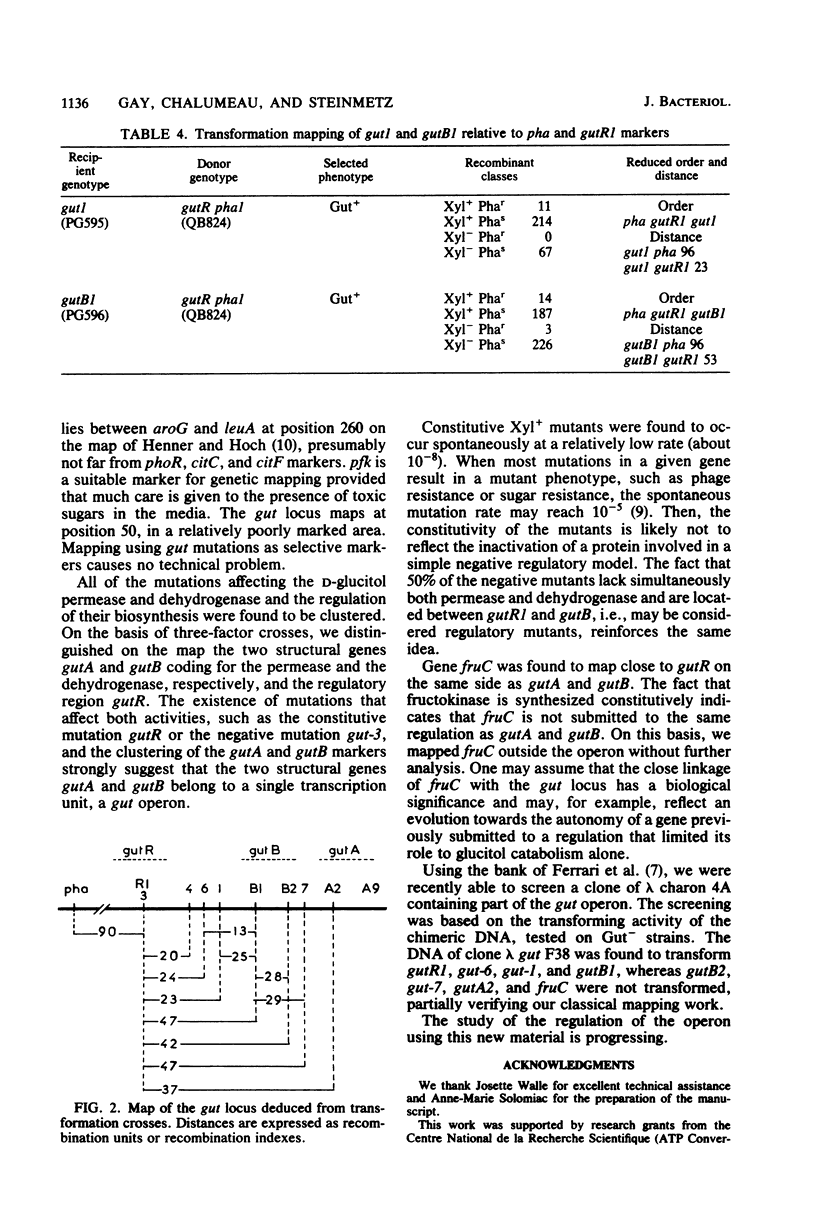
Mutations affecting the genes involved in B. subtilis D-glucitol catabolism were mapped either by PBS1-mediated transduction or DNA-mediated transformation. It was shown that the genes gutA and gutB coding for the D-glucitol permease and the D-glucitol dehydrogenase, respectively, and regulatory locus gutR are clustered in a gut operon localized between purB and dal close to the pha marker. A mutation affecting fructokinase activity (fruC) was mapped near the gut markers. The fruC gene does not belong to the operon. A mutation affecting phosphofructokinase activity (pfk) was mapped between the leuA and aroG markers.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chalumeau H., Delobbe A., Gay P. Biochemical and genetic study of D-glucitol transport and catabolism in Bacillus subtilis. J Bacteriol. 1978 Jun;134(3):920–928. doi: 10.1128/jb.134.3.920-928.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedonder R. A., Lepesant J. A., Lepesant-Kejzlarová J., Billault A., Steinmetz M., Kunst F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl Environ Microbiol. 1977 Apr;33(4):989–993. doi: 10.1128/aem.33.4.989-993.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delobbe A., Chalumeau H., Claverie J. M., Gay P. Phosphorylation of intracellular fructose in Bacillus subtilis mediated by phosphoenolpyruvate-1-fructose phosphotransferase. Eur J Biochem. 1976 Jul 15;66(3):485–491. doi: 10.1111/j.1432-1033.1976.tb10573.x. [DOI] [PubMed] [Google Scholar]
- Delobbe A., Chalumeau H., Gay P. Existence of two alternative pathways for fructose and sorbitol metabolism in Bacillus subtilis Marburg. Eur J Biochem. 1975 Feb 21;51(2):503–510. doi: 10.1111/j.1432-1033.1975.tb03950.x. [DOI] [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Hoch J. A. Isolation of Bacillus subtilis genes from a charon 4A library. J Bacteriol. 1981 Apr;146(1):430–432. doi: 10.1128/jb.146.1.430-432.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P., Delobbe A. Fructose transport in Bacillus subtilis. Eur J Biochem. 1977 Oct 3;79(2):363–373. doi: 10.1111/j.1432-1033.1977.tb11817.x. [DOI] [PubMed] [Google Scholar]
- Henner D. J., Hoch J. A. The Bacillus subtilis chromosome. Microbiol Rev. 1980 Mar;44(1):57–82. doi: 10.1128/mr.44.1.57-82.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesant-Kejzlarová J., Lepesant J. A., Walle J., Billault A., Dedonder R. Revision of the linkage map of Bacillus subtilis 168: indications for circularity of the chromosome. J Bacteriol. 1975 Mar;121(3):823–834. doi: 10.1128/jb.121.3.823-834.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesant J. A., Kunst F., Lepesant-Kejzlarová J., Dedonder R. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis Marburg. Mol Gen Genet. 1972;118(2):135–160. doi: 10.1007/BF00267084. [DOI] [PubMed] [Google Scholar]
- TAKAHASHI I. Transducing phages for Bacillus subtilis. J Gen Microbiol. 1963 May;31:211–217. doi: 10.1099/00221287-31-2-211. [DOI] [PubMed] [Google Scholar]


