In our paper “Coping with Viral Diversity in HIV Vaccine Design” [1], we presented several approaches to incorporate viral variability within vaccine immunogens, including judicious choice of natural strains. Most of our approaches included at least one collinear gene length corresponding to the Center-of-Tree (COT) sequence, which has near-optimal peptide coverage for a single gene. Inclusion of a COT sequence and optimizing the rest of the immunogen for coverage, as suggested in [2], yielded a construct (COT+) with the greatest coverage of peptide diversity, minimally sacrificing peptide coverage in comparison with unconstrained diversity optimization. Fischer et al. [3] introduced mosaics—a different approach to increasing coverage while maintaining collinearity using an optimization algorithm based on simulated recombination.
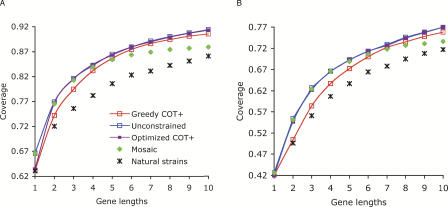
In their response to Nickle et al. [1], Fischer et al. [4] suggest that maintaining full collinearity of viral gene sequences with native viral proteins is the only tractable approach to producing immunogens inclusive of viral variability. This claim was based on the observation that mosaics had slightly higher coverage than COT+ at 3× and 4× strain lengths, despite the fact that all mosaic components are constrained to be collinear with the full gene. However, as we pointed out, a variety of optimization algorithms can be used to perform coverage optimization, with computationally intensive approaches typically yielding better results. Figure 1 compares the coverage of mosaics with COT+ constructs produced by two optimization algorithms—the simple greedy extension described in Nickle et al. [1], which can be implemented in hours and run in seconds on any modern personal computer, and the more complex combinatiorial optimization approach of [5] run for one day on a cluster of 300 PCs. We also include the coverage of a construct optimized without any collinearity constraints, derived using the Kirovski et al. [5] algorithm. The coverage of COT+ created by combinatorial optimization is greater than that of mosaics, especially at larger lengths where even the simple greedy algorithm surpasses the mosaic coverage. Furthermore, the optimized COT+ coverage is almost identical to the coverage of constructs optimized with no collinearity constraints, indicating that the price for imposing a constraint on the immunogen to include a single virus-like strain is small.
Figure 1. Comparison of Peptide Coverage Scores Achievable with Different Immunogen Formats and Algorithms.
In (A) and (B), we show the coverage for Gag and Nef, respectively, as a function of immunogen gene length for different immunogen formats. Natural strains: a cocktail of natural HIV sequences optimized for breadth of diversity by Gibbs sampling. Mosaics: a cocktail of constructs collinear with natural strains optimized by the code developed by Fischer et al. [3]. Greedy COT+: COT+ sequences were optimized by a simple greedy algorithm described by Jojic et al. [2,13] and Nickle et al. [1]. Optimized COT+: a stochastic combinatorial optimization of COT+ using the algorithm of Kirovski et al. [5], run on a 300 node Windows HPC cluster for one day. Unconstrained: a sequence optimized for coverage with the Kirovski algorithm without regard to gene collinearity. To emphasize that some of the constructs can be optimized at fractional lengths, those results are shown by lines (since points on the line are achievable), while the constructs consisting of an integer number of strains are shown as dots only.
Fischer and colleagues also argued that COT+ creates unnatural peptide sequences by concatenation. However, similar concatenation of their mosaics would have produced about 18 unnatural 9-mer peptides. Furthermore, the COT+ approach can be tuned to both penalize the introduction of unnatural peptides on concatenation, and to define the number of segments to be separately expressed, and thus reduce the requirement for concatenation.
Several additional inferences were made in the response by Fischer et al. that should be commented upon. First, COT+ may, of course, be optimized for arbitrary HIV clades or combinations of clades, but the publication of our paper in PLoS Computational Biology reflects our focus on approaches to immunogen design rather than on the production of an exhaustive series of constructs. Also, just as in the mosaic approach, COT+ can be optimized to exclude rare variants (referred to as smoothing in our paper).
Fischer et al. also discussed disappointing unpublished findings on the immunogenicity induced against Nef by a construct obtained by fusing a full-length Gag gene and the central portion of the Nef gene. However, these results can only be fairly assessed in light of what would be expected for the full-length Nef protein, and in the case of cellular immune responses, in the context of the same MHC specificities. However, these controls were not provided. We certainly agree that there are substantial challenges to the establishment of a multivalent CD8 response, yet multiple strategies have been and are being devised to overcome this important problem. For example, different groups have shown that CD8+ T cell responses can be successfully elicited against CD8+ T cell epitope strings when they are separated by short linker sequences and not in the context of the native protein, implying that they can be processed and presented in vivo [6–11].
