Abstract
The CRM domain is a recently recognized RNA binding domain found in three group II intron splicing factors in chloroplasts, in a bacterial protein that associates with ribosome precursors, and in a family of uncharacterized proteins in plants. To elucidate the functional repertoire of proteins with CRM domains, we studied CFM2 (for CRM Family Member 2), which harbors four CRM domains. RNA coimmunoprecipitation assays showed that CFM2 in maize (Zea mays) chloroplasts is associated with the group I intron in pre-trnL-UAA and group II introns in the ndhA and ycf3 pre-mRNAs. T-DNA insertions in the Arabidopsis thaliana ortholog condition a defective-seed phenotype (strong allele) or chlorophyll-deficient seedlings with impaired splicing of the trnL group I intron and the ndhA, ycf3-int1, and clpP-int2 group II introns (weak alleles). CFM2 and two previously described CRM proteins are bound simultaneously to the ndhA and ycf3-int1 introns and act in a nonredundant fashion to promote their splicing. With these findings, CRM domain proteins are implicated in the activities of three classes of catalytic RNA: group I introns, group II introns, and 23S rRNA.
INTRODUCTION
Catalytic ribonucleoprotein particles (RNPs) are at the core of several fundamental cellular processes, including protein synthesis, tRNA processing, and RNA splicing. In some catalytic RNPs, RNA subunits harbor the catalytic activity and proteins serve to enhance the assembly or stability of the catalytically active RNA structure (Weeks, 1997; Ban et al., 2000; Hsieh et al., 2004; Pyle and Lambowitz, 2006). The chloroplast RNA splicing and ribosome maturation (CRM) domain is found in four characterized proteins that assemble with catalytic RNA and was named to reflect the functions of these proteins: CRS1, CAF1, and CAF2, each harboring several CRM domains, are required for the splicing of different group II intron subsets in land plant chloroplasts and are bound to those introns in vivo (Till et al., 2001; Ostheimer et al., 2003); Escherichia coli YhbY is a stand-alone CRM domain that associates with assembling 50S ribosomal subunits (Barkan et al., 2007). Structural (Ostheimer et al., 2002) and biochemical (Barkan et al., 2007) data showed CRM domains to be RNA binding domains, and one multidomain CRM protein, CRS1, has been shown to bind RNA in a sequence-specific fashion (Ostersetzer et al., 2005). The Arabidopsis thaliana and rice (Oryza sativa) genomes include 16 and 14 genes, respectively, that encode proteins with one or more CRM domain (Barkan et al., 2007). Where examined, the functions of orthologous CRM proteins in monocots (e.g., maize [Zea mays]) and dicots (e.g., Arabidopsis) are conserved (Asakura and Barkan, 2006).
It is striking that all four of the CRM domain proteins characterized thus far function in concert with large catalytic RNAs (group II introns or 23S rRNA). To explore the functional repertoire of this recently recognized family of RNA binding proteins, we are using reverse genetics and RNA coimmunoprecipitation assays to establish the functions and RNA ligands of additional CRM domain proteins. Here, we show that a fourth CRM domain protein in plants, dubbed CFM2, functions in the splicing of specific chloroplast group II introns and that it is bound in vivo to those introns whose splicing it facilitates. CFM2 acts on both subgroup IIA and subgroup IIB introns; its subgroup IIB substrates also require the CRM domain proteins CAF1 and/or CAF2, and CFM2 and CAF1/2 are bound simultaneously to these introns. Thus, multiple CRM proteins act in a nonredundant fashion to promote the splicing of some group II introns. In addition, CFM2 is bound to and promotes the splicing of the single group I intron in land plant chloroplasts in the trnL-UAA gene. CRM domain proteins are now implicated in the metabolism of three classes of catalytic RNA (group I introns, group II introns, and 23S rRNA), and these are their only known RNA ligands. Thus, CRM domains may be particularly well suited to interact with catalytic RNAs in a manner that is permissive for their evolutionary decay into protein-dependent enzymes.
RESULTS
CFM2 Is Closely Related to CRS1 and Localizes to the Chloroplast Stroma
Predicted CRM domain proteins in rice and Arabidopsis comprise 14 orthologous groups that fall into four subfamilies (Barkan et al., 2007). We assigned the uncharacterized orthologous groups the names Crm Family Member 1 (CFM1) through CFM11. The orthologous group named CFM2 is in the same subfamily as the CRS1 orthologous group and includes one gene in Arabidopsis (At CFM2, At3g01370) and one in rice (Os Cfm2, Os04g39060). Partial sequence for a putative maize ortholog, Zm cfm2, was identified among maize EST and genomic DNA sequences, and these were used to obtain corresponding cDNAs (see Methods). Phylogenetic analysis (see Supplemental Figure 1 online) supports the orthology of the maize gene encoding these cDNAs with Os Cfm2 and At CFM2. An alignment among these three proteins is shown in Supplemental Figure 2A online.
CFM2 is closely related to CRS1, which binds to and facilitates the splicing of the group II intron in the chloroplast atpF gene (Jenkins et al., 1997; Till et al., 2001; Ostersetzer et al., 2005). The major difference between CFM2 and CRS1 is at the C terminus, where CFM2 includes a fourth CRM domain (Figure 1A; see Supplemental Figure 2B online). Maize, rice, and Arabidopsis CFM2 are all predicted to localize to chloroplasts by the TargetP (Emanuelsson and von Heijne, 2001) algorithm.
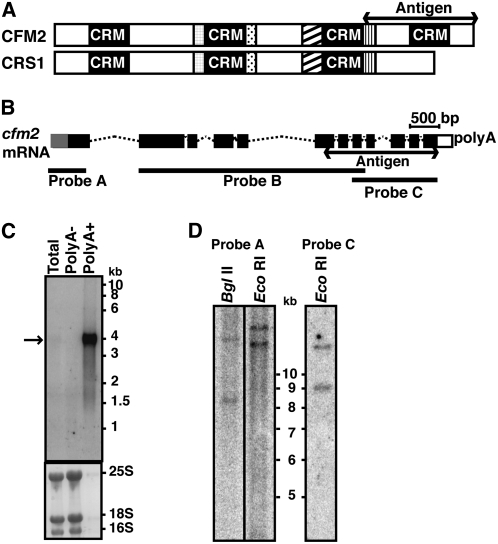
Figure 1.
The Maize cfm2 Gene.
(A) Domain organization of the CFM2 protein. Regions of similarity with CRS1 are represented with like shading. The fragment used for antisera generation is marked.
(B) Schematic of the major maize cfm2 mRNA. Exons are indicated with boxes and introns with dotted lines. Exons in black were represented in cDNAs. The gray exon segment at the 5′ end was inferred from genomic DNA sequence. The exons represented in the cDNA probes used for DNA and RNA gel blots are indicated.
(C) RNA gel blot hybridization showing cfm2 mRNA. Seedling leaf RNA (polyA+, polyA-, or total) was hybridized to cDNA probe B diagrammed in (B). The same blot stained with methylene blue is shown below, with rRNA bands marked.
(D) DNA gel blot hybridization suggesting the presence of two cfm2 genes in maize. Total leaf DNA digested with the indicated enzyme was hybridized with cDNA probes A or C, diagrammed in (B). The blot was washed at high stringency (65°C, 0.2× SSC). The genomic regions corresponding to these probes lack sites for the enzymes used to digest the DNA.
Three types of maize cfm2 cDNA were recovered, derived from RNAs with alternative polyadenylation sites. The largest cDNA encodes a protein that aligns throughout with predicted rice and Arabidopsis CFM2. In two cDNA variants, the use of alternative polyadenylation sites truncates the open reading frame such that it ends either within or after the third CRM domain (see Supplemental Figure 3 online). The proteins encoded by these mRNAs are predicted to be ∼112, 72, and 81 kD after removal of the transit peptide. However, RNA gel blot hybridization of seedling leaf mRNA detected only one transcript whose size matches the largest cDNA isoform (Figure 1C). Therefore, the relevance of the alternative cDNA forms to cfm2 function in leaf tissue is unclear. CFM2 is encoded by a single-copy gene in rice and Arabidopsis, but DNA gel blot data suggest the existence of two cfm2 genes in maize (Figure 1D). The sequences of the three maize cDNA isoforms are identical in their regions of overlap, so these cDNAs arise either from the same gene or from two nearly identical genes.
