Abstract
With the advent of engineered minichromosome technology in plants, an understanding of the properties of small chromosomes is desirable. Twenty-two minichromosomes of related origin but varying in size are described that provide a unique resource to study such behavior. Fourteen minichromosomes from this set could pair with each other in meiotic prophase at frequencies between 25 and 100%, but for the smaller chromosomes, the sister chromatids precociously separated in anaphase I. The other eight minichromosomes did not pair with themselves, and the sister chromatids divided equationally at meiosis I. In plants containing one minichromosome, the sister chromatids also separated at meiosis I. In anaphase II, the minichromosomes progressed to one pole or the other. The maize (Zea mays) Shugoshin protein, which has been hypothesized to protect centromere cohesion in meiosis I, is still present at anaphase I on minichromosomes that divide equationally. Also, there were no differences in the level of phosphorylation of Ser-10 of histone H3, a correlate of cohesion, in the minichromosomes in which sister chromatids separated during anaphase I compared with the normal chromosomes. These analyses suggest that meiotic centromeric cohesion is compromised in minichromosomes depending on their size and cannot be maintained by the mechanisms used by normal-sized chromosomes.
INTRODUCTION
Plant artificial chromosomes or engineered minichromosomes represent a potentially powerful research tool for understanding chromosome structure and function. Furthermore, they provide a means to assemble a collection of useful genes as an independent chromosome vector. Mammalian and Drosophila minichromosome analyses have been conducted (Heller et al., 1996; Harrington et al., 1997; Sun et al., 1997; Mills et al., 1999; Shen et al., 1999; Ebersole et al., 2000; Yang et al., 2000; Auriche et al., 2001). In mammals, minichromosomes can be assembled from constituent components or by removing the chromosome arms by telomere truncation, with subsequent or simultaneous addition of sites to accept additional DNA to the chromosome. The latter type of minichromosome provides a means to evaluate the loss of chromosome arms on mitotic and meiotic behavior. Engineered minichromosomes have been generated recently in plants (Yu et al., 2007), so a detailed understanding of the mitotic and meiotic properties of small chromosomes is integral to their eventual application.
The recovery of minichromosomes derived from the maize (Zea mays) B chromosome has been described (Zheng et al., 1999; Kato et al., 2005). This collection as well as subsequently recovered cases (this report) provides a unique resource for examining the effect of chromosome size on various meiotic processes that is not available in any other system. B chromosomes are nonessential, supernumerary chromosomes that are found in hundreds of plant and animal species (Jones and Rees, 1982).
The maize B chromosomes are maintained in populations because of their high frequency of nondisjunction at the second pollen mitosis, together with the fact that the sperm with two B chromosomes will preferentially fertilize the egg, rather than the polar nuclei, in the process of double fertilization (Roman, 1948; Carlson, 1969). B chromosomes are generally thought to be basically inert, in that no major genes have been identified on them and because they have no phenotypic effects on the plant, except in high numbers (Carlson, 1986).
Maize minichromosomes were originally generated by the breakage-fusion-bridge (BFB) cycle (McClintock, 1939, 1941) that was initiated in a modified translocation between the B chromosome and the short arm of chromosome 9 (9S) (Zheng et al., 1999). This translocation chromosome carries a mirror-image duplication of 9S that creates a dicentric condition upon intrachromosomal recombination in meiosis. The dicentrics form chromosome bridges that are broken in anaphase II. If the broken B-9 translocation chromosome undergoes nondisjunction at the second pollen mitosis, two broken chromosomes in the sperm are delivered to the zygote, the condition to establish the chromosome-type BFB cycle. During development, the B-9 chromosome continues to be reduced in size. Indeed, when the resulting plants were examined at pachynema, approximately one-third of the plants possessed minichromosomes (Zheng et al., 1999). The minichromosome structures and their transmission rates over several generations were determined (Kato et al., 2005).
Sister chromatid cohesion is important in meiosis I to hold the chromatids together for the faithful segregation of homologous pairs of chromosomes, but then centromere cohesion is released in meiosis II to allow sister separation. Studies primarily from yeast species have suggested that the enzyme separase releases sister chromatid cohesion at anaphase I, but the Shugoshin (SGO) protein protects the centromere so that cohesion is maintained until anaphase II (Nasmyth et al., 2000; Nasmyth, 2001; Kitajima et al., 2004; Watanabe, 2005a, 2005b). The multisubunit cohesin complex in meiosis differs from the one used in mitosis in that Rad21 is largely replaced by its meiotic counterpart, Rec8 (Nasmyth, 2001; Watanabe, 2005a, 2005b). The absence of first division (afd1) gene is the maize Rec8 homolog (Golubovskaya et al., 2006). The SGO protein has been suggested to play a major role in the protection of centromere cohesion in Drosophila and yeast (Kerrebrock et al., 1995; Kitajima et al., 2004; Marston et al., 2004; Vaur et al., 2005). Hamant et al. (2005) found that maize SGO1 is required for the maintenance of centromeric cohesion during meiosis and has no mitotic function. Normally, SGO is present in meiosis I but is degraded during meiosis II. However, if the sister chromatids separate at meiosis I, as in the afd1 mutant, SGO signals are not detected (Hamant et al., 2005).
Kaszas and Cande (2000) found that phosphorylation of histone H3 Ser-10 was correlated with the maintenance of sister chromatid cohesion. Phosphorylation of H3 is typically detected in pericentromeric regions of the mitotic chromosome and in meiosis (Zhang et al., 2005). In the afd1 mutant, phosphorylation is found only at pericentromeric regions in metaphase I. In the absence of sister association in meiosis II, no phosphorylation is observed in the pericentromeric regions. Kurihara et al. (2006) found that inhibition of Aurora kinase activity was related to diminished phosphorylation of Ser-10 of histone H3 in tobacco (Nicotiana tabacum) somatic cells and caused the delay and failure of sister chromatid separation. These results led to the hypothesis that SGO1 and Ser-10 phosphorylation are involved in the maintenance of sister centromere and chromatid cohesion.
