Abstract
Gibberellins (GAs) play many biological roles in higher plants. We collected and performed genetic analysis on rice (Oryza sativa) GA-related mutants, including GA-deficient and GA-insensitive mutants. Genetic analysis of the mutants revealed that rice GA-deficient mutations are not transmitted as Mendelian traits to the next generation following self-pollination of F1 heterozygous plants, although GA-insensitive mutations are transmitted normally. To understand these differences in transmission, we examined the effect of GA on microsporogenesis and pollen tube elongation in rice using new GA-deficient and GA-insensitive mutants that produce semifertile flowers. Phenotypic analysis revealed that the GA-deficient mutant reduced pollen elongation1 is defective in pollen tube elongation, resulting in a low fertilization frequency, whereas the GA-insensitive semidominant mutant Slr1-d3 is mainly defective in viable pollen production. Quantitative RT-PCR revealed that GA biosynthesis genes tested whose mutations are transmitted to the next generation at a lower frequency are preferentially expressed after meiosis during pollen development, but expression is absent or very low before the meiosis stage, whereas GA signal-related genes are actively expressed before meiosis. Based on these observations, we predict that the transmission of GA-signaling genes occurs in a sporophytic manner, since the protein products and/or mRNA transcripts of these genes may be introduced into pollen-carrying mutant alleles, whereas GA synthesis genes are transmitted in a gametophytic manner, since these genes are preferentially expressed after meiosis.
INTRODUCTION
Gibberellins (GAs) are tetracyclic diterpenoid compounds, some of which function as endogenous plant growth regulators. Phenotypic analyses of mutants with reduced GA production and responses to GA have revealed that active GAs play an essential role in many aspects of plant growth and development, including seed germination, leaf and stem elongation, flower induction and development, and fruit and seed development (Davies, 2005; Fleet and Sun, 2005). Reproductive development is one of the most important physiological events in which GAs are involved (Pharis and King, 1985; King and Evans, 2003). For example, Arabidopsis thaliana GA-deficient and GA-insensitive mutants have demonstrated that GA synthesis and signaling are important for flower induction and the development of flower organs, such as petals and stamens (Wilson et al., 1992; Goto and Pharis, 1999; Dill and Sun, 2001; Cheng et al., 2004). There is also evidence that active GA synthesis and signaling are essential for anther development. For example, the Arabidopsis mutant ga1 (a loss-of-function mutant in copalyl diphosphate synthase) has poorly developed anthers (Koornneef and van der Veen, 1980). In the anthers of this mutant, microsporogenesis occurs but the pollen grains are not viable (Goto and Pharis, 1999). Loss-of-function mutants in DELLA proteins, which are negative regulators in GA signaling, partially rescue the impaired development of floral organs in ga1, demonstrating that GA promotes these events by countering the function of DELLA proteins (Cheng et al., 2004; Tyler et al., 2004). Pollen development is also defective in the tomato (Solanum lycopersicum) GA-deficient mutants gib-1 (a loss-of-function mutant in copalyl diphosphate synthase) and gib-2 (a loss-of-function mutant in kaurenoic acid oxidase) (Nester and Zeevaart, 1988; Jacobsen and Olszewski, 1991). By contrast with At ga1, the development of the anthers in the tomato gib mutants is arrested at an earlier developmental stage. Similarly, a rice (Oryza sativa) loss-of-function mutant in GAMYB, which functions as a positive transacting factor in GA signaling in cereal aleurone cells, shows some defects in anther and pollen development (Kaneko et al., 2004). Murray et al. (2003) also reported that the overproduction of barley (Hordeum vulgare) GAMYB in transgenic barley causes abnormal anther development (decreased anther length, lighter anther color, lack of anther dehiscence, and male sterility). These observations demonstrate that defects in GA synthesis and GA signaling block anther and pollen development.
In addition, there have been some reports of the enhancement of pollen tube growth in vitro following GA application (Bhandal and Malik, 1979; Viti et al., 1990). Using transgenic Arabidopsis overexpressing a pea (Pisum sativum) cDNA encoding the GA-inactivating enzyme Ps GA2ox2, Swain and colleagues investigated the role of GAs in pollen tube growth (Singh et al., 2002; Swain et al., 2004). The growth of pollen tubes carrying the 35S-PsGA2ox2 transgene was lower than that of nontransformed pollen tubes. This reduced pollen tube growth in the 35S-PsGA2ox2 overproducers was partially rescued by GA treatment in vitro; by GA hypersensitive mutations, such as spy-5, sly1gar2, and rga; or by an RNA interference silencing construct targeting PsGA2ox2. Furthermore, the in vitro treatment of wild-type pollen tubes with the GA biosynthesis inhibitor uniconazole retarded pollen tube elongation. These observations strongly suggest that GAs are important for normal pollen tube growth, although the known GA biosynthesis and GA response mutants in various species have not been reported to exhibit an obvious pollen tube elongation phenotype (Singh et al., 2002).
Rice is a monocot model plant for which much information and material is available, including the whole-genome sequence, full-length cDNA clones, transformation systems, and many mutant collections. Taking advantage of these materials, we have performed extensive screens and characterization of GA-deficient and GA-insensitive mutants. In a search for GA-deficient mutants, we identified 18 mutants at six different loci that encode the GA metabolic enzymes ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO), ent-kaurenoic acid oxidase (KAO), GA 20-oxidase, and GA 3-oxidase (Sakamoto et al., 2004). Based on these mutants and the expression patterns of the corresponding genes in the wild type, we demonstrated that the enzymes that catalyze early steps in the GA biosynthetic pathway (CPS, KS, KO, and KAO) are encoded by single genes (Os CPS1, KS1, KO2, and KAO, respectively). Knockout mutations of these genes induce severe dwarf phenotypes and are severely defected in flower and seed development (Sakamoto et al., 2004). We also isolated three GA-insensitive mutants: gid1, which is defective in a soluble GA receptor (Ueguchi-Tanaka et al., 2005); gid2, which is defective in the degradation of a rice DELLA protein (SLR1) that is involved in GA signaling (Sasaki et al., 2003); and gamyb, which is defective in a positive transcription factor in GA signaling (Kaneko et al., 2004). We further isolated a mutant that constitutively responds to GA, slender rice1 (slr1; Ikeda et al., 2001; Itoh et al., 2002).
In the process of the genetic analysis of these rice GA-related mutants, we noticed a difference in the genetic frequency of transmission of mutant alleles (transmission frequency) of GA-deficient and GA-insensitive genes. For example, the frequency of a homozygous mutation of Os CPS1 (Os cps1) was less than a few percent in F2 segregants derived from self-pollinated F1 plants heterozygous in the allele, whereas that of the gid1 mutation almost fit Mendelian rules (see below). The differences in the transmission frequencies of GA-deficient and GA-insensitive mutations led us to speculate that a novel mechanism regulates GA signaling during the rice fertilization process. In this study, we attempted to determine why such differences between GA-deficient and GA-insensitive mutations occur. We isolated two new GA-deficient and GA-insensitive mutants of intermediate severity; plants of each line produce a few seeds following self-pollination. Analysis of these mutants revealed that the low transmission frequency of the GA-deficient mutation is caused by impaired pollen germination and pollen tube growth, whereas the GA-insensitive mutant is mainly defective in pollen developmental processes. Based on these observations, we discuss two different critical steps involved in GA signaling and GA biosynthesis in the process of pollen development.