The results of a transient expression assay involving the N-terminal 53 amino acids of Zm CFM2 fused to GFP suggested that CFM2 localizes to plastids (see Supplemental Figure 4 online). Immunoblots of leaf and chloroplast subfractions confirmed this to be the case: affinity-purified polyclonal antibodies raised to the CFM2 fragment diagrammed in Figure 1A detected two proteins that were highly enriched in isolated chloroplasts, in comparison with their concentration in leaf extract (Figure 2). Analysis of chloroplast subfractions showed that both proteins localize to the chloroplast stroma (Figure 2). Affinity-purified antibodies from two different rabbits gave identical results (data not shown), suggesting that both proteins detected are cfm2 gene products or arise from two highly similar genes. The size of the larger protein is consistent with that predicted for the phylogenetically conserved protein that includes all four CRM domains. The smaller protein (∼80 kD) is unlikely to be the product of either of the alternative polyadenylation variants detected as a cDNA because there is no more than a small overlap between the antigen and the open reading frame of either cDNA (see Supplemental Figure 3 online) and affinity purification using a nonoverlapping CFM2 fragment did not alter the ratio of the two proteins detected on immunoblots (data not shown). The CFM2 antibody did not detect Arabidopsis CFM2 (data not shown), suggesting that the ∼80-kD protein is more closely related to maize CFM2 than is At CFM2. These results and additional data shown below suggest that the ∼80-kD protein arises from an undetected cfm2 mRNA isoform, from a cfm2 coortholog, or from proteolysis of CFM2.
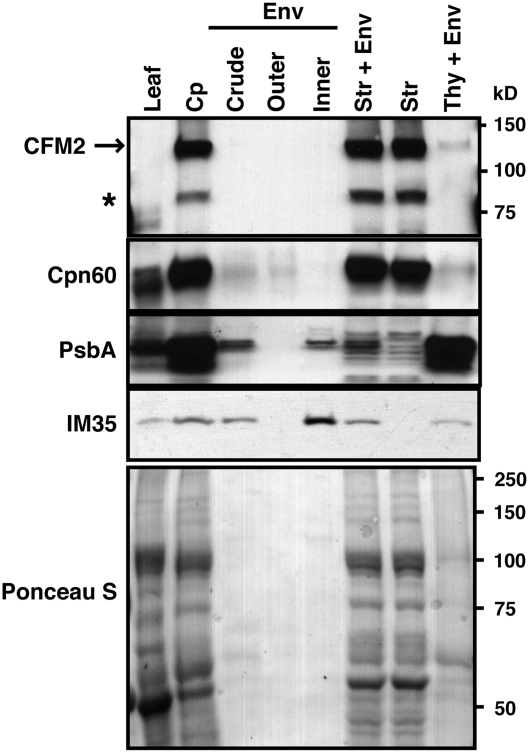
Figure 2.
Maize CFM2 Is Localized to the Chloroplast Stroma.
Chloroplasts (Cp) and chloroplast subfractions were from the fractionated chloroplast preparation described previously (Williams and Barkan, 2003). Equivalent proportions of each fraction were examined by immunoblot analysis using antisera specific for the protein indicated to the left. The same blot stained with Ponceau S is shown below. Cpn60, PsbA, and IM35 were used as markers for the stroma, thylakoid, and inner envelope, respectively. The major band detected with the CFM2 antibodies corresponds in size to the CFM2 protein with four CRM domains that is conserved with the gene models of the Arabidopsis and rice orthologs. The asterisk denotes a second protein detected by affinity-purified antibodies from two different rabbits immunized with the CFM2 antigen. Env, envelope; Str, stroma; Thy, thylakoid.
RNA Coimmunoprecipitation Assays Identified Group I and Group II Intron RNAs as CFM2 Ligands
CFM2 was anticipated to be an RNA binding protein because it is similar to the established RNA binding protein CRS1. To identify chloroplast RNAs that are associated with CFM2 in vivo, we used an RNA immunoprecipitation and chip hybridization (RIP-chip) assay as an initial screen: RNAs that coimmunoprecipitate with CFM2 from maize chloroplast extract were identified by hybridization to a tiling microarray of the maize chloroplast genome. Four immunoprecipitations were performed, two each with affinity-purified CFM2 antibodies from two different immunized rabbits; these antibodies precipitated both the ∼115- and 80-kD proteins detected on immunoblots of chloroplast extract (see Figure 4A). Analogous assays using antibodies to OE23 and OE16, chloroplast proteins that do not bind RNA, served as negative controls.
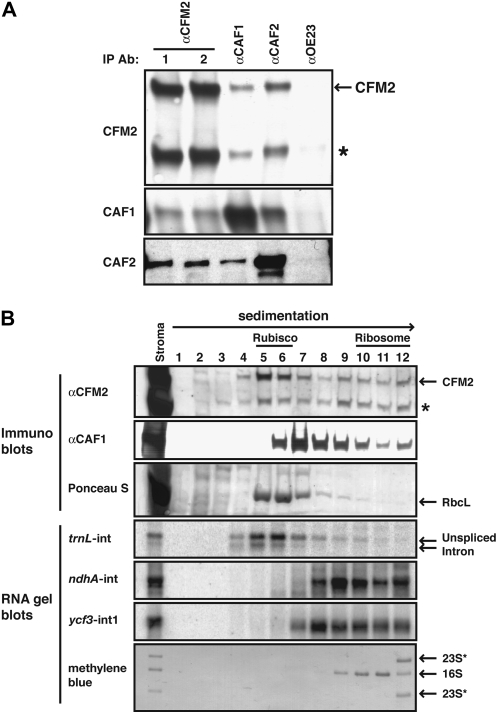
Figure 4.
CFM2 Is Found in Large RNPs That Include CAF1 and/or CAF2.
(A) Immunoblots showing coimmunoprecipitation of CFM2 with CAF1 and CAF2. Immunoprecipitations were performed with chloroplast stroma, using the antibody named at the top (IP Ab); antibodies from two different rabbits immunized with the CFM2 antigen were used in replicate assays (lanes 1 and 2). The presence of specific proteins in the immunoprecipitation pellets was determined by probing replicate blots with the antibodies listed to the left. All antibodies were affinity purified against their cognate antigen prior to use. An anti-OE23 immunoprecipitation served as a negative control. The asterisk marks the ∼80-kD protein detected with the CFM2 antisera.
(B) Cosedimentation of CFM2 with CAF1, CAF2, and intron RNAs. Stromal extract was sedimented in sucrose gradients under conditions in which particles greater than ∼60S pellet. An equal proportion of each fraction was analyzed by probing immunoblots with the antibody indicated to the left. The Ponceau S stained blot illustrates the position of Rubisco in the gradient; RbcL is the large subunit of Rubisco. Bottom panels show RNA gel blots of RNA purified from the same gradient fractions. The probes were intron specific and are indicated to the left. The methylene blue–stained blot illustrates the position of ribosomal subunits in the gradient: 16S rRNA marks 30S ribosomal subunits, and 23S* rRNAs mark 50S ribosomal subunits.
Figure 3A shows the degree to which each chloroplast RNA sequence was enriched in the immunoprecipitation pellets, plotted according to chromosomal position. Three strong and reproducible peaks of enrichment were observed, corresponding to the ycf3, trnL-UAA, and ndhA loci. The primary transcript from each of these loci includes introns: two group II introns in pre-ycf3 mRNA, one group II intron in pre-ndhA mRNA, and a group I intron in pre-trnL. Although each peak included several adjacent array fragments, the highest enrichment values within each peak corresponded to intron rather than exon sequence, supporting the notion that CFM2 is bound to intron RNAs, with flanking exons coimmunoprecipitated due to tethering to the same RNA molecule. Both ycf3 introns were enriched, but the peak fragments were within ycf3-int1, pointing to intron 1 as CFM2's ligand (Figure 3A). Statistical analysis (see Supplemental Table 1 online) shows the differential enrichment of sequences from the ycf3, ndhA, and trnL loci in CFM2 versus control immunoprecipitations to be highly significant.