In this study, we used a collection of minichromosomes to establish the impact of chromosome size on homolog pairing as well as the centromere cohesion properties of sister chromatids. The results indicate the interesting finding that chromosome size affects the properties of monopolar versus bipolar attachment of microtubules at meiosis I. Moreover, in the absence of substantial chromosome arm length, centromere cohesion at meiosis I fails despite the presence of SGO, which persists on the minichromosomes throughout meiosis I despite equational minichromosome behavior.
RESULTS
Formation of Minichromosomes
The minichromosome collection was generated in plants that were undergoing the chromosome type of BFB cycle. This process was initiated in the study of Zheng et al. (1999) using plants that contained one B-9-Dp9 chromosome (TB-9Sb-Dp9) together with two 9-B chromosomes. The B-9-Dp9 chromosome is a translocation between the supernumerary B chromosome and the short arm of chromosome 9 onto which a reverse duplication of 9S was recombined (Zheng et al., 1999) (Figures 1A and 1B). Meiotic analysis indicated that the progenitor B-9-Dp9 chromosome behaves normally at meiosis I, progressing to one pole (Figure 1C). When recombination occurs between the duplicated 9S regions, a dicentric chromosome is generated that will be broken at anaphase II (Figure 1D). Both centromeres of the dicentric chromosome are functional, which will cause the chromosome to be broken and rejoined repeatedly during the formation of the gametophyte and, following fertilization, during plant development. Minichromosomes will be stabilized if one of the two centromeres becomes inactive or a monocentric is formed by telomere addition to a broken end (Figure 2; see Supplemental Figure 1 online).
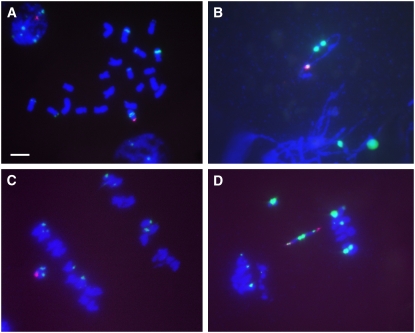
Figure 1.
Cytological Analysis of B-9-Dp-9 (TB-9Sb-Dp9), the Progenitor Translocation between the B Chromosome and Chromosome Arm 9S with a Reverse Duplication.
In all images, the B-specific sequence (ZmBs), is labeled in magenta and the 180-bp knob repeat is labeled in green.
(A) Root tip metaphase chromosomes. Bar = 10 μm.
(B) Pachynema.
(C) Anaphase I. Note the maintenance of sister chromatid cohesion in the B-9-Dp9 chromosome, the progenitor of the minichromosome collection.
(D) Anaphase II, showing the fragment and dicentric bridge formed as a result of recombination within the reverse duplication. The sister cell is not shown and the fragment is acentric.
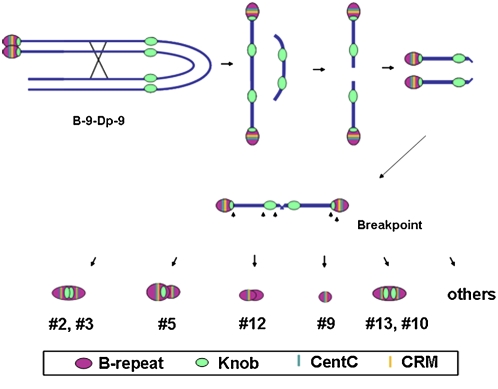
Figure 2.
Minichromosomes Produced by the Chromosome-Type BFB Cycle.
The B-9-Dp9 chromosome initiates the BFB cycle by crossing over with itself and releasing a terminal fragment; the broken ends fuse and continue the BFB cycle. With nondisjunction at the second pollen mitosis, two broken chromosomes can enter the zygote and initiate the chromosome type of BFB cycle that continues throughout plant development. Eventually, dicentric minichromosomes are stabilized by the inactivation of one centromere or the addition of a telomere to a broken chromosome end. Structural models of the smaller minichromosomes showing the distribution of ZmBs, Knob, CentC, and CRM sequences are depicted below.
The structure of the collection of minichromosomes was examined by fluorescence in situ hybridization (FISH) using probes for the B-specific sequence (Alfenito and Birchler, 1993) and the knob unit repeat (Peacock et al., 1981). In addition, they were probed with two maize centromere repeat units, CentC, and the retrotransposon, CRM (Jin et al., 2005). The normal B chromosome has centromere signals with the knob heterochromatin signal adjacent to it on the long arm (Lamb et al., 2005). All of the minichromosomes visualized by FISH contained the B chromosome centromere-specific sequences (ZmBs), indicating that the minichromosomes carry the B centromere. The structures of chromosomes 1 to 14 have been examined previously (Kato et al., 2004). The recovery of minichromosomes 15 to 22 is reported here (see Supplemental Figure 1 and Supplemental Table 1 online).
Meiotic Examination of Plants Containing One Minichromosome
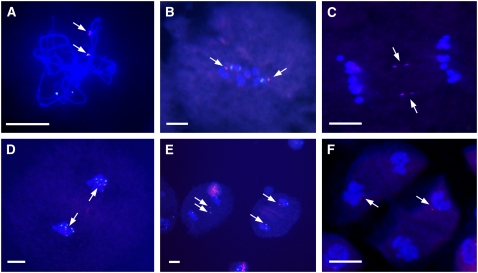
Tassel samples were collected from the various lines grown in the greenhouse to examine the meiotic behavior of a single copy of each member of the minichromosome collection. None of the small or tiny minichromosomes paired with chromosome 9 or the B chromosome at the pachytene stage (Figure 3A; see Supplemental Table 2 online). Eight minichromosomes (1, 4, 6, 11, 15, 16, 19, and 22) showed behavior typical of a B chromosome univalent and the progenitor chromosome (Figure 1C), which randomly moves to one pole during meiosis I, and then the sister chromatids separate at anaphase II. In contrast with the progenitor, the remaining 14 minichromosomes showed a different behavior, in that the sister chromatids separate at anaphase I. For example, minichromosome 3 was observed in all metaphase I cells (Figure 3B). In anaphase I, it usually lagged but divided equationally (Figure 3C). In telophase I, all of the examined cells contained one B repeat signal, indicating that despite lagging during meiosis I, the chromosome moved to the poles (Figure 3D). At meiosis II, the mini-Bs were observed in all of the metaphase II cells. In anaphase II, the sister chromatids of a given minichromosome progressed to one pole (Figure 3E). In tetrads at the end of meiosis, most contained B-specific signals (Figure 3F).