RESULTS
Differences in the Segregation Frequencies of GA-Deficient and GA-Signaling Mutants
Table 1 shows the segregation ratios of a GA-deficient mutant and a GA-insensitive mutant at the F2 generation stage of self-pollinated F1 plants carrying heterozygous alleles. The GA-deficient mutant tested was Os cps1-1, which contains a severe mutation in the gene encoding CPS, and the GA-signaling mutant was gid1-3, a severe mutation in the GA receptor, because these mutants show the typical severe phenotypes of GA-related mutants (Sakamoto et al., 2004; Ueguchi-Tanaka et al., 2005). The F2 segregation of the GID1 locus almost fit the expected ratio, 1 (GID1/GID1):2 (GID1/gid1):1 (gid1/gid1), indicating that the gid1 mutation is transmitted to the next generation in a normal manner. By contrast, the segregation of the cps1 locus was apparently distorted, with the frequency of the cps/cps genotype ∼3% and that of the CPS1/CPS1 genotype >40% (Table 1). Such low transmission of the mutant allele in the next generation was observed not only in cps1-1 but also in other GA-deficient mutants (see below).
Table 1.
Segregation of F2 Plants Derived from Self-Pollinated F1 Plants Heterozygous at the GID1 (GA Receptor) Locus or the Os CPS1 (GA Synthesis Enzyme) Locus
| GID1/gid1-3 × GID1/gid1-3 Plant Number | Genotype
|
||||
|---|---|---|---|---|---|
| gid1-3/gid1-3 | GID1/gid1-3 | GID1/GID1 | Total | χ2 (1:2:1) | |
| 1 | 22 (31.9%) | 32 (46.4%) | 15 (21.7%) | 69 | 1.78NS |
| 2 | 22 (24.4%) | 47 (52.2%) | 21 (23.3%) | 90 | 0.20NS |
| 3 | 13 (21.7%) | 29 (48.3%) | 18 (30.0%) | 60 | 0.90NS |
| CPS1/cps1-1 × CPS1/cps1-1 Plant Number | Genotype
|
||||
| cps1-1/cps1-1 | CPS1/cps1-1 | CPS1/CPS1 | Total | χ2 (1:2:1) | |
| 1 | 3 (2.8%) | 56 (51.9%) | 49 (45.4%) | 108 | 39.33a |
| 2 | 3 (3.1%) | 50 (52.6%) | 42 (44.2%) | 95 | 32.28a |
Significant at the 0.5% level.
Numbers in parentheses indicate the percentage relative to the total number of plants. NS, not significant.
Isolation and Characterization of the GA-Deficient Mutant rpe1
To study the mechanism of the distorted transmission of GA deficiency mutations, we isolated a new GA-deficient mutant of intermediate severity, since previously isolated GA-deficient mutants barely develop floral organs. From >100 semidwarf mutants, we selected one mutant that was produced using ethylene imine treatment. The frequency of the dwarf phenotype in the F2 progenies of heterozygous plants did not agree with the expected 3:1 ratio (P < 0.005): the phenotype was found in ∼6% of the F2 plants (Table 2). To investigate whether the male or female gametophyte is defective in this mutant, we artificially crossed the mutant with the cultivar from which it was derived, Fujiminori, using emasculated flowers. When the mutant was used as the female parent (pollen receiver), we found that 70 to 80% of the seeds were fertile, and fertile seeds were produced at a similar frequency following artificial self-pollination of the original cultivar (Table 3). However, when the mutant was used as a male parent, only 6 to 17% of the seeds were fertile, indicating that this mutation caused a defect in the male gametophyte. The mutant exhibited reduced pollen germination and elongation (see below). We designated this mutant reduced pollen elongation1 (rpe1).
Table 2.
Segregation of F2 Plants Derived from Self-Pollinated F1 Plants Heterozygous at the RPE1 Locus
| RPE1/rpe1 × RPE1/rpe1 Plant Number | Phenotype
|
|||
|---|---|---|---|---|
| Wild Type | rpe1 | Total | χ2 (3:1) | |
| 1 | 182 (93.8%) | 12 (6.2%) | 194 | 35.63a |
| 2 | 190 (93.5%) | 13 (6.4%) | 203 | 36.45a |
| 3 | 258 (93.8%) | 17 (6.2%) | 275 | 50.94a |
Significant at the 0.5% level.
Numbers in parentheses indicate the percentage relative to the total number of plants.
Table 3.
Reciprocal Crossing of rpe1 and Wild-Type Plants
| WT(♀) × WT(♂) | Fertilea | Infertilea | Total |
|---|---|---|---|
| 1 | 15 (78.9%) | 4 (21.1%) | 19 |
| rpe1(♀)× WT(♂) | |||
| 1 | 10 (76.9%) | 3 (23.1%) | 13 |
| 2 | 17 (68%) | 8 (32.0%) | 25 |
| WT(♀)× rpe1(♂) | |||
| 1 | 1 (6.3%) | 15 (93.8%) | 16 |
| 2 | 4 (14.3%) | 24 (85.7%) | 28 |
| 3 | 2 (16.7%) | 10 (83.3%) | 12 |
Flowers that set seed were judged as fertile, and those that failed were judged as infertile.
Numbers in parentheses indicate the percentage relative to the total number of spikelets. The percentage of success of artificial crossing of rice is ∼80%. WT, wild type.
Next, we confirmed that the abnormal phenotypes of rpe1 are caused by a defect in GA biosynthesis. Using 30 dwarf F2 plants of a cross between rpe1 (japonica) and Kasalath (indica), we analyzed the linkage between the RPE1 locus and molecular markers corresponding to four GA synthesis genes, Os CPS1, KS1, KO2, and KAO, all of which have already been mapped and characterized (Sakamoto et al., 2004). GA synthesis requires multiple steps catalyzed by six different kinds of enzymes, but we did not examine the genes encoding two enzymes that catalyze later GA biosynthesis steps, GA20 oxidase and GA3 oxidase, because previous studies had revealed that defects in these enzymes do not affect fertility (Sakamoto et al., 2004). The Os KAO molecular marker was completely linked with the rpe1 mutation, suggesting that RPE1 is identical to Os KAO. We then examined the sequence of Os KAO in the rpe1 mutant and found one nucleotide change in exon 2 that causes a conserved amino acid, Leu, to be replaced with Phe (see Supplemental Figure 1A online).