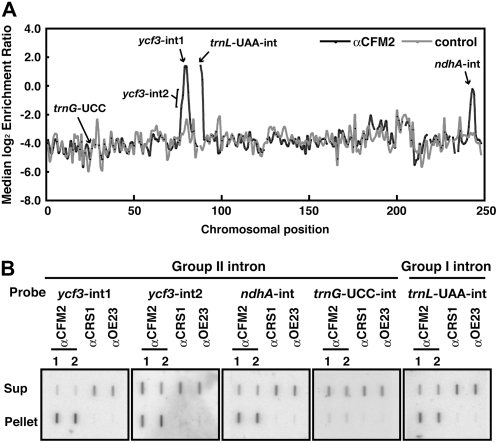
Figure 3.
Identification of CFM2's RNA Ligands in Coimmunoprecipitation Assays.
(A) Summary of RIP-chip data. The median log2-transformed enrichment ratios (F635/F532) for four replicate CFM2 RIP-chip assays (two with each of two CFM2 antisera) are plotted according to chromosomal position (black line). The median ratios for two negative control immunoprecipitations (one with antibody to OE16 and one with antibody to OE23) are shown for comparison (gray line). Fragments for which fewer than two replicate spots per array yielded an F532 signal above background were excluded and appear as gaps in the curve.
(B) Validation of CFM2's RNA ligands by slot blot hybridization. RNA purified from the pellets and supernatants of immunoprecipitations with antisera to the indicated proteins was hybridized to the indicated intron-specific probes. Immunoprecipitations with CRS1 and OE23 antisera and probing for the trnG-UCC intron served as negative controls. The two anti-CFM2 immunoprecipitations used antibodies from different immunized rabbits. The same fraction of the pellet and supernatant samples was applied to the membrane. Differences in the maximal enrichment observed for ycf3-int1, ndhA-int, and trnL-int are not unexpected because the fraction of the RNA pool bound to CFM2 could differ in each case.
The RIP-chip data were validated by slot blot hybridization of RNAs purified from immunoprecipitation pellets and supernatants (Figure 3B). Both ycf3 introns coimmunoprecipitated with CFM2, but intron 1 was substantially more enriched than intron 2, as in the RIP-chip assays. The ndhA and trnL-UAA introns were also strongly enriched in CFM2 immunoprecipitation pellets. The group II intron in trnG-UCC, which did not emerge as a positive from the RIP-chip assays (Figure 3A), was likewise not enriched in CFM2 immunoprecipitations when assayed by slot blot hybridization. Taken together, these results provide strong evidence that CFM2 is associated with ycf3-int1, the ndhA intron, and the trnL-UAA intron RNAs in chloroplast extract. The RIP-chip data suggest further that CFM2 does not bind any additional ligands.
CFM2 Is Found in Large RNP Complexes That Contain CAF1 and/or CAF2
The RNA ligands detected for CFM2 overlap with those of the CRM domain splicing factors CAF1 and CAF2, which include the ndhA and ycf3-int1 introns among their substrates (Ostheimer et al., 2003). Coimmunoprecipitation and cosedimentation assays were used to determine whether CFM2 resides in stromal RNPs that include CAF1 and/or CAF2 (Figure 4). Both the ∼115- and ∼80-kD proteins detected with the CFM2 antisera coimmunoprecipitated with CAF1 and CAF2 from chloroplast stroma (Figure 4A). Furthermore, when stromal extract was size-fractionated by sedimentation through sucrose gradients, both proteins detected by the CFM2 antisera were found in large particles whose sizes overlap those of CAF1- and CAF2-containing particles (Figure 4B; for CAF2, see Watkins et al., 2007). Unlike CAF1 and CAF2, however, CFM2 was found in two peaks: one cosediments with ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) (∼500 kD) and the second was between Rubisco and 30S ribosomal subunits. The ratio of the two proteins detected with the CFM2 antisera differed in these two peaks, with the larger protein dominating in the ∼500-kD peak, but not in the more rapidly sedimenting peak. RNA gel blot hybridization of RNA extracted from the gradient fractions showed that trnL intron sequences cosedimented with the ∼500-kD particles, whereas ndhA intron and ycf3-int1 RNAs cosedimented with the higher molecular weight peak. Taken together, the coimmunoprecipitation and cosedimentation data show that CFM2 is associated with CAF1, CAF2, and specific intron RNAs in large RNP complexes in the chloroplast stroma. The similar behavior of the two proteins detected by the CFM2 antibodies in both the coimmunoprecipitation and sedimentation assays strongly suggests that both proteins are cfm2 gene products or the products of closely related cfm2 coorthologs.
Mutations in At CFM2 Disrupt the Splicing of Arabidopsis Orthologs of Introns That Coimmunoprecipitate with Zm CFM2
To determine the role of CFM2 in the metabolism of the RNAs with which it coimmunoprecipitates, we sought loss-of-function cfm2 mutants. Previously, we recovered maize crs1, caf1, and caf2 mutants in reverse-genetic screens of our large collection of nonphotosynthetic maize mutants (http://pml.uoregon.edu). However, a comprehensive search of this collection for insertions in maize cfm2 yielded only one mutant, which had an insertion 70 nucleotides upstream of the start codon and did not have a visible phenotype. Based on the distribution of mutant alleles recovered from this collection to date (data not shown), it is at or near saturation for genes whose disruption causes a chlorophyll-deficient phenotype; thus, our failure to recover such alleles for cfm2 suggests that disruption of cfm2 may not cause such a phenotype. Taken together with the DNA gel blot data suggesting the presence of a second cfm2 gene in maize (Figure 1D), it seems possible that maize has two functionally redundant cfm2 genes.
Because a reverse-genetic approach in maize was problematic, we sought mutations in the single Arabidopsis cfm2 ortholog as an alternative. The splicing functions of maize CRS1, CAF1, and CAF2 are conserved in their Arabidopsis orthologs (Asakura and Barkan, 2006), so it seemed likely that the functions of CFM2 would also be conserved. T-DNA insertions in At CFM2 were obtained from the SALK and SAIL insertion collections (Sessions et al., 2002; Alonso et al., 2003). At cfm2-1 and At cfm2-2 have T-DNA insertions in exons 5 and 11, respectively (Figure 5A; see Supplemental Figure 5A online). Self-pollination of At cfm2-2/+ plants yielded ∼25% mutant seedlings with albino cotyledons and pale green leaves (Figures 5B and 5C) that die soon after germination when grown in soil. All eight of the chlorophyll-deficient plants and none of the 38 normal plants examined were homozygous for the At cfm2-2 insertion, indicating that the mutation causing this phenotype is linked to At cfm2 (data not shown). By contrast, siliques resulting from self-pollinating At cfm2-1/+ plants segregated ∼25% chlorophyll-deficient seeds that shriveled during seed maturation (Figure 5B, top). The normal seeds from these siliques gave rise to normal-appearing plants; of the 37 such plants examined by PCR, none were homozygous for the At cfm2-1 insertion, indicating that the shriveled seed phenotype is linked to At cfm2-1. These shriveled seeds germinate poorly; some can be observed to germinate on Murashige and Skoog (MS) medium but these yield severely stunted roots (visible only under a microscope) and little or no cotyledon tissue (data not shown). In those rare cases where cotyledons and leaves become visible to the naked eye, they are albino and they grow very slowly (Figure 5C). PCR genotyping of such plants showed that this phenotype is linked to the At cfm2-1 insertion (see Supplemental Figure 5B online). These results show that both the stunted albino seedling and defective seed phenotypes are linked to the At cfm2-1 insertion.