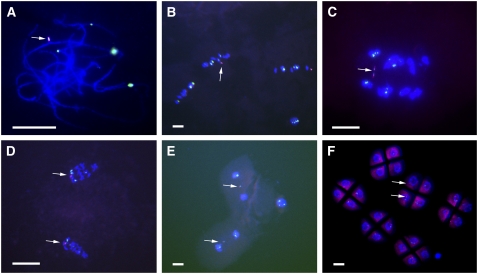
Figure 3.
Cytological Analysis of One Copy of Minichromosome 3.
The ZmBs is labeled in magenta, and the knob is labeled in green.
(A) Pachynema.
(B) Metaphase I.
(C) Anaphase I.
(D) Telophase I.
(E) Telophase II.
(F) Tetrads.
Note sister chromatid separation in anaphase I. The minichromosome lags in meiosis II but typically is incorporated into two of the four microspore nuclei in the tetrad. Note that minichromosome 3 has two B-specific repeat arrays, which will both label with the ZmBs probe. Arrows indicate the minichromosomes. Bars = 20 μm.
We also examined the α-tubulin localization for a minichromosome that exhibited equational division at meiosis I. Figure 4 shows that univalent minichromosome 9 forms a bioriented spindle, as typically occurs only in mitosis and meiosis II. These results confirm the mechanical basis of the equational division of this minichromosome at meiosis I.
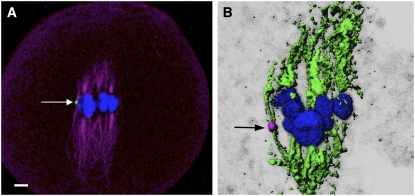
Figure 4.
α-Tubulin Immunolocalization on Minichromosome 9.
Minichromosome 9 shows a biorientation; the two sister chromatids will move to opposite poles in meiosis I.
(A) Green indicates the B repeat; blue indicates 4′,6-diamidino-2-phenylindole (DAPI); magenta indicates α-tubulin. The arrow indicates the minichromosome. Bar = 10 μm.
(B) Magenta indicates the ZmBs; green indicates α-tubulin; blue indicates DAPI. The arrow indicates the minichromosome.
Effect of Chromosome Size on Homolog Pairing
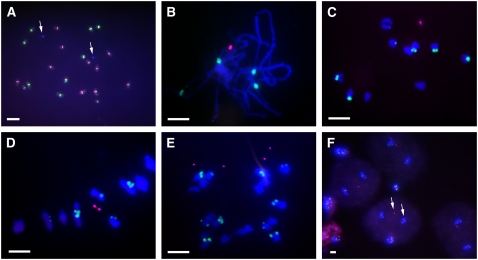
The larger minichromosomes tended to pair with each other when two copies were present in the same cell (see Supplemental Table 3 online). Fourteen of the minichromosomes further reduced in size were found to pair with each other when two copies were present during meiosis I. The frequency varied from 25 to 100% (see Supplemental Table 3 online). The other small or tiny minichromosomes did not pair at the pachytene stage (Figure 5A). For example, two minichromosomes 3 were observed in all of the examined metaphase I cells (Figure 5B) as univalents. In anaphase I, the two minichromosomes usually lagged but divided equationally (Figure 5C). In telophase I, all of the cells contained two B repeat signals (typical of minichromosome 3), indicating that the minichromosome eventually progressed to the poles (Figure 5D). In meiosis II, the minichromosomes were observed in anaphase II cells and the minichromosomes progressed to one or the other pole (Figures 5E and 5F).
Figure 5.
Cytological Analysis of Two Copies of Minichromosome 3.
ZmBs is labeled in magenta; the knob is labeled in green.
(A) Pachynema.
(B) Metaphase I.
(C) Anaphase I.
(D) Telophase I.
(E) Telophase II.
(F) Tetrads.
Note the absence of homolog pairing in pachynema and the separation of sister chromatids at anaphase I. Arrows indicate minichromosomes. Bars = 10 μm.
It is interesting that the very small minichromosome 9 can pair when two copies are present per cell (Figures 6A to 6C). The two minichromosomes 9 were observed in all of the examined metaphase I cells (Figure 6D) as bivalents. In anaphase I, the two minichromosomes usually lagged but divided equationally (Figure 6E). In telophase I, the minichromosomes eventually progressed to the poles (Figure 6F). By way of comparison, the minichromosome 3 described above is larger than minichromosome 9, but the homologs do not pair. Other minichromosomes, such as 18, are larger than minichromosome 9 but otherwise are similar in structure and also could not form bivalents (see Supplemental Figure 2 online). In parallel with minichromosome 9, sister chromatid separation of minichromosome 18 at meiosis I is equational.
Figure 6.
Cytological Analysis of Two Copies of Minichromosome 9.
(A) Somatic chromosomes of a plant with two minichromosomes 9 revealed by ZmBs (blue), CentC (green), and CRM (magenta).
(B) Pachytene stage showing two minichromosomes 9 paired with each other (magenta).
(C) Diakinesis.
(D) Metaphase I.
(E) Anaphase I. Sister chromatids have separated and progressed to the poles.
(F) Anaphase II. Note the presence of homolog pairing but the separation of sister chromatids at meiosis I.
Arrows indicate the minichromosomes. Bars = 10 μm.