To confirm that KAO corresponds to the rpe1 locus, we performed a complementation test. A 9.4-kb DNA fragment containing the entire KAO sequence, including ∼3.0 and ∼1.0 kb of the 5′- and 3′-flanking regions, respectively, was introduced into rpe1 by Agrobacterium tumefaciens–mediated transformation. The dwarf phenotype of all of the plants that were resistant to hygromycin, the selection marker used for transformation, was rescued (see Supplemental Figure 1B online). We had previously isolated knockout alleles of KAO that exhibit a severe dwarf phenotype and barely develop floral organs (Sakamoto et al., 2004). These observations indicate that the rpe1 mutant phenotype is caused by a partial defect in the functioning of KAO.
rpe1 Develops Normal Flowers but Shows Impaired Pollen Germination and Elongation
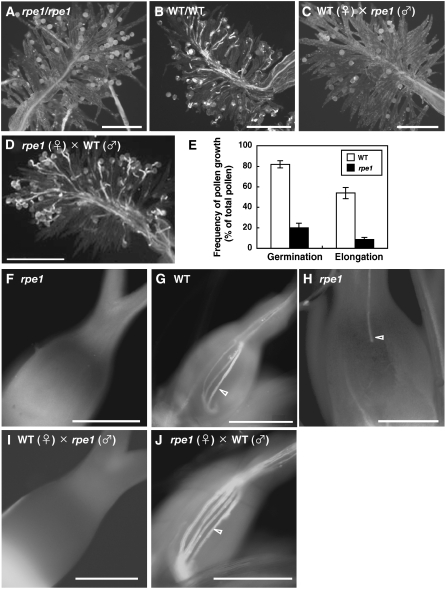
rpe1 exhibits a typical GA-related dwarf phenotype (Figure 1A). The mutant develops normal flowers with normal pistils and stamens (Figures 1B to 1D). Although the pollen viability and the number of mature pollen grains in rpe1 are similar to those of the wild type (Figure 1E, Table 4), the seed viability is ∼40% compared with the viability of the wild type of >90% (Table 4). We examined the germination and elongation of pollen by staining pollinated stigmas with aniline blue (Figure 2). At 30 min after artificial self-pollination, ∼80 and 50% of the wild-type pollen grains had germinated and elongated (Figures 2B and 2E). By contrast, ∼20% of the rpe1 pollen grains had germinated, and <10% had elongated (Figures 2A and 2E). Impaired pollen germination and elongation were also observed following pollination of wild-type stigma with rpe1 pollen (Figure 2C), whereas wild-type pollen germinated normally and elongated on the rpe1 stigma (Figure 2D), indicating that the impaired pollen germination and elongation is due to a defect in rpe1 pollen. At 2 h after pollination, several elongated wild-type pollen tubes were observed in the ovary, and some had reached the wild-type ovule (arrowhead in Figure 2G), but no elongated rpe1 pollen tubes were seen in the mutant ovary (Figure 2F). However, after 4 h, elongating pollen tubes were sometimes observed to reach the ovule in the mutant ovary (arrowhead in Figure 2H). This indicates that the rpe1 mutation does not completely abolish pollen germination or elongation but does severely disturb these processes. This observation is also consistent with the semifertility of rpe1 (Table 2). We confirmed by reciprocal crossing that this impaired pollen elongation is due to the rpe1 mutation (Figures 2I and 2J).
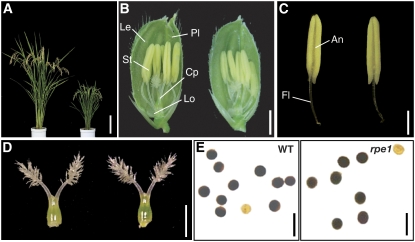
Figure 1.
Phenotype of the rpe1 Mutant.
(A) Gross morphology of wild-type Fijiminori (left) and the rpe1 mutant (right) at the ripening stage. Bar = 30 cm.
(B) Flowers of the wild type (left) and rpe1 (right). Cp, carpel; Le, lemma; Lo, lodicle; Pl, palea; St, stamen. Bar = 2 mm.
(C) Mature stamens of the wild type (left) and rpe1 (right). An, anther; Fl, filament. Bar = 1 mm.
(D) Pistils of the wild type (left) and rpe1 (right). Bar = 1 mm.
(E) Pollen grains stained with I2-KI solution. Wild type (left) and rpe1 (right). Pollen grains staining black were judged as viable, and those staining yellow or light red were judged as sterile. Bars = 100 μm.
Table 4.
Pollen and Seed Viability in the rpe1 Mutant at the Mature Stage
| Wild Type (Fujiminori) | rpe1 | Wild Type (T65) | Slr1-d3 | |
|---|---|---|---|---|
| Viable pollen (%)a | 91.61 ± 7.04 | 88.57 ± 6.52 | 90.5 ± 0.84 | 29.20 ± 4.51* |
| Number of pollen grains | 4317 ± 257 | 4300 ± 427 | 4875 ± 730 | 4592 ± 474 |
| Seed viability (%)b | 92.30 ± 0.97 | 42.39 ± 2.38* | 82.0 ± 1.27 | 56.0 ± 3.03* |
Pollen grains staining black with I2-KI solution were judged as viable, and those staining yellow or light red were considered sterile. The percentage of viable pollen was calculated relative to the total pollen counted. *Significantly different from the wild type (P < 0.01).
Seed viability represents the percentage of spikelet number that set seed per total number.
Figure 2.
Germination and Elongation of rpe1 Pollen.
(A) to (D) and (F) to (J) Growth of pollen of rpe1 ([A], [C], [F], [H], and [I]) and the wild type ([B], [D], [G], and [J]) on rpe1 ([A], [D], [F], [H], and [J]) or wild-type stigmas ([B], [C], [G], and [I]). Pollen grains were stained with aniline blue at 30 min ([A] to [D]), 2 h ([F], [G], [I], and [J]), or 4 h (H) after artificial pollination. Arrowheads indicate pollen tube elongation. Bars = 200 μm.
(E) Germination and elongation frequencies of wild-type and rpe1 pollen. Open and closed bars represent the frequencies of wild-type and rpe1 pollen germination or elongation, respectively. Germination and elongation frequencies were estimated by staining with aniline blue at 30 min and 2 h after self-fertilization, respectively.
GA Is Essential for the Germination and Elongation of Rice Pollen
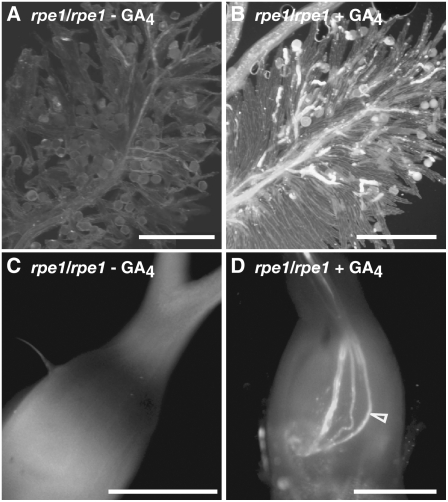
We confirmed that the impaired germination and elongation of rpe1 pollen is caused by a deficiency of GA. Pretreatment of rpe1 stigmas with 10−7 M GA4 before self-pollination resulted in pollen germination and elongation (Figures 3A and 3B), and some pollen tubes had reached the ovule by 2 h after pollination, as observed with wild-type pollen (cf. Figures 3D with 3C and 2G).
Figure 3.
Effect of GA on Pollen Tube Elongation.
Growth of pollen from rpe1 ([A] to [D]) on its own stigmas that were left treated with mock ([A] and [C]) or with GA4 ([B] and [D]). Pollen grains were stained with aniline blue at 30 min ([A] and [B]) or 2 h ([C] and [D]) after artificial pollination. Emasculated wild-type flowers with stamens removed were pretreated with GA4 for 30 min before pollination. Arrowhead indicates pollen elongation. Bars = 200 μm.