Figure 5.
At CFM2 Insertion Mutants.
(A) T-DNA insertions in At CFM2 (At3g01370). Exons and introns are indicated with black rectangles and thin lines, respectively. The positions of the T-DNA insertions in At cfm2-1 (SALK_000703) and At cfm2-2 (SAIL_67_E05) are indicated by filled triangles. Arrows show the primers used for RT-PCR (D). LB, T-DNA left border.
(B) Phenotypes conditioned by At CFM2 mutations. Siliques resulting from self-pollination of an At cfm2-1/+ plant are shown at top. White seeds in a young silique and brown shriveled seeds in an older silique are indicated with arrows. These shriveled seeds germinate poorly; those that germinate yield stunted roots and either no shoot (data not shown) or albino, stunted shoots as shown in (C). Shown below are the phenotypes of a homozygous At cfm2-2 mutant and a heteroallelic plant harboring both mutant alleles. Plants were grown for 17 d on MS medium containing 2% sucrose in cycles of 14 h light/10 h dark. Bars = 5 mm.
(C) Phenotype of rare At cfm2-1 homozygote that developed leaves. Plants were grown for 25 d on MS medium containing 2% sucrose in cycles of 10 h light/14 h dark. An At cfm2-2 homozygote grown in parallel is shown for comparison. Bars = 5 mm
(D) RT-PCR analysis of At CFM2 mRNA in At CFM2 mutants. Two hundred nanograms of total leaf RNA from plants with the indicated genotype (homozygous unless otherwise indicated) was analyzed by RT-PCR using the primer pairs indicated to the left and diagrammed in (A). At crs1-1 mutants were analyzed to show the level of At CFM2 mRNA in a different nonphotosynthetic mutant. Dilutions of the wild-type sample were analyzed to illustrate the relationship between signal intensity and input RNA level. RT-PCR of actin 2 mRNA was used as an internal control. The PCR products were detected by staining with ethidium bromide. –RT, control reaction lacking reverse transcriptase.
Complementation tests were performed by crossing At cfm2-1/+ and At cfm2-2/+ plants; these yielded segregating yellow green seedlings (∼1:3 ratio) with a phenotype intermediate between those segregating in each of the parental lines (Figures 5B and 5C). The chlorophyll-deficient progeny of these crosses harbor both mutant alleles, whereas their normal siblings do not (see Supplemental Figure 5B online), demonstrating that their phenotype results from the disruption of At CFM2. Taken together, these results provide strong evidence that both the defective seed and stunted albino seedling phenotypes are conditioned by the At cfm2-1 mutation. By contrast, a pale-green, seedling-lethal phenotype is conditioned by the At cfm2-2 mutation. The differing severities of these phenotypes correlates as one would anticipate from the positions of the T-DNA insertions: the At cfm2-1 insertion disrupts an exon in the middle of the gene and is likely to cause a severe loss of gene function, whereas the At cfm2-2 insertion disrupts sequences close to the 3′ end of the gene and could allow the expression of a truncated protein that retains partial activity. The accumulation of At CFM2 RNA sequences in these mutants support this scenario: At CFM2 RNA sequences between the two insertions accumulate to near normal levels in At cfm2-2 homozygotes but not in At cfm2-1 homozygotes (see primers A+C in Figure 5D), whereas RNA sequences downstream of both insertions are severely reduced in both mutant lines (see primers B+D in Figure 5D).
We were unable to assay At CFM2 protein levels because the antisera were raised against maize CFM2 and did not detect At CFM2. The amount of leaf tissue that could be obtained from the rare At cfm2-1 homozygotes that developed leaves was very limited, so most of the molecular analyses described below involved At cfm2-2 homozygotes and the At cfm2-1/At cfm2-2 progeny of complementation crosses.
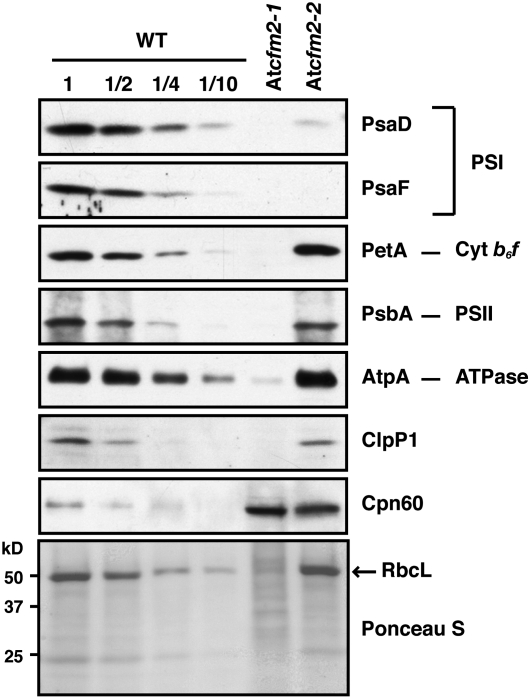
The accumulation of each photosynthetic enzyme complex that includes plastid-encoded subunits was monitored by immunoblot analysis of leaf extract using antibodies to representative core subunits (Figure 6). The abundance of the photosystem I subunits PsaD and PsaF was reduced >10-fold in At cfm2-2 mutants, whereas the other proteins monitored were reduced less than twofold. These results indicate that the weak At cfm2-2 allele disrupts the expression of one or more genes involved in the biogenesis of photosystem I. The ycf3 pre-mRNA, with which CFM2 associates, encodes a protein that is necessary for photosystem I accumulation (Boudreau et al., 1997; Ruf et al., 1997). The specific depletion of photosystem I proteins in At cfm2-2 mutants is consistent with a role for CFM2 in ycf3 expression. The protein deficiencies in At cfm2-1 homozygotes were more global and severe, suggesting a global plastid gene expression defect in these plants.
Figure 6.
Immunoblot Analysis of Subunits of Photosynthetic Complexes in At CFM2 Mutants.
Total proteins from seedling leaves (10 μg or dilutions as indicated) were analyzed on immunoblots by probing with antibodies specific for the proteins indicated to the right. The Ponceau S–stained filter at the bottom illustrates the relative loading of the samples and the level of RbcL, the large subunit of Rubisco. PSI, photosystem I.
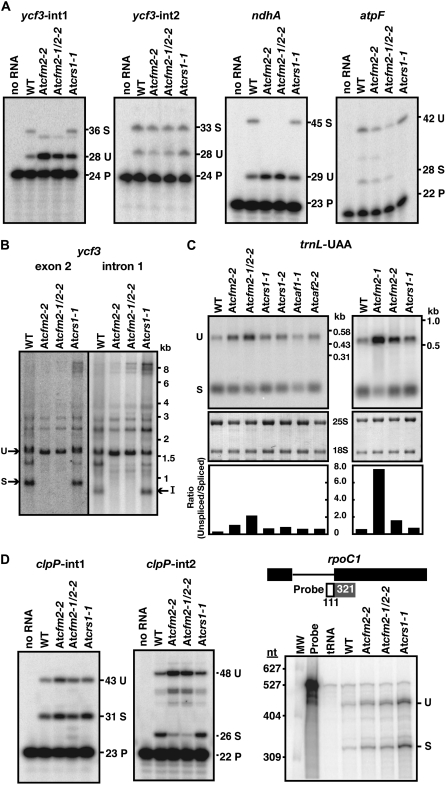
Given the similarity between CFM2 and the group II intron splicing factors CRS1, CAF1, and CAF2, it seemed likely that CFM2 facilitates the splicing of the group II introns with which it associates. To test this possibility, the ratio of spliced to unspliced RNAs from the chloroplast ycf3 and ndhA genes was measured with poisoned-primer extension assays. In these assays, a radiolabeled oligonucleotide complementary to exon sequences near a 3′-splice junction was used to prime a reverse transcription reaction in the presence of a dideoxynucleotide that terminates cDNA synthesis after different distances on spliced and unspliced RNA templates. Little if any spliced RNA was detected from ycf3-int1 or ndhA in At cfm2 mutants, whereas the unspliced precursors accumulated to increased levels (Figure 7A). RNA gel blot hybridizations showed that both fully spliced ycf3 mRNA and excised ycf3-int1 intron accumulate to reduced levels in At CFM2 mutants (Figure 7B), providing further evidence that the defect lies in splicing rather than in stabilization of the spliced mRNA. The splicing of two other group II introns, ycf3-int2 and atpF, was unaffected (Figure 7A), indicating that these mutations do not cause global splicing defects. Furthermore, the ycf3 and ndhA RNAs were spliced normally in At crs1-1 mutants, which lack the chloroplast ATP synthase, demonstrating that the splicing defects in At CFM2 mutants do not result from compromised photosynthesis. The splicing defects in At CFM2 mutants, together with the intron coimmunoprecipitation data from maize, provide strong evidence that CFM2 promotes the splicing of the ycf3-int1 and ndhA group II introns and that it does so through direct interaction with intron RNA. The ycf3 splicing defect can account for the photosystem I deficiency conditioned by the weak At cfm2-2 mutant allele.