Nondisjunction of Minichromosomes Is Lost but Can Be Restored by the Presence of Normal B Chromosomes in the Cell
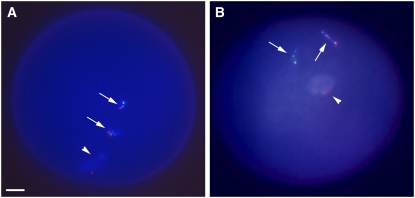
We performed pollen FISH analysis to examine the somatic stability and nondisjunction properties of the minichromosomes. FISH on maize pollen carrying one normal B chromosome shows nondisjunction at the second pollen mitosis, such that one sperm has two B chromosomes (as a united signal) and the other has none (Han et al., 2007). This nondisjunction, as noted above, is part of the accumulation mechanism of the B chromosome and requires the tip of the B chromosome long arm to be present for its action. Pollen FISH patterns of the 22 minichromosome lines showed ZmBs signal in both the two-sperm nuclei and the vegetative nucleus, indicating that nondisjunction had not occurred at the second pollen mitosis (Figure 7; see Supplementary Table 4 online). This result is expected, given the need for the tip of the B chromosome long arm to be present for nondisjunction (Carlson, 1986). When normal B chromosomes were combined with each minichromosome, all minichromosomes recovered the nondisjunction function, because the B long arm tip was now present again in the cell.
Figure 7.
Pollen FISH Analysis of Nondisjunction for Minichromosome 3.
Pollen FISH was performed with the ZmBs probe (magenta) and knob (green) on microsporocytes at the three-cell stage and counterstained with DAPI (blue).
(A) Pollen from a plant containing one minichromosome 3. In the absence of the B chromosome long arm, minichromosome 3 disjoins properly and each nucleus (vegetative [arrowhead] and two sperm [arrows]) has magenta signals. Bar = 10 μm.
(B) Pollen from plants containing one B chromosome and one minichromosome 3. ZmBs signal (magenta) is seen in only one of the two sperm. The normal B and the minichromosome are both observed in the vegetative nucleus. The minichromosome undergoes nondisjunction when a normal B chromosome is present.
The Timing of Histone H3 Phosphorylation in Minichromosomes
In plants, cell cycle–dependent phosphorylation of histone H3 has been described (Houben et al., 2007). We examined the distribution of this modification in meiotic cells containing minichromosomes derived from the B chromosome. As expected, the only signal present is localized to the nucleolar organizing region during the pachytene stage of prophase I (Kaszas and Cande, 2000). During later stages of meiosis, histone H3 is highly phosphorylated at Ser-10 throughout the entire chromosome at metaphase I but is restricted to the pericentromeric regions at metaphase II (Kaszas and Cande, 2000).
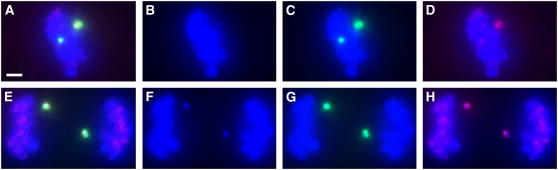
We anticipated that the minichromosomes would show single chromatids resulting from the equational division of univalents at anaphase I to have no H3 phosphorylation, because in rye (Secale cereale) univalents (Manzanero et al., 2000; Houben et al., 2007) or in the maize afd1 mutant (Kaszas and Cande, 2000), the staining is restricted to the pericentromeric regions during metaphase I and anaphase I, with no staining during meiosis II. However, for minichromosomes 3, 5, and 9, there is no change of distribution of phosphorylated histone H3 at Ser-10 between the minichromosome and the normal maize chromosomes. For example, there is staining at metaphase II and anaphase II of minichromosome 3 (Figures 8A and 8E).
Figure 8.
Distribution of Phosphorylated Histone H3 in Plants Containing Two Minichromosomes 3.
The DAPI-stained chromosomes are blue; the polyclonal antibody raised against phosphorylated Ser-10 to histone H3 is magenta; the ZmBs is green. (A) to (D) show metaphase II, and (E) to (H) show anaphase II. The sister cells are not shown. There is no difference in the distribution of phosphorylated histone H3 at Ser-10 between the normal chromosomes and the minichromosome. (A) and (E) show merged images, (B) and (F) show DAPI, (C) and (G) show B repeat plus DAPI, and (D) and (H) show histone phosphorylation plus DAPI. Bar = 10 μm.
Distribution of Maize SGO on Minichromosomes during Meiosis
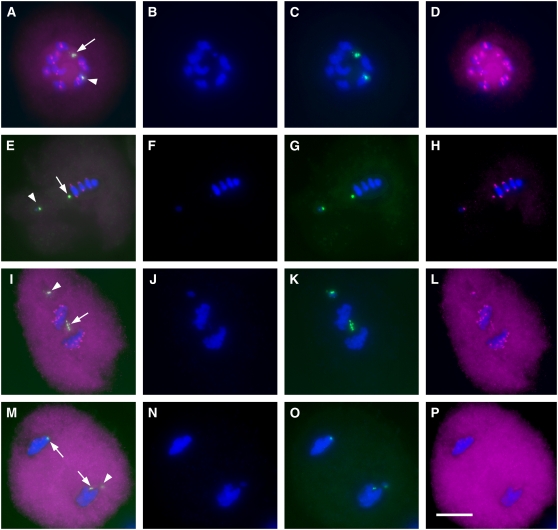
We investigated the localization of the SGO1 protein on minichromosomes during meiosis using immunocytochemistry and FISH. For minichromosomes 3, 5, and 9, there is strong staining of SGO1 in anaphase I, even though the sister chromatids divided equationally (Figure 9). In order to compare the localization of SGO1 between normal B chromosomes and minichromosomes, plants containing one B chromosome and one minichromosome 3 were selected in the progeny of a cross between parents with B chromosomes and minichromosomes. We first performed the immunolocalization of SGO1 and then probed with ZmBs to identify the B chromosome and minichromosome 3. There was strong staining of SGO1 on the minichromosome 3 centromere at the diakinesis, metaphase I, and anaphase I stages (Figures 9A, 9E, and 9I). Single chromatids resulting from the equational division of minichromosome 3 univalents at anaphase I showed stronger SGO1 signals (Figure 9I). Previous results showed one set of centromere sequences of minichromosome 3 to be inactive (Han et al., 2006). However, interestingly, both active and inactive centromere sequences colocalized with SGO1 signal at anaphase I.