Finally, despite 25 years of AIDS research and intensive yet uniformly failed efforts to develop an AIDS vaccine, the scientific community is poorly positioned to determine which, if any, approach to vaccine immunogen design will prove successful. Thus, arguing over methodologies developed with the same goal of incorporating variability has little significance as long as we do not know whether maximizing variability or inclusion of the entire full-length viral proteins are valid strategies. It may very well be that removing certain epitopes could be a more judicious approach than an overall epitope maximization strategy [12]. Indeed, the flexibility afforded by the COT+ approach, which is not limited to full-length proteins, may well prove superior to immunization with full-length viral protein immunogens.
Footnotes
David C. Nickle, Rosetta Inpharmatics, Seattle, Washington, United States of America
Morgane Rolland, James I. Mullins (jmullins@u.washington.edu), University of Washington School of Medicine, Seattle, Washington, United States of America
Nebojsa Jojic, David Heckerman, Darko Kirovski, Microsoft Research, Redmond, Washington, United States of America
Vladimir Jojic, Stanford University, Stanford, California, United States of America
Sergei Kosakovsky Pond, University of California San Diego, La Jolla, California, United States of America
Funding: The authors received no specific funding for this article.
Competing Interests: The authors have declared that no competing interests exist.
References
- Nickle DC, Rolland M, Jensen MA, Pond SL, Deng W, et al. Coping with viral diversity in HIV vaccine design. PLoS Comput Biol. 2007;3:e75. doi: 10.1371/journal.pcbi.0030075. doi: 10.1371/journal.pcbi.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jojic N, Jojic V, Frey B, Meek C, Heckerman D. Using epitomes to model genetic diversity: Rational design of HIV vaccine cocktails; Proceedings of the 19th Annual Conference on Neural Information Processing Systems; 5–10 December 2005;; Whistler, British Columbia, Canada: 2005. [Google Scholar]
- Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- Fischer W, Liao HX, Haynes BF, Letvin NL, Korber B. Coping with viral diversity in HIV vaccine design: A response to Nickle et al. PLoS Comput Biol. 2007;4:e15. doi: 10.1371/journal.pcbi.0040015. doi: 10.1371/journal.pcbi.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirovski D, Heckerman D, Jojic N. Combinatorics of the vaccine design problem: Definition and an algorithm. Microsoft Research Technical Report MSR-TR-2007–2148. 2007.
- Dorrell L. Therapeutic immunization for the control of HIV-1: Where are we now? Int J STD AIDS. 2006;17:436–441. doi: 10.1258/095646206777689035. [DOI] [PubMed] [Google Scholar]
- Goonetilleke N, Moore S, Dally L, Winstone N, Cebere I, et al. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J Virol. 2006;80:4717–4728. doi: 10.1128/JVI.80.10.4717-4728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbramanian RA, Kuroda MJ, Charini WA, Barouch DH, Costantino C, et al. Magnitude and diversity of cytotoxic-T-lymphocyte responses elicited by multiepitope DNA vaccination in rhesus monkeys. J Virol. 2003;77:10113–10118. doi: 10.1128/JVI.77.18.10113-10118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbramanian RA, Charini WA, Kuroda MJ, Seaman M, Chhay H, et al. Expansion after epitope peptide exposure in vitro predicts cytotoxic T lymphocyte epitope dominance hierarchy in lymphocytes of vaccinated mamu-a*01+ rhesus monkeys. AIDS Res Hum Retroviruses. 2006;22:445–452. doi: 10.1089/aid.2006.22.445. [DOI] [PubMed] [Google Scholar]
- De Groot AS, Marcon L, Bishop EA, Rivera D, Kutzler M, et al. HIV vaccine development by computer assisted design: The GAIA vaccine. Vaccine. 2005;23:2136–2148. doi: 10.1016/j.vaccine.2005.01.097. [DOI] [PubMed] [Google Scholar]
- Thomson SA, Jaramillo AB, Shoobridge M, Dunstan KJ, Everett B, et al. Development of a synthetic consensus sequence scrambled antigen HIV-1 vaccine designed for global use. Vaccine. 2005;23:4647–4657. doi: 10.1016/j.vaccine.2005.04.045. [DOI] [PubMed] [Google Scholar]
- Rolland M, Nickle DC, Mullins JI. HIV-1 group M conserved elements vaccine. PLoS Pathogens. 2007;3:e157. doi: 10.1371/journal.ppat.0030157. doi: 10.1371/journal.ppat.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jojic N, Frey B, Kannan A. Epitomic analysis of appearance and shape. Washington (D.C.): IEEE Computer Society; 2003. pp. 34–43. [Google Scholar]