Figure 7.
Chloroplast RNA Splicing Defects in At CFM2 Mutants.
(A) Poisoned-primer extension assays monitoring splicing of chloroplast group II introns. Total leaf RNA (10 μg) from wild-type, At cfm2-2, At cfm2-1/At cfm2-2, and At crs1-1 seedlings was analyzed. The length in nucleotides of each predicted product is indicated. The size of the unlabeled product in the ycf3-int1 assay matches that predicted for the product from an unspliced RNA template in which the last nucleotide of the intron (a cytidine residue) is edited. U, unspliced; S, spliced; P, primer.
(B) RNA gel blots confirming a ycf3-int1 splicing defect in At CFM2 mutants. Total leaf RNA (5 μg) was probed with exon 2 or intron 1 specific probes, as indicated. I, excised intron; S, spliced mRNA; U, unspliced precursor that includes intron 1.
(C) RNA gel blots demonstrating trnL-UAA splicing defect in At CFM2 mutants.
Leaf RNA (1 μg) from seedlings with the indicated genotypes was analyzed by RNA gel blot hybridization using a probe that includes both exon and intron sequences. The same blots stained with methylene blue are shown below; 25S and 18S are cytosolic rRNAs. The ratio of unspliced-to-spliced RNA was quantified with a phosphor imager and is shown in the bar graphs. U, unspliced; S, spliced.
(D) Analysis of the splicing of chloroplast introns found in Arabidopsis but not in maize. Poisoned-primer extension assays were used to monitor clpP-int1 and clpP-int2 splicing, as described in (A). The unlabeled band in the clpP-int2 assays may represent a structure-induced reverse transcriptase stop within the intron, as its abundance is proportional to that of the full-length product from an unspliced template. A ribonuclease protection assay using the diagrammed probe was used to monitor rpoC1 splicing. U, unspliced; S, spliced; P, primer.
RNA gel blot hybridization was used to monitor the effects of At CFM2 mutations on the splicing of the group I intron in pre-trnL-UAA (Figure 7C), which also coimmunoprecipitates with CFM2. Quantification of the data (Figure 7C, bottom) showed that the levels of spliced trnL-UAA and its unspliced precursor correlate in opposite ways with the At CFM2 genotype: the more severe the disruption of At CFM2, the greater the loss of spliced trnL-UAA and the greater the overaccumulation of unspliced precursor. The ratio of unspliced to spliced trnL-UAA in At cfm2-1/At cfm2-2 mutants and in the rare At cfm2-1 homozygotes that developed leaves was ∼4- and ∼15-fold greater, respectively, than the ratio in other photosynthetically impaired mutants with lesions in CRM domain splicing factors (At caf1-1, At caf2-2, and At crs1-1). Ribonuclease protection assays yielded similar results (see Supplemental Figure 6A online). These results together with the coimmunoprecipitation and cosedimentation of trnL intron RNA with CFM2 provide strong evidence that CFM2 promotes trnL-UAA splicing.
At CFM2 Is Required for the Splicing of the Chloroplast clpP-int2 Intron, Which Is Not Found in Maize
The Arabidopsis plastid genome encodes three group II introns that are absent in maize: two in the clpP gene (clpP-int1 and clpP-int2) and one in the rpoC1 gene. Interactions between these introns and CFM2 could not have been detected in the coimmunoprecipitation assays because they used maize chloroplast extract, and analogous experiments were not performed with Arabidopsis because the Zm CFM2 antisera do not detect At CFM2. To determine whether At CFM2 influences the splicing of these three introns, their splicing was assayed in At CFM2 mutants (Figure 7D). A poisoned-primer extension assay showed that RNAs spliced for clpP-int2 accumulate to reduced levels and the unspliced precursor accumulates to increased levels in At CFM2 mutants, supporting a role for CFM2 in clpP-int2 splicing (Figure 7D). RNA gel blots likewise showed the loss of fully spliced clpP mRNA and increased accumulation of precursor forms in At CFM2 mutants (see Supplemental Figure 6B online). By contrast, clpP-int1 and the rpoC1 intron are spliced normally in At cfm2 mutants (Figure 7D).
DISCUSSION
Results presented here show that the CRM domain protein CFM2 is associated with two group II introns and one group I intron in large RNPs in maize chloroplasts, that it is required for the efficient splicing of the orthologs of these introns in Arabidopsis, and that it also promotes the splicing of the clpP-int2 group II intron, which is found in Arabidopsis but not in maize. These findings expand the substrate repertoire of CRM domain proteins to include group I introns. That three classes of large, highly structured catalytic RNA (23S rRNA, group I introns, and group II introns) are associated with CRM domain proteins and that these are their only known RNA ligands suggests that this domain is particularly well suited to interact with RNAs of this nature.
The group I intron in trnL-UAA is an ancient intron that is thought to have been present in the common ancestor of cyanobacteria and acquired by plastids via vertical transmission (Xu et al., 1990; Simon et al., 2003). Whereas cyanobacterial trnL introns self-splice efficiently in vitro, the orthologous intron in diverse eukaryotic algae perform only the first splicing step in vitro, and the land-plant orthologs appear to be incapable of any self-splicing (Xu et al., 1990; Simon et al., 2003). Thus, self-splicing capacity seems to have been lost in a step-wise fashion after chloroplasts took up residence in eukaryotic cells. The degeneration of this intron was presumably enabled by acquisition of compensatory interactions with host-encoded proteins. CFM2 is the only protein known to enhance the splicing of the trnL intron. The expansion of the CRM domain family from a single-domain YhbY-like progenitor into the diverse single and multidomain protein family found in land plants occurred after the divergence of the chlorophyte algae (Barkan et al., 2007). This may have been a key event that led to the relinquishment of self-splicing capacity by the trnL-UAA intron.
In plants harboring the At cfm2-2 allele, the splicing of the trnL group I intron is only slightly affected, whereas the splicing of CFM2's group II intron ligands is strongly affected (Figure 7). The At cfm2-2 insertion is near the 3′ end of the gene and mRNA upstream of this insertion accumulates to near normal levels in plants harboring this allele (Figure 5D); thus, At cfm2-2 may give rise to a truncated protein lacking the fourth CRM domain. The differing sensitivities of At CFM2's group I and II intron ligands to this allele is consistent with the possibility that the group I splicing activity does not require the fourth CRM domain, whereas the group II splicing activity does. Alternatively, CFM2's function in trnL splicing may be partially redundant with that of other proteins. An activity of this nature is unlikely to reside in the CRM domain proteins CRS1, CAF1, or CAF2 because these do not coimmunoprecipitate with trnL intron RNA (Figure 3B; Schmitz-Linneweber et al., 2005; our unpublished data). However, two abundant chloroplast proteins with RRM RNA binding domains were reported to coimmunoprecipitate with unspliced trnL-UAA RNA from tobacco chloroplast extract (Nakamura et al., 1999) and are candidates for harboring this type of activity.