Figure 9.
Immunolocalization Analysis of SGO1 in Plants Containing One B Chromosome (Arrowheads) and One Minichromosome 3 (Arrows).
Both the normal B chromosome and the mini-B exhibit SGO signal at anaphase I. The DAPI-stained chromosomes are blue; maize SGO antibody (SGO1) is magenta; the ZmBs is green. (A) to (D) show diakinesis, (E) to (H) show metaphase I, (I) to (L) show anaphase I (sister chromatids start to separate; SGO1 signals are still present), and (M) to (P) show telophase I. (A), (E), (I), and (M) show merged images; (B), (F), (J), and (N) show DAPI; (C), (G), (K), and (O) show B repeat plus DAPI; and (D), (H), (L), and (P) show SGO1 plus DAPI. Arrows in (M) indicate the sister chromatids, and the arrowhead indicates the full-sized B chromosome that maintained sister chromatid cohesion and progressed to one pole. Bar = 10 μm.
DISCUSSION
We examined the mitotic and meiotic behavior of 22 minichromosomes that originated in plants that were undergoing the chromosomal type of BFB cycle. All of the minichromosomes have been confirmed to be derived from the B-A chromosome by the presence of B-specific repeat signals at their centromeric region, thus allowing a comparison of the effects of size among chromosomes of related origin. The amount of detectable knob heterochromatin adjacent to the centromere varied from none to increased amounts relative to the progenitor chromosome. Minichromosomes 2, 3, 5, 10, and 13 each contain two sets of centromere sequences, but one set is inactive (Han et al., 2006). Minichromosome 5 has two distinct centromeric structures; the larger centromere has a stronger B repeat signal with knob heterochromatin, while the smaller one does not. All of the remaining minichromosomes possess some representation of knob sequences and B repeat signals. The simplest scenario to explain the structure of the small and tiny minichromosomes is that breakage occurred near or within the centromere and the broken ends healed without fusion (see Supplemental Figure 1 online).
B chromosome nondisjunction at the second microspore division can be detected by pollen FISH (Shi et al., 1996; Rusche et al., 1997, 2001; Han et al., 2007). All of the minichromosomes have lost the nondisjunction function, as anticipated by the loss of factors at the tip of the B chromosome long arm, which is required for the centromeric region to undergo nondisjunction (Han et al., 2007). However, the addition of a normal B chromosome to the genotype reestablished this property. These findings indicate that engineered mini-B chromosomes (Yu et al., 2007) of virtually any length could be manipulated in terms of dosage by the addition of normal B chromosomes to the genotype.
The tiny minichromosomes consisting of basically the centromeric region present an interesting circumstance to examine the parameters of homolog pairing. Because of the highly repetitive sequences on all chromosomes in maize, pairing must eventually rely on unique combinations of sequences (Pawlowski et al., 2004). Meiotic analysis of minichromosomes indicated that below a certain threshold, chromosome size is not necessarily correlated with pairing. For example, the extremely small minichromosome 9 in two copies exhibited a very high frequency of homolog pairing. However, larger examples, such as minichromosome 3 or 18, did not pair at all during meiosis I. Thus, the specific features of each minichromosome likely determine their pairing fate.
Several univalent minichromosomes were found to divide equationally at meiosis I. Tubulin immunostaining results indicated that these minichromosomes exhibit biorientation of the kinetochore. This behavior is in contrast with that of normally paired homologs in a bivalent state, which separate reductionally from each other. Interestingly, full sized univalent B chromosomes seldom equationally divide at meiosis I (Carlson and Roseman, 1992) (Figure 9). The failure of sister chromatid cohesion at meiosis I was roughly correlated with the size of the chromosome and not necessarily with homolog pairing. The similar behavior of a tiny-fragment chromosome derived from an A chromosome (McClintock, 1978; Maguire, 1987) and a minichromosome composed of a portion of chromosome 10 (Brock and Pryor, 1996) indicates that precocious sister centromere separation is not unique to small B chromosomes. Because our collection is derived from a common progenitor, a role for chromosome size is implied.
Sister chromatid cohesion is important in meiosis I to hold the chromatids together for the segregation of homologous pairs of chromosomes, but then centromere cohesion is released in meiosis II to allow sister separation. Phosphorylation of histone H3 has been found to correlate with the maintenance of such cohesion (Kaszas and Cande, 2000). Phosphorylation typically is found at pericentromeric regions in mitotic chromosomes and during meiosis II. By contrast, there is a uniform distribution along the meiotic chromosomes at metaphase I. Recently, phosphoserines on the specific centromeric histone H3 versus the canonical histone H3 were reported to delineate the centromeric and pericentromeric regions of the chromosome with regard to the functions of chromosome segregation and cohesion (Zhang et al., 2005).
Single chromatids resulting from equational division of rye univalents at anaphase I have been reported to show no H3 phosphorylation (Houben et al., 2007). For minichromosomes, despite the fact that they separate early in meiosis I and thus have an absence of sister association in meiosis II, phosphorylation of histone H3 is maintained in meiosis II. In the same cells, maize A chromosomes, which divided reductionally in meiosis I, and minichromosomes, which divided equationally, have the same distribution of H3 phosphorylation at Ser-10.
The SGO protein ensures centromeric cohesion during meiosis I in Drosophila and yeast (Kerrebrock et al., 1995; Rabitsch et al., 2004) by protecting it from the action of the separase pathway that otherwise dissolves sister chromatid cohesion at anaphase I (Kitajima et al., 2004). In mutants lacking the SGO1 protein, sister chromatids proceed to the same pole, suggesting that monopolar attachment is intact, but they separate precociously during anaphase I. Because centromere cohesion is required for bipolar attachment at metaphase II and this function is missing, the sister chromatids are distributed randomly in the second meiotic division (Watanabe, 2005a, 2005b).