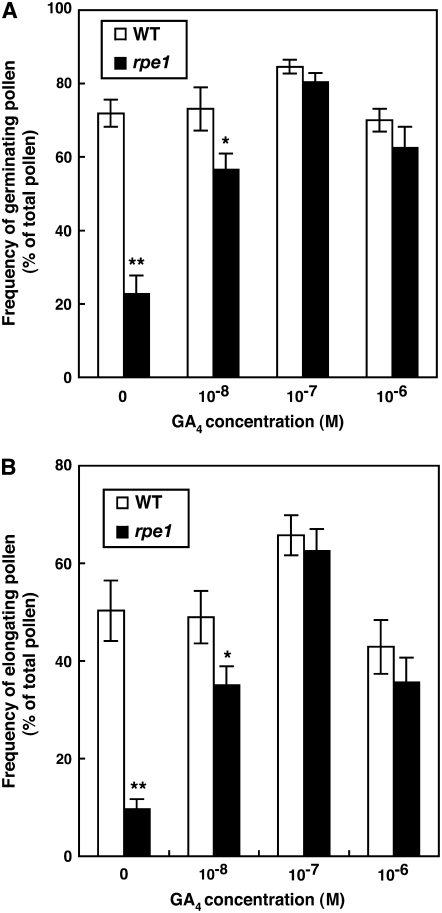
We also investigated the GA dose dependency of pollen germination and elongation. Both events were increased by an increase in the GA4 concentration to 10−7 M and then decreased by treatment with 10−6 M GA4 (Figure 4). With 10−7 M GA4, the frequencies of germinating and elongating pollen in the mutant were similar to those in the wild type, indicating that the impaired germination and elongation of pollen in rpe1 depends completely on a defect in GA synthesis. Interestingly, in both wild-type and mutant flowers treated with 10−6 M GA4, the frequencies of both events were lower than those observed following treatment with 10−7 M GA4. This result shows that a higher GA4 concentration has an inhibitory effect on these events (see Discussion). Similar observations of high GA concentrations inhibiting wild-type pollen tube elongation have been reported (Singh et al., 2002).
Figure 4.
Dose-Dependent Effect of GA4 on Pollen Germination and Elongation.
Frequencies of pollen germination (A) and elongation (B) were estimated by counting the germinated and elongated pollen grains stained with aniline blue after 30 min or 2 h, respectively, on artificially pollinated wild-type stigmas treated with various concentrations of GA4. The GA4 treatment procedure was the same as in Figure 3. Open and closed bars represent the frequencies for wild type and rpe1 pollen, respectively. * and ** indicate significant differences at the 5 and 1% levels, respectively, as judged using the Student's t test.
Isolation and Characterization of a New GA-Insensitive Mutant, Slr1-d3
The above results with rpe1 clearly demonstrate that a defect in GA synthesis causes impaired pollen germination and elongation. We next attempted to observe the reproductive process in a GA-insensitive mutant. One mild GA-insensitive dwarf mutant from our collection of mutants, gid1-8, which is partially defective in the GID1 gene, shows a semidwarf phenotype and has normally developed floral organs (Figure 5). However, as this mutant has normal fertility, it was unsuitable for the above purpose. Another GA-insensitive mutant, gid1-7, has a more severe phenotype than gid1-8, as it is completely defective in pollen development (Figure 5); therefore, this mutant was also unsuitable for our experiment. Thus, we performed a screen for a new GA-insensitive mutant with semidwarfism and semifertility and found one candidate that fulfilled these conditions.
Figure 5.
Phenotypes of GA-Insensitive Mutants with Different Levels of Impairment.
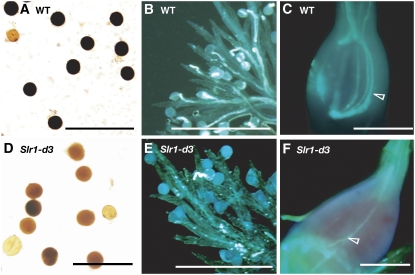
Gross morphology at the ripening stage. Flower, stamen, and pistil of the wild type (T65), gid1-8, Slr1-d3, and gid1-7 (left to right). Wild-type and gid1-8 produce fertile flowers, Slr1-d3 produces semifertile flowers, and gid1-7 does not produce fertile flowers.
This new mutant has a semidwarf phenotype with a height similar to that of rpe1 at the heading stage (Figure 5). The mutant develops flowers with normal stamens and pistils (Figure 5). Molecular characterization of this mutant revealed that the mutation is located on the SLR1 locus and results in an amino acid substitution of Phe for Leu-99 in its TVHYNP domain (see Supplemental Figure 2 online). The SLR1 locus encodes a DELLA protein, a suppressor protein in GA signaling, and its GA-dependent degradation is essential for GA action in rice (Ikeda et al., 2001; Itoh et al., 2002). In Arabidopsis, wheat (Triticum aestivum), maize (Zea mays), and rice, some lines with mutations in DELLA genes that cause in-frame deletions or amino acid exchanges in conserved domains, such as the DELLA and TVHYNP domains, produce mutant proteins that constitutively suppress GA action, even under high GA conditions (Peng et al., 1997, 1999; Itoh et al., 2002). Similarly, the semidwarf phenotype of the new mutant was inherited in a semidominant fashion. These results indicate that the mutant carries a gain-of-function allele of the SLR1 locus whose product functions constitutively to partially suppress GA action. We named this mutant Slr1-d3 and used homozygous plants for further studies.
Slr1-d3 is semifertile, even though it develops normal flowers with morphologically normal stamens and pistils (Figure 5). The anthers of Slr1-d3 appear normal and produce a similar number of pollen grains per spikelet as those of the cultivar from which it was derived, T65 (Table 4). However, the viability of the mutant pollen is lower than that of wild-type pollen (Figures 6A and 6D, Table 4). Interestingly, however, when we examined the germination and elongation of Slr1-d3 pollen at 30 min after pollination by the same way as rpe1, the frequencies were much lower than those of the wild type, while the elongation was almost similar to those of the wild type and much faster than those of rpe1 (cf. Figures 6E with 2A and 6B). Two hours after artificial self-pollination, an elongated pollen tube was sometimes observed to reach the ovule in self-pollinated Slr1-d3 ovary, but elongated pollen tubes were observed more frequently in the wild-type ovary (cf. Figures 6C and 6F). These findings suggest that the semifertility of the Slr1-d3 mutant is mainly caused by a high frequency of nonviable pollen rather than by impaired pollen tube growth.
Figure 6.
Viability, Germination, and Elongation of Slr1-d3 Pollen.
(A) and (D) Viability of wild-type T65 (A) and Slr1-d3 (D) pollen. Pollen grains were stained with I2-KI solution. Pollen grains staining black were judged as viable, and those staining yellow or light red were considered sterile.
(B), (C), (E), and (F) Growth of pollen of the wild type ([B] and [C]) and Slr1-d3 ([E] and [F]) on their own stigmas. Pollen was stained with aniline blue after 30 min ([B] and [E]) or 2 h ([C] and [F]). Artificial pollination was performed by hand. Arrowheads indicate pollen elongation.
Bars = 200 μm.
Why Do rpe1 and Slr1-d3 Show Different Phenotypes in the Pollination Process?