CFM2 is unusual among proteins shown to promote ribozyme activity in that it is highly specific in its effects, and yet its natural substrates include RNAs from multiple structural classes: subgroup IIB introns, a subgroup IIA intron, and a group I intron. The yeast DEAD box protein MSS116 also promotes the splicing of both group I and group II introns. However, MSS116 is nonspecific, influencing the splicing of all of the group I and group II introns in yeast mitochondria as well as other RNA processing events and the translation of several mRNAs (Huang et al., 2005). MSS116 does not bind RNA with sequence specificity and its functions in vivo and in vitro can be substituted by a nonorthologous DEAD box protein from a different organism (Huang et al., 2005; Solem et al., 2006; Halls et al., 2007). It seems likely that MSS116 interacts weakly with most or all RNAs and that these interactions disrupt inhibitory RNA structures or stabilize on-pathway folding intermediates. By contrast, CFM2 shows striking specificity for several RNA ligands in vivo. By extrapolation from studies involving the closely related protein CRS1 (Ostersetzer et al., 2005), we expect that CFM2 has intrinsic high affinity and specificity for its in vivo ligands. Thus, the mechanistic basis for the influence of CFM2 on RNA activity is likely to differ from that for MSS116. It seems likely that each of CFM2's RNA ligands includes a small sequence or structural motif that is recognized by CFM2; shared motifs among these ligands do not emerge from simple alignments, but this is not surprising given the large size of these introns. Detailed in vitro studies will be required to pinpoint the motifs that are recognized by CFM2.
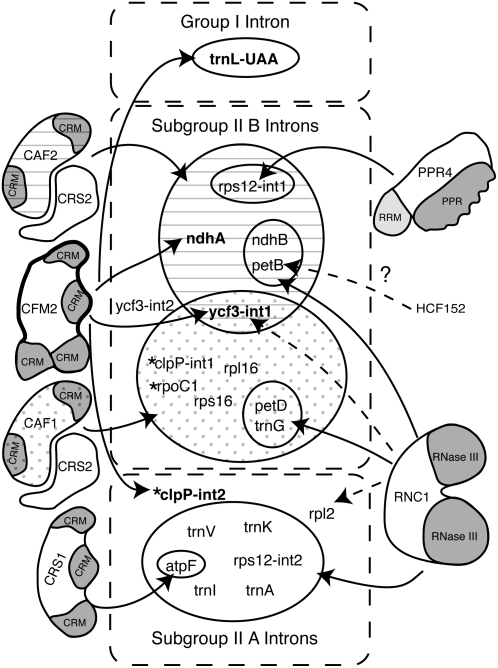
The nucleus-encoded proteins that have been shown to promote the splicing of chloroplast introns in land plants are summarized in Figure 8. CFM2 is the fourth nucleus-encoded protein identified that promotes the splicing of the ycf3-int1 and ndhA group II introns. The ndhA intron requires a heterodimeric complex containing the CRM domain protein CAF2 and the peptidyl-tRNA hydrolase homolog CRS2, whereas ycf3-int1 requires both a CAF2/CRS2 and CAF1/CRS2 complex (Jenkins et al., 1997; Ostheimer et al., 2003; Asakura and Barkan, 2006; Ostheimer et al., 2006). CAF1 and CAF2 are bound to these introns in native extract (Ostheimer et al., 2003; Schmitz-Linneweber et al., 2005), and both proteins coimmunoprecipitate with CFM2 (Figure 4). Thus, CFM2 and one or more CAF/CRS2 complex are simultaneously bound to the same intron. The genetic and biochemical evidence together show that the CFM2 and CAF members of the CRM domain family play distinct roles in the splicing of their shared intron ligands. It seems likely that CFM2 functions similarly to the closely related protein CRS1. CRS1 binds to its atpF intron ligand through recognition of elements in intron domains 1 and 4 that are predicted to be on the periphery of the folded intron; CRS1 binding reorganizes the RNA such that intron elements expected to be at the catalytic core become less accessible to solvent (Ostersetzer et al., 2005). Preliminary in vitro studies with recombinant CRS2/CAF complexes suggest they interact with intron elements closer to the catalytic center (M. Rojas and A. Barkan, unpublished data). A current working model is that CFM2 acts analogously to CRS1 and promotes intron folding by binding peripheral intron elements, whereas the CRS2/CAF complexes play a distinct and possibly more fundamental role close to the intron's catalytic center.
Figure 8.
Chloroplast Splicing Factors and Their Intron Targets in Land Plants.
The group II introns in land plant chloroplasts are divided into subgroups IIA and IIB according to Michel et al. (1989). Those found in Arabidopsis but not in maize are marked with asterisks. Nucleus-encoded splicing factors, shown to the outside, are annotated with their conserved domains. Solid arrows mark interactions supported both by splicing defects in mutant backgrounds and by RNA coimmunoprecipitation data. Dashed arrows mark interactions supported by either genetic or coimmunoprecipitation data, but not both. Where analyzed (this work; Asakura and Barkan, 2006), the functions of these proteins are conserved between maize and Arabidopsis. A role for the plastid-encoded protein MatK in subgroup IIA splicing has been proposed but has not been confirmed (reviewed in Bonen and Vogel, 2001). HCF152 is an Arabidopsis protein that is required for the accumulation of spliced petB RNA but not for the accumulation of excised intron (Meierhoff et al., 2003). Results are summarized from this work and from Jenkins et al. (1997), Vogel et al. (1999), Till et al. (2001), Ostheimer et al. (2003), Schmitz-Linneweber et al. (2005, 2006), and Watkins et al. (2007).
In addition to its role in splicing orthologs of Zm CFM2's intron ligands, At CFM2 promotes the splicing of clpP-int2, which is not found in maize. A member of the pentatricopeptide repeat family was shown recently to participate in the processing of chloroplast clpP pre-mRNA in moss, but this protein does not have an obvious ortholog in the nuclear genomes of vascular plants (Hattori et al., 2007). The defective seed phenotype conditioned by the strong mutant allele At cfm2-1 can be accounted for by the failure to express clpP, which is essential for cell viability in tobacco (Kuroda and Maliga, 2003). The splicing of clpP-int2 is not disrupted in either At CAF1 or At CAF2 mutants (Asakura and Barkan, 2006), so CFM2 acts independently of CRS2/CAF complexes to promote clpP-int2 splicing, much as CRS1 acts independently of CRS2/CAF complexes to promote atpF splicing. These protein requirements correlate with the structural classifications of these introns: both clpP-int2 and the atpF intron are in subgroup IIA, whereas the CRS2/CAF-dependent introns are in subgroup IIB (Michel et al., 1989). Most subgroup IIA introns in maize chloroplasts also require the RNase III domain protein RNC1 for their splicing (Watkins et al., 2007), so it seems plausible that At RNC1 promotes clpP-int2 splicing in conjunction with At CFM2. Elucidation of the mechanistic basis for the distinct protein requirements of chloroplast introns in subgroups IIA and IIB will require reconstitution of protein-facilitated splicing in vitro, an effort that is facilitated by recent progress in identifying the protein components of chloroplast intron RNPs.