To analyze the equational division of minichromosomes at meiosis I, we examined the distribution of SGO. During meiosis I, there was no change in SGO distribution on the minichromosomes compared with the normal maize chromosomes (Figure 9). The sister chromatids of the smallest minichromosomes separate at anaphase I; thus, their SGO1 signals were expected to disappear at this stage. However, it was surprising to find that sister chromatid centromeres had very strong signals at anaphase I (Figure 9). These results suggest that maize SGO cannot protect centromeric cohesion from the separase pathway during meiosis I for very small chromosomes. Apparently, other factors come into play that are dependent on chromosome size. In fission yeast, a role for pericentromeric heterochromatin has been postulated for the proper establishment of cohesion at centromeres (Bernard et al., 2001; Nonaka et al., 2002). Thus, for these very small chromosomes that are missing substantial pericentromeric regions, centromere cohesion establishment might be compromised.
Our analysis reveals several previously unknown properties of minichromosomes. First, for smaller chromosomes, sister chromatids tend to separate at meiosis I in contrast with normal-sized chromosomes. This phenomenon occurs even when homologous minichromosome pairs are present and therefore is not necessarily due to a lack of homolog pairing. This result suggests that the proper establishment of cohesion of sister centromeres at meiosis I is due in part to a certain length of the chromosome. Second, very small chromosomes, when present in two copies, can exhibit homolog pairing at meiosis I, depending on structural characteristics that are not yet known. Interestingly, those minichromosomes that do not pair with each other when two of them are present in a cell will also not pair with a normal-sized B chromosome. Third, meiotic chromosome cohesion behaves differently for minichromosomes than expected from previous results analyzing univalents. The protection of centromere cohesion by SGO is not maintained for small chromosomes. The reason for this difference is unknown, but chromosome size appears to play a role in determining its behavior, perhaps due to missing pericentromeric regions that are needed for the proper establishment of centromere cohesion (Bernard et al., 2001; Nonaka et al., 2002).
The knowledge gained about the properties of minichromosomes revealed in this study will guide the development of artificial chromosomes and engineered minichromosomes. Any applications of minichromosomes must accommodate the precocious sister chromatid separation as well as the pairing properties of each construct. Despite the unusual segregation properties of small chromosomes, their transmission from generation to generation is still at workable frequencies. However, selection procedures for pollen containing minichromosomes will be necessary for complete fidelity of transmission (Yu et al., 2007).
METHODS
Plant Materials
Twenty-two minichromosome lines from maize (Zea mays) were scored for mini-B number by FISH on root tip spreads; they were then were grown in the greenhouse or at the Genetics Farm at the University of Missouri-Columbia. Male inflorescences at the meiotic stage were fixed in ethanol:acetic acid (3:1, v/v) on ice for 2 h, transferred to 70% ethanol, and stored at −20°C. Fresh pollen was fixed in ethanol:acetic acid (3:1, v/v) at −20°C overnight, transferred to 70% ethanol, and stored at −20°C.
DNA Probe Preparation
For meiotic analysis, the ZmBs probe (Alfenito and Birchler, 1993) was labeled with Texas red-5-dUTP (2 ng/μL) and the knob-specific sequence (Peacock et al., 1981) was labeled with fluorescein-12-dUTP, both by a modified version of the nick translation method described by Kato et al. (2004). In order to detect the structure of the minichromosome centromere, ZmBs was labeled with coumarin-5-dUTP (2 ng/μL), CentC (satellite repeat) was labeled with fluorescein-12-dUTP (2 ng/μL), and CRM (maize centromeric retrotransposon) was labeled with Texas red-5-dUTP (2 ng/μL).
FISH
Mitosis
Somatic preparations and FISH screening were performed according to Kato et al. (2004).
Meiosis
Slides of various stages were collected as described by Gao et al. (1999), UV cross-linked for 2 min, washed in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) (three times for 5 min each), rinsed in 70, 95, and 100% ethanol for 5 min each, and air-dried for 30 min. After application of 6 μL of probe solution (4 ng/μL of each probe in 2× SSC and 1× TE [10 mM Tris-Cl and 1 mM EDTA] buffer, previously denatured for 5 min in boiling water and then placed on ice), the slides were heated for 5 min at 100°C and then incubated at 55°C overnight in a humid chamber. After hybridization, the slides were washed in 2× SSC and mounted in Vectashield mounting medium (containing 1.5 μg/mL DAPI; Vector Laboratories). The FISH images were examined using a Zeiss Universal microscope, captured with a MagnaFire cooled charge-coupled device camera, and processed with Photoshop 7.0.
Pollen FISH
Pollen was quickly rinsed in 2× SSC and washed with 2× SSC three times, each for 5 min. The samples were then washed with 10 mM HCl for 10 min, treated with 1× pepsin (50 μg/mL) for 10 min at 37°C, and washed with 2× SSC two times, each for 5 min. A 10-μL probe mixture was applied to treated pollen and incubated for 6 min at 80°C in a hybridization mixture of 50% formamide, 2× SSC, and 10 ng/μL of each probe (B repeat and knob). Following denaturation, the pollen was incubated overnight at 37°C. Pollen from maize variety B73, a line without B chromosomes, served as a control. Detection and visualization were performed as described (Han et al., 2007).