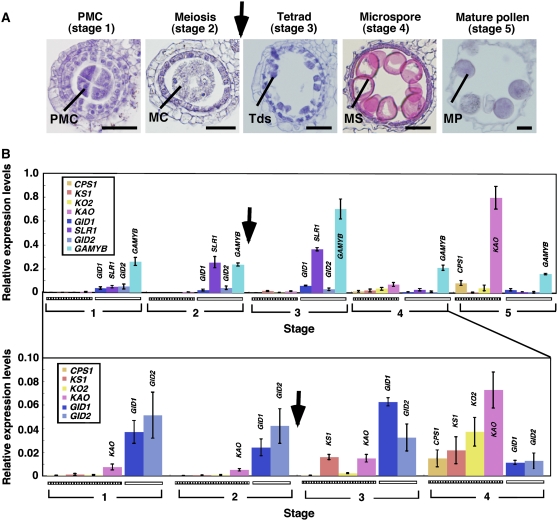
We next examined the expression profiles of GA-signaling and GA synthesis genes at various anther developmental stages. As mentioned above, rice knockout mutants in genes encoding enzymes that catalyze early steps in the GA synthesis pathway (Os cps1, ks1, ko2, and kao) show severe defects in flower and seed development, whereas those catalyzing later steps (sd1 and d18) show normal flower and seed development (Itoh et al., 2001; Ashikari et al., 2002; Sasaki et al., 2002, Sakamoto et al., 2004). Thus, we focused on expression analysis of four of the genes that encode early enzymes in the pathway: CPS1, KS1, KO2, and KAO. We also analyzed the expression of four GA signaling–related genes: GID1, GID2, SLR1, and GAMYB. Based on previous observations by Itoh et al. (2005), we divided the process of rice anther development into five stages: establishment of pollen mother cells (stage 1), meiosis (stage 2), tetrad (stage 3), microspore (stage 4), and mature pollen (stage 5) (Figure 7A). We performed real-time RT-PCR analysis using RNA isolated from whole anthers at various stages but not from pollen or pollen mother cells because of difficulty of isolation of some amounts of RNAs. The results of real-time RT-PCR revealed that all of the GA synthesis–related genes examined were predominantly expressed at stage 3 or later, which corresponds to the postmeiotic stage (Figure 7B). High level expression of CPS1 and KAO was seen in the mature pollen stage (stage 5), whereas expression level of KS1 was lower at stage 5 than that at stage 4. High level expression of CPS1 in mature pollens has also been observed by in situ hybridization analysis (Fukuda et al., 2004). At the premeiotic stage, however, no (CPS1, KS1, and KO2) or low (KAO) expression was observed (Figure 7B). By contrast, all of the GA-signaling genes tested were expressed at high level at the premeiotic stage, and their expression was rapidly decreased after stage 3 (Figure 7B).
Figure 7.
Expression Pattern of GA Synthesis and GA-Signaling Genes in the Process of Anther Development.
(A) Anther development of wild-type rice at various stages. Each developmental stage was scored according to the length of the lemma: pollen mother cell stage (stage 1, lemma 2 mm), meiosis stage (stage 2, 2 to 3 mm), tetrad stage (stage 3, 3 to 5 mm), microspore stage (stage 4, 5 to 8 mm), and mature pollen stage (stage 5, ∼8 mm). Arrow indicates the timing of cytokinesis of meiosis. PMC, pollen mother cell; MC, meiocyte; Tds, tetrads; MS, microspore; MP, mature pollen. Bars = 25 μm.
(B) Quantitative RT-PCR analysis of GA biosynthesis and GA-signaling genes at various anther developmental stages. The numbers 1 to 5 correspond to the anther developmental stages. Relative mRNA level was determined by normalizing the PCR threshold cycle number of each gene with that of the Actin1 reference gene, and data were the average of three replicates. The top panel represents the relative expression levels of all genes tested at stages 1 to 5. The bottom panel shows the enlarged view of the expression levels of genes with lower expression at stages 1 to 4. The striped and open boxes represent the GA synthesis and GA-signaling genes, respectively. Arrows indicate the timing of cytokinesis of meiosis. Error bars represent se of three biological replicates.
We also examined the segregation ratio in progenies of heterozygous plants of GA-deficient (cps1, ks1, ko2, and kao) and GA-insensitive (gid1-1, gid2-2, slr1-1, Slr1-d3, and gamyb-2) mutants (Table 5). As previously mentioned, the segregation of all of the GA-insensitive mutants almost fit the expected ratio with one exception of Slr1-d3 (see Discussion). However, the segregation of all of the GA-deficient mutants examined was apparently distorted. Interestingly, the segregation frequencies of the GA-deficient mutants varied widely. For example, that of kao was ∼8%, whereas that of cps1 was 2 to 3% and that of ko2-1 was <1%. We also confirmed the statistical significance of these segregation frequencies in GA-deficient mutants using Tukey's method (Table 5). This statistical analysis confirmed that the transmission of the kao-1 mutation was significantly higher than that of cps1-1, cps1-2, and ko2 mutations and that of ko2-1 was significantly lower than that of kaos and ks1-2.
Table 5.
Genetic Analysis of GA-Deficient and GA-Signaling Mutants
| Wild-Type Phenotype | Mutant Phenotype | Total | Mutant (%) | χ2 (3:1) | |
|---|---|---|---|---|---|
| cps1-1 | 544 | 13 | 557 | 2.3cd | 151.41* |
| cps1-2 | 429 | 13 | 442 | 2.9bcd | 113.53* |
| ks1-1 | 255 | 10 | 265 | 3.8abcd | 62.55* |
| ks1-2 | 475 | 26 | 501 | 5.2abc | 103.81* |
| ko2-1 | 237 | 2 | 239 | 0.8d | 73.14* |
| ko2-2 (d35) | 485 | 82 | 567 | 14.5 | 33.02* |
| kao-1 | 265 | 23 | 288 | 8.0a | 43.56* |
| kao-3 | 178 | 15 | 193 | 7.8ab | 29.64* |
| gid1-1 | 240 | 75 | 315 | 23.8 | 0.18NS |
| gid2-2 | 195 | 80 | 275 | 29.1 | 2.24NS |
| slr1-1 | 227 | 71 | 298 | 23.8 | 0.16NS |
| Slr1-d3 | 410** | 81 | 491 | 16.5 | 18.48* |
| gamyb-2 | 67 | 25 | 92 | 27.2 | 0.13NS |
The letters a to d denote statistically significant differences (P < 0.05) according to the χ2 goodness-of-fit test followed by the Tukey's wholly significant difference test. NS, not significant; *, significant at the 0.5% level; **, included wild (SLR1/SLR1) and mild dwarf (Slr1-d3/SLR1) phenotypes.
There is a correlation between the transmission frequency of these mutations and their expression patterns during the process of pollen development. For example, KAO, whose mutation showed the highest transmission among these mutants, was expressed most rapidly during the developmental process in these genes, whereas CPS1 and KO2, whose mutations showed lower transmission, were expressed only at late stages (Figure 7B). The correlation between the expression pattern and the transmission frequency suggests that gene expression around the meiotic stage is critical in determining the transmission frequencies of mutant alleles of GA synthesis genes (see Discussion). Furthermore, it is noteworthy that the transmission frequency of a mild allele of kao, rpe1 (Table 2), is similar to those of corresponding null alleles, kao-1 and kao-3 (Table 5). This suggests that the difference in the KAO enzymatic activities of the mild allele (rpe1) and the strong alleles (kao-1 and -3) is irrelevant in the process of pollen germination and elongation, even though such differences clearly reflect a difference in the severities of their dwarfism. Similarly, a mild allele of ko2, d35Tan-Ginbozu (ko2-2), which is a well-known semidwarf mutant of a high-yielding variety and therefore exhibits very good fertility (Itoh et al., 2004), showed a lower segregation ratio (∼15%) in its F2 progeny. This suggests that even a mild mutation that is thought to induce no abnormality except dwarfism also causes a defect in pollen germination and elongation. In other words, pollen germination and elongation might be very sensitive to the GA level, as are leaf and stem elongation (see Discussion).