METHODS
Sequence Analysis of Maize cfm2 cDNAs
The nucleotide sequence of Os Cfm2 (Os04g39060) was used to identify closely related maize (Zea mays) cDNAs at the POGs (http://plantrbp.uoregon.edu/) and Plant GDB (http://www.plantgdb.org/) databases. Corresponding maize cDNAs from the Arizona full-length cDNA collection (accession numbers DR794454.1, DT948003.1, DY686659.1, DY619028.1, and DV518296.1) were obtained and sequenced. All of these cDNAs were truncated at the 5′ end. To obtain more upstream sequence, additional cDNAs were amplified by nested PCR from a maize seedling leaf cDNA library (inbred line B73) with the following gene-specific and vector primers: Cfm2, 1st PCR, nsw472crm2 (5′-TGCACCTTCGTAGCGACCAT-3′) and 5′AD+ (5′-TCACTACAGGGATGTTTAATACCACTAC-3′), nested PCR, tk5415cfm23′ (5′-CTCCCAGTCAGACATGGACC-3′) and 5′AD (5′-AGGGATGTTTAATACCACTAC-3′); Cfm2 Alt1, 1st PCR, ac3384cfm2R (5′-ACAAAGGCGGCAGCAAGTTT-3′) and 5′AD+, nested PCR, YA2166Cfm2 3′ (5′-AGGGAAAGGTTGAGGAGAATG-3′) and 5′AD. PCR products were cloned and sequenced. The sequence encoding the N-terminal 97 amino acids was inferred from genomic sequence (GSS Contig ZmGSStuc11-12-04.114819.1; accession numbers CG268073, CG268062, and CW008631). The cDNA sequences matched the genomic sequences perfectly, consistent with their originating from the same gene.
Plant Material
Maize seedlings (Z. mays inbred line B73; Pioneer HiBred) were grown in soil in a growth chamber under a 16-h-light (28°C, 400 μE m−2 s−1)/8-h-dark (26°C) cycle. They were harvested between 7 and 10 d after planting and used for preparation of chloroplasts, stromal extract, genomic DNA, and polyadenylated RNA.
Arabidopsis thaliana insertion lines were obtained from the Salk Institute Genomic Analysis Laboratory: SALK_000703 (At cfm2-1), SAIL_67_E05 (At cfm2-2), and SALK_026861 (At crs1-1). Arabidopsis plants used for RNA extraction were grown on MS plates (0.3% [w/v] Phytagel [Sigma-Aldrich], 1× MS salts [Sigma-Aldrich], 1× Gamborg's B5 vitamin [Sigma-Aldrich], and 2% [w/v] sucrose, pH 5.7). Plants were generally grown under 14-h-light (200 μE m−2 s−1; 23°C)/10-h-dark (21°C) cycles. However, tissue from the rare At cfm2-1 homozygotes that yielded leaves came from seeds germinated and grown under short-day conditions (10 h light/14 h dark).
Transient Expression of CFM2-GFP in Onion Epidermal Cells
Sequences encoding the N-terminal 53 amino acids of maize CFM2 were amplified by nested PCR from maize leaf DNA (inbred line B73) with the following primers: 1st PCR, YAcfm2-F4(NheI) (5′-CCTGCTAGCATGCTCCTCTCATTCTCCCCG-3′) and ac79crm2 (5′-CGCCTCATGTTCATAGCCCA-3′); nested PCR, YAcfm2-F5(SpeI) (5′-CGTACTAGTATGCTCCTCTCATTCTCCCCG-3′) and YAcfm2-R10(KpnI) (5′-CCTGGTACCCGGAGATGCGGCGGAGCGCGGA-3′). The PCR product was digested with SpeI and KpnI and cloned into pOL-LT that was kindly supplied by Ian Small. Biolistic transformation of onion cells and visualization of GFP localization in epidermal tissue were performed as described previously (Barkan et al., 2007).
Production of Anti-CFM2 Antisera
The C-terminal 324 amino acids of maize CFM2 were expressed in Escherichia coli as a His-tagged fusion protein from a pET-28b(+) vector (Novagen). Antibody raised to this fragment was predicted to be specific for CFM2, as its closest match in rice (Oryza sativa; other than Os CFM2) shows only 31% identity over a 101–amino acid region. The expression plasmid was introduced into BL21 Star (DE3) cells (Invitrogen) and induced by the addition of 1 mM isopropyl-1-thio-β-d-galactopyranoside for 4 h at 37°C. The recombinant CFM2 fragment was purified on a Talon metal affinity column (Clontech) according to the instruction manual. Polyclonal antisera were generated in two rabbits at the University of Oregon antibody facility. Antisera were affinity purified against the antigen that had been used for the immunizations coupled to a HiTrap NHS-activated column (GE Healthcare Life Science).
Chloroplast Fractionation and Protein Analysis
Total leaf protein from Arabidopsis was extracted from 2- to 3-week-old seedlings grown on MS medium containing 2% sucrose, as described previously for maize (Barkan, 1998). The maize chloroplast subfractions used here were the same preparation as those described previously (Williams and Barkan, 2003). Antisera against IM35, Cpn60, and ClpP1 were kindly provided by Danny Schnell (University of Massachusetts), Harry Roy (Rensselaer Polytechnic Institute), and Zach Adam (Hebrew University of Jerusalem), respectively. The other antibodies were described previously (Roy and Barkan, 1998).
RIP-Chip and Slot-Blot Hybridization Analyses
RIP-chip and slot-blot hybridization assays of RNAs that coimmunoprecipitate with CFM2 were performed and analyzed as described previously (Schmitz-Linneweber et al., 2005). Each immunoprecipitation used affinity-purified antibodies and 100 μL of stromal extract (∼1 mg of stromal protein). Four biological replicates were performed: two each involving anti-CFM2 antibodies from each of the two immunized rabbits. For the slot blot assays, 1/24th of each pellet sample and 1/24th of each supernatant sample was applied to each slot.
Arabidopsis DNA Extraction and PCR Amplification
Arabidopsis DNA isolation and the At CRS1, At CAF1, and At CAF2 mutants used as controls were described previously (Asakura and Barkan, 2006). T-DNA insertions were confirmed by PCR amplification using T-DNA left border primer LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) for At cfm2-1 or LB (SAIL) (5′-TTCATAACCAATCTCGATACAC-3′) for At cfm2-2. Gene-specific primers were as follows: Atcfm2-1, 3g013-5′ (5′-GACTGGAGGGACTCTGATATCTCGGG-3′) and 3g013-3′ (5′-TAGTATGCCACCACTTTCTGCTTCCA-3′); At cfm2-2, Atcfm2-1 (5′-GTCGAGGACAGCGCAACCAATGAAACTTG-3′) and Atcfm2-2 (5′-CTTTAGCTTTGCGATGACTTCTTGCAC-3′).
Arabidopsis RNA Analysis
RNA was extracted from the leaf tissue of plants grown for 2 to 3 weeks on MS medium containing 2% sucrose using TriZol reagent (Gibco BRL) according to the manufacturer's instructions. RNA gel blot hybridizations and ribonuclease protection assays were performed as described previously (Jenkins et al., 1997). The probes for trnL-UAA were amplified by PCR and RT-PCR from Arabidopsis genomic DNA or RNA with the primers YA At trnL F (5′-GGGGATATGGCGGAATTGGTA-3′) and YA At trnL R (5′-ACTTGAACCCTCACGATTTTA-3′). Poisoned primer extension assays were performed using 10 μg of leaf RNA as described previously (Asakura and Barkan, 2006).
For RT-PCR analysis of At CFM2 mRNA, 200 ng of total leaf RNA was annealed with 5 μM reverse gene-specific primers in 10 μL of annealing buffer (20 mM Tris-HCl, pH 8.5, and 150 mM NaCl) at 95°C for 30 s and cooled on ice. Reverse transcription was initiated by the addition of a 10-μL solution containing 5 units of AMV reverse transcriptase (Promega), 2 mM deoxynucleotide triphosphate, 20 units of RNAsin ribonuclease inhibitor (Promega), and Promega AMV buffer. Reactions were incubated at 48°C for 1 h. Two microliters of each RT reaction was used as template in subsequent PCR reactions (30 μL). Primers used for RT-PCR were as follows: At CFM2 3′ region, Atcfm2-1 (5′-GTCGAGGACAGCGCAACCAATGAAACTTG-3′) and Atcfm2-2 (5′-CTTTAGCTTTGCGATGACTTCTTGCAC-3′); At CFM2, central region, atCfm2F2 (5′-TTATGAGCGGCCTCAATGTCTA-3′ and atCfm2R2 (5′-ATGGGTCTACTTCACCTTCGC-3′); Actin2, YAAtactin2F (5′-GAAGCTCTCCTTTGTTGCTGTT-3′) and YAAtactin2R (5′-TTAGAAACATTTTCTGTGAACGA-3′).