Immunolocalization in Meiotic Cells: Antibody
Monoclonal rabbit antibody raised against histone H3 phosphorylated at Ser-10 and α-tubulin antibody were from Upstate. Zm SGO1 antibody was provided by Zac Cande (University of California-Berkeley). Tassels were fixed and stored as described by Kaszas and Cande (2000). Anthers at different stages were collected and then cut open to release the meiocytes into 10 μL of buffer A (80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, and 15 mM PIPES buffer, pH 7.0) plus 0.32 M sorbitol on a glass slide followed by the immediate addition of 5 μL of activated acrylamide stock. The slides were rotated for a few seconds, and a cover glass (18 × 18 mm) was placed over the sample for 30 min or longer in a moisture box, then removed with a razor blade and transferred to 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) for 5 min. The slides were treated for 2 to 3 h in 1% Triton X-100 (1× PBS and 1 mM EDTA) and then washed twice with 1× PBS, for 5 min each. About 100 μL of diluted antibodies (in 3% BSA, 1× PBS, and 0.1% Tween 20) was added to the pads. The incubation was conducted overnight at room temperature. Samples were then washed in 1× PBS, 0.1% Tween 20, and 1 mM EDTA three times, each for 10 min. The secondary antibody (goat anti-rabbit antibody labeled by Texas red) was added and allowed to bind for 3 to 4 h at 37°C. After washing the slides in 1× PBS three times, each for 5 min, samples were stained with DAPI (1.5 μg/mL; Vector Laboratories). After screening the slides, those with good spreads were washed in 1× PBS three times, each for 10 min. Then, 10% formaldehyde was applied for 10 min followed by washing in 1× PBS. Denatured DNA probes (15 μL) were applied to the slide and sealed with rubber cement. The spreads on the slides were denatured on a PCR block at 94°C for 5 min followed by overnight incubation at 37°C. The tubulin images were taken as a confocal z-stack, and a flat projection of the three-dimensional image was created with the Surpass viewer of Imaris version 4.5.2 (Bitplane).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Different Sizes of Minichromosomes Derived from TB-9Sb-Dp9.
Supplemental Figure 2. Cytological Analysis of Two Copies of Minichromosome 18.
Supplemental Table 1. Characterization of Minichromosomes Revealed by FISH using ZmBs, Knob, CentC, and CRM as Probes.
Supplemental Table 2. Meiotic Analysis of Plants Containing One Minichromosome.
Supplemental Table 3. Meiotic Analysis of Plants Containing Two Minichromosomes.
Supplemental Table 4. Transmission Rates Revealed by Pollen FISH and Selfing.
Supplementary Material
Acknowledgments
We thank Z. Cande and R. Wang for kindly providing the SGO antibody. We thank G. Esteban Fernandez and B. Troutwine from the Molecular Cytology Core, University of Missouri-Columbia, for help with image acquisition. This work was supported by the National Science Foundation (Grants DBI0421671 and DBI0423898) and the USDA (Grant 2002-01280).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: James A. Birchler (birchlerj@missouri.edu).
Online version contains Web-only data.
References
- Alfenito, M.R., and Birchler, J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auriche, C., Donini, P., and Ascenzioni, F. (2001). Molecular and cytological analysis of a 5.5 Mb minichromosome. EMBO Rep. 2 102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, P., Maure, J.-F., Partridge, J.F., Genier, S., Javerzat, J.-P., and Allshire, R.C. (2001). Requirement of heterochromatin for cohesion at centromeres. Science 294 2539–2542. [DOI] [PubMed] [Google Scholar]
- Brock, R.D., and Pryor, A.J. (1996). An unstable minichromosome generates variegated oil yellow maize seedlings. Chromosoma 104 575–584. [DOI] [PubMed] [Google Scholar]
- Carlson, W.R. (1969). Factors affecting preferential fertilization in maize. Genetics 62 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, W.R. (1986). The B chromosome of maize. CRC Crit. Rev. Plant Sci. 3 201–226. [Google Scholar]
- Carlson, W.R., and Roseman, R.R. (1992). A new property of the maize B chromosome. Genetics 131 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole, T.A., Ross, A., Clark, E., McGill, N., Schindelhauer, D., Cooke, H., and Grimes, B. (2000). Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet. 9 1623–1631. [DOI] [PubMed] [Google Scholar]
- Gao, Z., Han, F., He, M., Ma, Y., and Xin, Z. (1999). Characterization of genomes and chromosomes in a partial amphiploid of wheat-wheatgrass Zhong 2 using fluorescence in situ hybridization (FISH) and chromosome pairing analysis. Acta Bot. Sin. 41 25–28. [Google Scholar]
- Golubovskaya, I.N., Hamant, O., Timofejeva, L., Wang, C.R., Braun, D., Meeley, R., and Cande, W.Z. (2006). Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 119 3306–3315. [DOI] [PubMed] [Google Scholar]
- Hamant, O., Golubovskaya, I., Meeley, R., Fiume, E., Timofejeva, L., Schleiffer, A., Nasmyth, K., and Cande, W.Z. (2005). A rec8-dependent plant shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr. Biol. 15 948–954. [DOI] [PubMed] [Google Scholar]
- Han, F., Lamb, J.C., and Birchler, J.A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 103 3238–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, F., Lamb, J.C., Yu, W., Gao, Z., and Birchler, J.A. (2007). Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. Plant Cell 19 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, J.J., Van Bokkeln, G., Mays, R.W., Gustashaw, K., and Willard, H.F. (1997). Formation of de novo centromeres and construction of first generation human artificial chromosomes. Nat. Genet. 15 345–355. [DOI] [PubMed] [Google Scholar]
- Heller, R., Brown, K.E., Burgtorf, C., and Brown, W.R.A. (1996). Mini-chromosomes derived from the human Y chromosome by telomere directed chromosome breakage. Proc. Natl. Acad. Sci. USA 93 7125–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben, A., Demidov, D., Caperta, A.D., Karimi, R., Agueci, F., and Vlasenko, L. (2007). Phosphorylation of histone H3 in plants—A dynamic affair. Biochim. Biophys. Acta 1769 308–315. [DOI] [PubMed] [Google Scholar]
- Jin, W., Lamb, J.C., Vega, J.M., Dawe, R.K., Birchler, J.A., and Jiang, J. (2005). Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 17 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R.N., and Rees, H. (1982). B Chromosomes. (London: Academic Press).