DISCUSSION
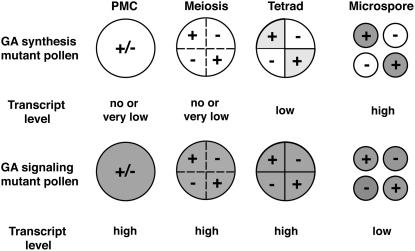
In this study, we attempted to reveal the molecular mechanisms of differences in the transmission frequencies of GA-deficient and GA-insensitive mutations in the F2 segregation stage. For this purpose, we isolated new mutants, the GA-deficient rpe1 and the GA-insensitive Slr1-d3. Phenotypic analyses of these mutants, as well as genetic analyses of previously isolated GA-related mutants, demonstrated that even though both GA-deficient and GA-insensitive mutations affect pollen activity and consequently induce a male-sterile phenotype, GA-deficient mutants are mainly defective in pollen germination and elongation and GA-insensitive mutants are mainly defective in pollen development. Expression analysis revealed that the expression of genes involved in GA signaling, such as GID1, GID2, SLR1, and GAMYB, actively occurs at the premeiosis stage in the pollen developmental process, whereas the expression of genes involved in GA synthesis, such as CPS1, KS1, KO2, and KAO, occurs preferentially after meiosis (Figure 7B). Taking these observations together, we predict that differences in the transmission frequencies of GA-deficient and GA-insensitive mutations depend on differences in the expression levels of these genes in the pollen developmental process (Figure 8). The GA-signaling genes are actively expressed in anthers, specifically in pollen mother cells and the tapetum (Kaneko et al., 2003, 2004; Tsuji et al., 2006), before meiosis, and the mRNA and/or protein products of these genes might be introduced into pollen carrying mutant alleles. Consequently, the transmission of the GA-signaling genes occurs in a sporophytic manner. By contrast, the GA synthesis genes are preferentially expressed after meiosis; therefore their transmission depends on the genotypes of each pollen grain in a gametophytic manner. The correlation between the mRNA levels of the GA synthetic genes in the pollen developmental process and their transmission frequencies supports this hypothesis.
Figure 8.
Model to Explain Differences in the Transmission Frequencies of GA-Deficient and GA-Insensitive Mutations.
Pollen carrying GA-deficient mutations, which are poorly transmitted, does not synthesize GA, which is essential for pollen tube germination and elongation, because the expression of these genes occurs after meiosis. By contrast, GA signaling functions normally in pollen carrying GA-insensitive mutations because the GA-signaling genes are actively expressed just before meiosis; therefore, their products should be transported into pollen carrying mutant alleles. +/− indicates a heterozygous genotype of pollen mother cells, whereas + and − indicate the wild-type and mutant alleles, respectively, of pollen cells. Gray shading indicates the expression of GA synthesis or GA-signaling genes in pollen mother cells or pollen.
De Novo Synthesis of GA Is Essential for Pollen Germination and Elongation
Examination of the rpe1 phenotype demonstrated that pollen germination and elongation essentially depend on the de novo synthesis of GA in rice (Figure 2). The dependence of pollen germination and elongation on GA was also supported by experiments with exogenous treatments of GA4 (Figure 3). Swain and colleagues reported that GA plays a physiological role in Arabidopsis pollen tube growth, based on experiments using a transgenic line carrying a cDNA encoding the pea GA-inactivating enzyme GA2 oxidase2 under the control of the 35S promoter (Singh et al., 2002; Swain et al., 2004). The authors observed that the growth of pollen tubes carrying the transgene was lower than that of normal pollen, and the impaired pollen tube growth was partially reversed by GA application in vitro or by combination with mutations that cause an increased GA response, such as spy-5, sly1gar2, or rga (Singh et al., 2002; Swain et al., 2004). The inhibitory effect of uniconazole on pollen tube growth in vitro also supported their hypothesis (Singh et al., 2002). Although some studies of the role of GA in pollen tube growth were discussed based on results of the application of GA on pistils or the in vitro treatment of pollen (Bhandal and Malik, 1979; Viti et al., 1990), the observations by Swain and colleagues were the first report that directly suggested the importance of GA in pollen tube elongation. Curiously, however, no GA mutants previously identified in pea, Arabidopsis, or other species have been reported to show impaired pollen development (Swain et al., 1997). In this regard, Singh et al. (2002) discussed the possibility that many of these mutants may have relatively mild changes in pollen tube elongation that cannot be easily identified. This is not the case with the rice GA-deficient mutants, however. The four rice GA-deficient mutants with defects in early steps of the GA biosynthetic pathway show distorted pollen transmission frequencies, even with a mild mutation, d35Tan-Ginbozu. As d35Tan-Ginbozu was used in the production of a high-yielding variety, Tan-Ginbozu, this single recessive mutation was thought to cause no abnormal phenotypes other than the semidwarf phenotype (Itoh et al., 2004). However, this study shows that the d35Tan-Ginbozu mutation causes not only a semidwarf phenotype but also impaired fertility. This observation indicates that pollen tube elongation, as well as shoot elongation, is one of the most sensitive biological events and reflects the endogenous GA level. This idea is supported by the observation that the transmission frequency of an intermediate allele of the kao mutation, rpe1, is similar to those of the null alleles kao-1 and kao-3. This result indicates that the remaining activity of the KAO enzyme produced by rpe1, which can cause partial shoot and stem elongation and also supports almost complete flower organ development (Figure 1), does not function in pollen tube elongation. In other words, the acceptable value of GA synthetic activity should be high enough for normal pollen elongation, and even mild defects in GA synthesis severely affect GA activity. The reason why GA-deficient mutants of other species do not exhibit an obvious impaired pollen tube elongation phenotype has not yet been elucidated. One possibility is that these GA-deficient mutations are inherited in a sporophytic manner because of the expression of these genes in the premeiotic stage, as in the rice genes involved in GA signaling.