Sucrose Gradient Analysis
Three hundred microliters of maize stromal extract (∼3 mg of protein) supplemented with 120 units of RNAsin (Promega) was fractionated by sedimentation through a 10 to 30% sucrose gradient containing KEX buffer [30 mM HEPES-KOH, pH 8, 150 mM KOAc, 10 mM Mg(OAc)2, and 5 mM DTT] in a Beckman SW55.1 rotor at 50,000 rpm for 4 h at 4°C. RNA was extracted from 300 μL of each fraction by adding 50 μL of 5% SDS and 0.2 M EDTA, followed by phenol/chloroform extraction and ethanol precipitation. An equal proportion of each fraction was used for immunoblot and RNA gel blot analysis. Intron-specific probes for the RNA gel blots were PCR amplified from cloned chloroplast DNA fragments with the following primers: ndhA (5′-CTAGCAATATCTCTACGTGTGATTCG-3′) and (5′-CATAGTCGATGATAACATCACAG-3′); ycf3-int1 (5′-ACTTATTATAGAGATGGTGCGATTTGA-3′) and (5′-GGATTGAGCCAACATCCGTTA-3). Probes were radiolabeled of random hexamer priming in the presence of [α-32P]dCTP.
Accession Numbers
Sequence data for CFM2, CFM2 Alt1, and CFM2 Alt2 cDNAs can be found in the GenBank/EMBL/DDBJ data libraries under accession numbers EU047713, EU047714, and EU047715. The RIP-chip data have been deposited at MIAME-Express under accession numbers E-MEXP-743 and E-MEXP-784 for the CFM2 and control data sets, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Unrooted Tree Demonstrating Orthology among Maize, Rice, and Arabidopsis CFM2.
Supplemental Figure 2. Multiple Sequence Alignments of CFM2 Proteins and CRM Domains.
Supplemental Figure 3. Maize cfm2 cDNA Variants.
Supplemental Figure 4. Chloroplast Localization of CFM2-GFP in Onion Epidermal Cells.
Supplemental Figure 5. T-DNA Insertions in At CFM2.
Supplemental Figure 6. Confirmation of Splicing Defects in At CFM2 Mutants.
Supplemental Table 1. Top-Ranking Fragments in CFM2 RIP-Chip Assays.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the Arabidopsis T-DNA insertion mutants and Christian Schmitz-Linneweber for overseeing the sequencing of maize cfm2 cDNAs. We thank Ian Small for providing plasmids for GFP fusion protein analysis and Danny Schnell, Harry Roy, and Zach Adam for providing antibodies. We also thank Rosalind Williams-Carrier, Margarita Rojas, Tiffany Kroeger, Kenneth Watkins, Jana Prikryl, Susan Belcher, Lisa Smolenska, and Christian Schmitz-Linneweber for technical advice and Kenneth Watkins for comments on the manuscript. This work was supported by Grant DBI-0421799 to A.B. from the National Science Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alice Barkan (abarkan@uoregon.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Asakura, Y., and Barkan, A. (2006). Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 142 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P., and Steitz, T. (2000). The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289 905–920. [DOI] [PubMed] [Google Scholar]
- Barkan, A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297 38–57. [Google Scholar]
- Barkan, A., Klipcan, L., Ostersetzer, O., Kawamura, T., Asakura, Y., and Watkins, K. (2007). The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA 13 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen, L., and Vogel, J. (2001). The ins and outs of group II introns. Trends Genet. 17 322–331. [DOI] [PubMed] [Google Scholar]
- Boudreau, E., Takahashi, Y., Lemieux, C., Turmel, M., and Rochaix, J. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16 6095–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., and von Heijne, G. (2001). Prediction of organellar targeting signals. Biochim. Biophys. Acta 1541 114–119. [DOI] [PubMed] [Google Scholar]
- Halls, C., Mohr, S., Del Campo, M., Yang, Q., Jankowsky, E., and Lambowitz, A.M. (2007). Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 365 835–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori, M., Miyake, H., and Sugita, M. (2007). A pentatricopeptide repeat protein is required for RNA processing of clpP pre-mRNA in moss chloroplasts. J. Biol. Chem. 282 10773–10782. [DOI] [PubMed] [Google Scholar]
- Hsieh, J., Andrews, A.J., and Fierke, C.A. (2004). Roles of protein subunits in RNA-protein complexes: lessons from ribonuclease P. Biopolymers 73 79–89. [DOI] [PubMed] [Google Scholar]
- Huang, H.R., Rowe, C.E., Mohr, S., Jiang, Y., Lambowitz, A.M., and Perlman, P.S. (2005). The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. USA 102 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, B., Kulhanek, D., and Barkan, A. (1997). Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, H., and Maliga, P. (2003). The plastid clpP1 protease gene is essential for plant development. Nature 425 86–89. [DOI] [PubMed] [Google Scholar]
- Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., and Schuster, G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15 1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, F., Umesono, K., and Ozeki, H. (1989). Comparative and functional anatomy of group II catalytic introns - A review. Gene 82 5–30. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Ohta, M., Sugiura, M., and Sugita, M. (1999). Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett. 460 437–441. [DOI] [PubMed] [Google Scholar]
- Ostersetzer, O., Watkins, K., Cooke, A., and Barkan, A. (2005). CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer, G., Barkan, A., and Matthews, B. (2002). Crystal structure of E. coli YhbY: A representative of a novel class of RNA binding proteins. Structure 10 1593–1601. [DOI] [PubMed] [Google Scholar]
- Ostheimer, G., Williams-Carrier, R., Belcher, S., Osborne, E., Gierke, J., and Barkan, A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J. 22 3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer, G.J., Rojas, M., Hadjivassiliou, H., and Barkan, A. (2006). Formation of the CRS2–CAF2 group ll intron splicing complex is mediated by a 22 amino acid motif in the C-terminal region of CAF2. J. Biol. Chem. 281 4732–4738. [DOI] [PubMed] [Google Scholar]
- Pyle, A., and Lambowitz, A. (2006). Group II introns: Ribozymes that splice RNA and invade DNA. In The RNA World, R. Gesteland, T. Cech, and J. Atkins, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 469–506.
- Roy, L.M., and Barkan, A. (1998). A secY homologue is required for the elaboration of the chloroplast thylakoid membrane and for normal chloroplast gene expression. J. Cell Biol. 141 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf, S., Kossel, H., and Bock, R. (1997). Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell 17 2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Williams-Carrier, R.E., Williams-Voelker, P.M., Kroeger, T.S., Vichas, A., and Barkan, A. (2006). A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 Pre-mRNA. Plant Cell 18 2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, D., Fewer, D., Friedl, T., and Bhattacharya, D. (2003). Phylogeny and self-splicing ability of the plastid tRNA-Leu group I Intron. J. Mol. Evol. 57 710–720. [DOI] [PubMed] [Google Scholar]
- Solem, A., Zingler, N., and Pyle, A.M. (2006). A DEAD protein that activates intron self-splicing without unwinding RNA. Mol. Cell 24 611–617. [DOI] [PubMed] [Google Scholar]
- Till, B., Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2001). CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., Boerner, T., and Hess, W. (1999). Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 27 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, K., Kroeger, T., Cooke, A., Williams-Carrier, R., Friso, G., Belcher, S., van Wijk, K.J., and Barkan, A. (2007). A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19 2606–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, K.M. (1997). Protein-facilitated RNA folding. Curr. Opin. Struct. Biol. 7 336–342. [DOI] [PubMed] [Google Scholar]
- Williams, P., and Barkan, A. (2003). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36 675–686. [DOI] [PubMed] [Google Scholar]
- Xu, M.-Q., Kaathe, S., Goodrich-Blair, H., Nierzwicki-Bauer, S., and Shub, D. (1990). Bacterial origin of a chloroplast intron: Conserved self-splicing group I introns in cyanobacteria. Science 250 1566–1569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.