- Kaszas, E., and Cande, W.Z. (2000). Phosphorylation of histone H3 is correlated with changes in the maintenance of sister chromatid cohesion during meiosis in maize, rather than the condensation of the chromatin. J. Cell Sci. 113 3217–3226. [DOI] [PubMed] [Google Scholar]
- Kato, A., Lamb, J.C., and Birchler, J.A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., Zheng, Y.Z., Auger, D.L., Phelps-Durr, T., Bauer, M.J., Lamb, J.C., and Birchler, J.A. (2005). Minichromosomes derived from the B chromosome of maize. Cytogenet. Genome Res. 109 156–165. [DOI] [PubMed] [Google Scholar]
- Kerrebrock, A.W., Moore, D.P., Wu, J.S., and Orr-Weaver, T.L. (1995). Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 83 247–256. [DOI] [PubMed] [Google Scholar]
- Kitajima, T.S., Kawashima, S.A., and Watanabe, Y. (2004). The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427 510–517. [DOI] [PubMed] [Google Scholar]
- Kurihara, D., Matsunaga, S., Kawabe, A., Fujimoto, S., Noda, M., Uchiyama, S., and Fukui, K. (2006). Aurora kinase is required for chromosome segregation in tobacco BY-2 cells. Plant J. 48 572–580. [DOI] [PubMed] [Google Scholar]
- Lamb, J.C., Kato, A., and Birchler, J.A. (2005). Centromere associated sequences are present throughout the maize B chromosome. Chromosoma 113 337–349. [DOI] [PubMed] [Google Scholar]
- Maguire, M.P. (1987). Meiotic behavior of a tiny fragment chromosome that carries a transposed centromere. Genome 29 744–747. [DOI] [PubMed] [Google Scholar]
- Manzanero, S., Arana, P., Puertas, M., and Houben, A. (2000). The chromosomal distribution of phosphorylated histone H3 differs between plants and animals at meiosis. Chromosoma 109 308–317. [DOI] [PubMed] [Google Scholar]
- Marston, A.L., Tham, W.H., Shah, H., and Amon, A. (2004). A genome-wide screen identifies genes required for centromeric cohesion. Science 303 1367–1370. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1939). The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA 25 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1941). The stability of broken ends of chromosomes in Zea mays. Genetics 26 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1978). Mechanisms that rapidly reorganize the genome. In Stadler Symposium, Vol. 10. G.P. Redei, ed (Columbia, MO: University of Missouri). pp. 25–48.
- Mills, W., Critcher, R., Lee, C., and Farr, C. (1999). Generation of an 2.4 Mb human centromere based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Hum. Mol. Genet. 8 751–761. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (2001). Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35 673–745. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., Peters, J., and Uhlman, F. (2000). Splitting the chromosome: Cutting the ties that bind sister chromatids. Science 288 1379–1384. [DOI] [PubMed] [Google Scholar]
- Nonaka, N., Kitajima, T., Yokobayashi, S., Xiao, G., Yamamoto, M., Grewal, S.I.S., and Watanabe, Y. (2002). Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4 89–93. [DOI] [PubMed] [Google Scholar]
- Pawlowski, W.P., Golubovskaya, I.N., Timofejeva, L., Meeley, R.B., Sheridan, W.F., and Cande, W.Z. (2004). Coordination of meiotic recombination, pairing and synapsis by PHS1. Science 303 89–92. [DOI] [PubMed] [Google Scholar]
- Peacock, W.J., Dennis, E.S., Rhoades, M.M., and Pryor, A.J. (1981). Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch, K.P., Gregan, J., Schleiffer, A., Javerzat, J.P., Eisenhaber, F., and Nasmyth, K. (2004). Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 14 287–301. [DOI] [PubMed] [Google Scholar]
- Roman, H. (1948). Directed fertilization in maize. Proc. Natl. Acad. Sci. USA 34 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, M.L., Mogensen, H.L., Cabound, A., Faure, J.E., Rougier, M., Keim, P., and Dumas, C. (2001). B chromosomes of maize (Zea) are positioned nonrandomly within sperm nuclei. Sex. Plant Reprod. 13 231–234. [Google Scholar]
- Rusche, M.L., Mogensen, H.L., Shi, L., Keim, P., Rougier, M., Chabound, A., and Dumas, C. (1997). B chromosome behavior in maize pollen as determined by a molecular probe. Genetics 147 1915–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, M.H., Mee, P.J., Nichols, J., Yang, J., Brook, F., Gardner, R.L., Smith, A.G., and Brown, W.R.A. (1999). A structurally defined minichromosome vector for the mouse germ line. Curr. Biol. 10 31–34. [DOI] [PubMed] [Google Scholar]
- Shi, L., Zhu, T., Mogensen, H.L., and Keim, P. (1996). Sperm identification in maize by fluorescence in situ hybridization. Plant Cell 8 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X.P., Wahlstrom, J., and Karpen, G. (1997). Molecular structure of a functional Drosophila centromere. Cell 91 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaur, S., Cubizolles, F., Plane, G., Genier, S., Rabitsch, P.K., Gregan, J., Nasmyth, K., Vanoosthuyse, V., Hardwick, K.G., and Javerzat, J. (2005). Control of shugoshin function during fission-yeast meiosis. Curr. Biol. 15 2263–2270. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y. (2005. a). Sister chromatid cohesion along arms and at centromeres. Trends Genet. 21 405–412. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y. (2005. b). Shugoshin: Guardian spirit at the centromere. Curr. Opin. Cell Biol. 17 590–595. [DOI] [PubMed] [Google Scholar]
- Yang, J.W., Pendon, C., Yang, J., Haywood, N., Chand, A., and Brown, W.R.A. (2000). Human mini-chromosomes with minimal centromeres. Hum. Mol. Genet. 9 1891–1902. [DOI] [PubMed] [Google Scholar]
- Yu, W., Han, F., Gao, Z., Vega, J.M., and Birchler, J.A. (2007). Construction and behavior of engineered minichromosomes in maize. Proc. Natl. Acad. Sci. USA 104 8924–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Li, X., Marshall, J.B., Zhong, C.X., and Dawe, R.K. (2005). Phosphoserines on maize centromeric histone H3 and histone H3 demarcate the centromere and pericentromere during chromosome segregation. Plant Cell 17 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y.Z., Roseman, R.R., and Carlson, W.R. (1999). Time course study of the chromosome-type breakage-fusion-bridge cycle in maize. Genetics 153 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.