Function of the GID1/DELLA-Mediated GA Perception System in Pollen Tube Elongation
The semifertility of the Slr1-d3 homozygous mutant was mainly caused by impaired pollen development instead of impaired pollen tube elongation (Figure 6). However, the requirement for GA synthesis in rice pollen for pollen tube elongation directly indicates that the GA-signaling pathway should work in pollen. Why then is the critical step in the fertilization process different between the GA-signaling and GA-deficient mutants? One possible explanation is that the GID1/DELLA-mediated GA-signaling pathway is not essential for pollen tube elongation and that an alternative, unknown, GA-signaling pathway functions in this process. However, this possibility is unlikely, at least in Arabidopsis, because of the following observations. The impaired pollen elongation of transgenic Arabidopsis ectopically expressing a gene encoding pea GA2ox2 is rescued by gar2, a gain-of-function mutant of the GA signaling–specific F-box protein gene (SLY1), and by rga, a loss-of-function mutant of an Arabidopsis DELLA protein (Singh et al., 2002), both of which cause the GA-hypersensitive phenotype as a result of changes in the sensitivity of the GID1/DELLA-mediated GA-signaling pathway. The lack of functional versions of three DELLA proteins, RGL1, RGL2, and RGA, completely restored petal and stamen development in ga1-3, a severe GA-deficient Arabidopsis mutant, and permitted normal seed set (Cheng et al., 2004), supporting the above idea. Furthermore, in rice, we also observed that the transmission of Slr1-d3, the partially dominant allele of slr1, was significantly lower than the theoretical value (<20%, Table 5). A dominant-negative form of SLR1 protein, Slr1-d3, expressed postmeiosis would be expected to affect pollens carrying the Slr1-d3 allele rather than those carrying the wild allele. The relatively mild effect of the Slr1-d3 allele on the segregation ratio rather than GA-deficient mutations can be discussed as follows. All pollen grains from the heterozygous plants (+/Slr1-d3) will have some carryover Slr1-d3 mRNA. Consequently, the additional copies of Slr1-d3 mRNA in pollens may only have a minor additional inhibitory effect on pollen germination and growth. These observations and discussion support the idea that the GID1/DELLA-dependent GA-signaling pathway also functions in pollen tube elongation in rice.
In addition, the inhibitory effect of a high concentration of GA4 on pollen germination and elongation (Figure 4) suggests that an alternative GA-signaling pathway might be involved in these events because other GA-responding events mediated by the GID1/DELLA perception system do not show such an inhibitory effect at high concentrations. Actually, when we consider the molecular mechanism of the GID1/DELLA system, in which GA induces the degradation of DELLA proteins by collaboration with the GA signal-specific SCF complex, it is difficult to believe that overdosing with GA can cause an inhibitory effect in this system. Such an inhibitory effect of high concentrations of GA on pollen tube growth was also observed in Arabidopsis (Singh et al., 2002). Taking all these observations together, we predict a possible scenario for the involvement of GA in rice pollen germination and elongation: de novo GA synthesis is necessary for these events, and the synthesized GA is perceived by the GID1/DELLA-mediated GA perception system in pollen cells, as in other organs, whereas an alternative pathway might function to suppress GA signaling at higher GA levels. Further study is required to confirm an alternative pathway of GA signaling in pollen.
METHODS
Isolation of the rpe1 and Slr1-d3 Mutants
The rice (Oryza sativa) mutants rpe1 and Slr1-d3 were initially isolated in a screen for dwarf rice by treatment with ethylene imine and N-methyl-N-nitrosourea, respectively. Rice seeds were surface-sterilized and soaked for 3 d, sown in artificial soil, and then grown for 20 to 30 d. The seedlings were then transplanted into a rice field and transferred to a glasshouse at the heading stage to examine pollen growth.
Genotype Analysis
To examine the segregation between the homozygous GID1/GID1, the heterozygous GID1/gid1-3, and the homozygous gid1-3/gid1-3 and between the homozygous CPS1/CPS1, the heterozygous CPS1/cps1-1, and the homozygous cps1-1/cps1-1, DNA was extracted from leaves of 14-d-old seedlings derived from GID1/gid1-3 and CPS1/cps1-1 plants (see below for the DNA extraction procedure). The primers used for PCR amplification are listed in Supplemental Table 1 online. PCR amplification was then performed using 1 μg of DNA in a total volume of 10 μL. The PCR conditions were an initial incubation at 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s, with a final extension for 7 min at 72°C. To observe the band patterns that revealed the homozygous and heterozygous genotypes, 10 μL of each PCR product was separated by electrophoresis in agarose gels (2.5% agarose in TBE buffer).
Treatment with GA and a GA Inhibitor
Treatments with GA were performed for 30 min before artificial pollination. Three microliters of solutions containing 10−9, 10−8, 10−7, or 10−6 M GA were applied directly to the insides of emasculated spikelets.
Viable Pollen Assays
To evaluate pollen viability, six anthers before flowering were removed from a spikelet and placed on a glass slide. The anthers were crushed into a fine powder and stained with 10 μL of 1% (v/v) of I2 in 3% (v/v) KI, and 1 μL was sampled to observe fertile and infertile pollen using a light microscope. Pollen grains that were round in shape and stained black were judged as viable or living pollen, and sterile or dead pollen was stained yellow or light red.
Examination of Pollen Tube Growth
Aniline blue staining was performed as described by Ryan et al. (1998) and Singh et al. (2002) with modifications.
Pollen Germination
Rice flowers were emasculated and artificially pollinated by hand. After 30 min, the pistils were removed and directly stained with aniline blue on a glass slide for a few minutes before observation by UV microscopy (Olympus; U-TV 0.5 XC). Germinated pollen refers to pollen grains attached to the stigma whose pollen tube growth can be detected by UV fluorescence.
Pollen Elongation
At 2 or 4 h following artificial pollination, rice pistils were excised and fixed in 3:1 ethanol:acetic acid for 30 min and then softened in 1 n KOH for 30 min at 55°C using a block heater. The pistils were washed in distilled water for a few minutes and stained with 0.1% aniline blue in K3PO4 buffer, pH 8.5, for 2 h at room temperature. The pistils were rinsed briefly in distilled water and mounted in 50% glycerol. The samples were visualized by UV microscopy. Elongated pollen refers to germinated pollen whose filament tubes elongate in a time-dependent manner.
DNA Sequence Analysis
To examine the nucleotide sequence in the rice GA biosynthetic gene Os KAO (accession number AK069429), DNA was extracted from leaves of 2-week-old seedlings of the rpe1 mutant and the wild type from which it was derived (cv Fujiminori). DNA extraction was performed using an ISOPLANT extraction kit (Nippon Gene). PCR was then performed using the primers listed in Supplemental Table 1 online. The PCR conditions were 94°C for 5 min followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The amplified DNA fragments were then separated by electrophoresis on a 0.7% (w/v) low-melting-point agarose gel, and the fragments were sequenced directly with appropriate primers.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted using the RNeasy plant mini kit (Qiagen) from wild-type rice anthers at the various stages as described in Figure 7A. First-strand cDNA was synthesized from ∼1 μg of total RNA with an oligo(dT) primer and Omniscript RT kit (Qiagen). Each transcript for GA-related genes was quantified by a real-time PCR analysis using 10% of the resulting cDNA as a template. Real-time PCR was performed with the LightCycler system (Roche) and the QuantiTect SYBR Green PCR kit (Qiagen). For this analysis, a linear standard curve and threshold cycle number versus log (designated transcript level) were constructed using a series of dilutions of each PCR product (10−17, 10−18, 10−19, and 10−20 M); the levels of the transcript in all unknown samples were determined according to the standard curve. The Actin1 gene was used as an internal standard for normalizing cDNA concentration variations. Data were the average of three replicates. The sequences of primer pairs are listed in Supplemental Table 1 online.
Histological Analysis
Plant materials fixed in formalin:acetic acid:70% ethanol (1:1:18) were dehydrated through a graded ethanol series and embedded in Paraplast Plus (Sherwood Medical). Microtome sections (10 μm thick) were stained with 0.2% hematoxylin.
Complementation Test
A 9.4-kb genomic DNA fragment containing the full-length rice KAO gene was separately isolated by digesting a BAC clone with SmaI and XhoI and with XhoI and DraI. The two isolated DNA fragments were each inserted into the pBluescript II SK+ vector between the SmaI and XhoI sites or the XhoI and DraI sites, respectively. The vectors containing the inserts were redigested with SmaI and XhoI or with XhoI and XbaI, respectively. The two insert fragments were then ligated and fused into pBluescript II SK+ between the SmaI and XbaI sites. Finally, the DNA fragment (9.4 kb) was subcloned between the SmaI and XbaI sites in a binary vector containing kanamycin and hygromycin resistance genes, pBI-Hm12 (provided by H. Hirano). The insert containing the full-length KAO was transformed into the rpe1 mutant using Agrobacterium tumefaciens (Hiei et al., 1994).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: At GAI (At1g14920), At KAO1 (At1g05160), At KAO2 (At2g32440), At RGA (At2g01570), At RGL1 (At1g66350), At RGL2 (At3g03450), At RGL3 (At5g17490), Br RGA1 (AY928549), Hv D3 (Q9AXH9), Hv SLN1 (Q8W127), Os CPS1 (AK100333), Os GID1 (AK074026), Os GID2 (AB100246), Os GAMYB (AK102841), Os KAO (AK069429), Os KO2 (AK066285), Os KS1 (AY347878), Os SLR1 (BAE96289), Zm D3 (AAC49067), Zm D8 (Q9ST48), and Te Rht-D1b (Q9ST59).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mutation Site of the rpe1 Allele.
Supplemental Figure 2. Diagram of the Structure of the DELLA Proteins and the Mutation Site of Slr1-d3 and Alignment of SLR1 and Other DELLA Proteins at the TVHYNP Domain.
Supplemental Table 1. List of Primer Sequences.
Supplementary Material
Acknowledgments
We thank Kazuhiro Kobayashi and Yoshiaki Harushima for suggestions regarding the examination of pollen tube elongation and Hitomi Kihara, Mayuko Kawamura, and Hiroko Ohmiya for technical assistance. This study was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (19570037 to M.U.-T., 18075003 to M.W., and 18107001 and 18075006 to M.M.), the Ministry of Agriculture, Forest, and Fisheries of Japan (Green Technology Project IP1003; M.M. and M.A.), the Target Protein Project (M.M.), and research fellowships from the Japan Society for the Promotion of Science for Young Scientists (K.A. and K.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Makoto Matsuoka (makoto@nuagr1.agr.nagoya-u.ac.jp).
Online version contains Web-only data.
References
- Ashikari, M., Sasaki, A., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Datta, S., Ishiyama, K., Saito, T., Kobayashi, M., Khush, G.S., Kitano, H., and Matsuoka, M. (2002). Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘Green Revolution’. Breed. Sci. 52 143–150. [Google Scholar]
- Bhandal, I.S., and Malik, C.P. (1979). Effect of gibberellic acid, (2-chloroethyl)phosphoric acid, actinomycin-D and cycloheximide on the activity and leaching of some hydrolases in pollen suspension cultures of Crotalaria juncea. Physiol. Plant. 45 297–300. [Google Scholar]
- Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D.E., Cao, D., Luo, D., Harberd, N.P., and Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (2005). Plant Hormones. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Dill, A., and Sun, T.-P. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet, C.M., and Sun, T.-P. (2005). A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85. [DOI] [PubMed] [Google Scholar]
- Fukuda, A., Nemoto, K., Chono, M., Yamaguchi, S., Nakajima, M., Yamagishi, J., Maekawa, M., and Yamaguchi, I. (2004). Expression pattern of the coparyl diphosphate synthase gene in developing rice anthers. Biosci. Biotechnol. Biochem. 68 1814–1816. [DOI] [PubMed] [Google Scholar]
- Goto, N., and Pharis, R.P. (1999). Role of gibberellin in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana. Can. J. Bot. 77 944–954. [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Tatsumi, T., Sakamoto, T., Otomo, K., Toyomasu, T., Kitano, H., Ashikari, M., Ichihara, S., and Matsuoka, M. (2004). A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54 533–547. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sentoku, N., Kitano, H., Matsuoka, M., and Kobayashi, M. (2001). Cloning and functional analysis of two gibberellin 3β-hydoroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., Kitano, H., and Nagato, Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46 23–47. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1991). Characterization of the arrest in anther development associated with gibberellin deficiency of the gib-1 mutant of tomato. Plant Physiol. 97 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, M., Inukai, Y., Ueguchi-Tanaka, M., Itoh, H., Izawa, T., Kobayashi, Y., Hattori, T., Miyao, A., Hirochika, H., Ashikari, M., and Matsuoka, M. (2004). Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, M., Itoh, H., Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Ashikari, M., and Matsuoka, M. (2003). Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35 104–115. [DOI] [PubMed] [Google Scholar]
- King, R.W., and Evans, L.T. (2003). Gibberellins and flowering of grasses and cereals: prizing open the lid of the “florigen” black box. Annu. Rev. Plant Biol. 54 307–328. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.). Theor. Appl. Genet. 58 257–263. [DOI] [PubMed] [Google Scholar]
- Murray, F., Kalla, R., Jacobsen, J., and Gubler, F. (2003). A role for HvGAMYB in anther development. Plant J. 33 481–491. [DOI] [PubMed] [Google Scholar]
- Nester, J.E., and Zeevaart, J.A.D. (1988). Flower development in normal tomato and a gibberellin-deficient (ga-2) mutant. Am. J. Bot. 75 45–55. [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). Green revolution genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Pharis, R.P., and King, R.W. (1985). Gibberellins and reproductive development in seed plants. Annu. Rev. Plant Physiol. 36 517–568. [Google Scholar]
- Ryan, E., Grierson, I.C., Cavell, A., Steer, M., and Dolan, L. (1998). TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol. 138 49–58. [Google Scholar]
- Sakamoto, T., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., Ishiyama, K., Saito, T., Kobayashi, M., Khush, G.S., Kitano, H., and Matsuoka, M. (2002). A mutant gibberellin-synthesis gene in rice. Nature 416 701–702. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Singh, D.-P., Jermakow, A.M., and Swain, S.M. (2002). Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14 3133–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Muller, A.J., and Singh, D.P. (2004). The gar2 and rga alleles increase the growth of gibberellin-deficient pollen tubes in Arabidopsis. Plant Physiol. 134 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Reid, J.B., and Kamiya, Y. (1997). Gibberellins are required for embryo and seed development in pea. Plant J. 12 1329–1338. [Google Scholar]
- Tsuji, H., Aya, K., Ueguchi-Tanaka, M., Shimada, Y., Nakazono, M., Watanabe, R., Nishizawa, N.K., Gomi, K., Shimada, A., Kitano, H., Ashikari, M., and Matsuoka, M. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47 427–444. [DOI] [PubMed] [Google Scholar]
- Tyler, L., Thomas, S.G., Hu, J., Dill, A., Alonso, J.M., Ecker, J.R., and Sun, T.-P. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.-Y., Hsing, Y.C., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Viti, R., Bartolini, S., and Vitagliano, C. (1990). Growth regulators on pollen germination in olive. Acta Hortic. 286 227–230. [Google Scholar]
- Wilson, R., Heckman, J.W., and Somerville, C. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.