Abstract
The plant hormone cytokinin regulates many aspects of growth and development. Cytokinin signaling involves His kinase receptors that perceive cytokinin and transmit the signal via a multistep phosphorelay similar to bacterial two-component signaling systems. The final targets of this phosphorelay are a set of Arabidopsis thaliana Response Regulator (ARR) proteins containing a receiver domain with a conserved Asp phosphorylation site. One class of these, the type-A ARRs, are negative regulators of cytokinin signaling that are rapidly transcriptionally upregulated in response to cytokinin. In this study, we tested the role of phosphorylation in type-A ARR function. Our results indicate that phosphorylation of the receiver domain is required for type-A ARR function and suggest that negative regulation of cytokinin signaling by the type-A ARRs most likely involves phosphorylation-dependent interactions. Furthermore, we show that a subset of the type-A ARR proteins are stabilized in response to cytokinin in part via phosphorylation. These studies shed light on the mechanism by which type-A ARRs act to negatively regulate cytokinin signaling and reveal a novel mechanism by which cytokinin controls type-A ARR function.
INTRODUCTION
Two-component signaling systems are used by prokaryotic and eukaryotic organisms to sense and respond to changes in the environment (Stock et al., 2000; West and Stock, 2001). In a canonical two-component system, a stimulus is perceived by a sensor kinase, which autophosphorylates on a conserved His residue in the kinase domain. The signal is transmitted by transfer of the phosphoryl group to a conserved Asp residue on the receiver domain of a response regulator. Variations of the simple two-component system involve intermediate elements in the phosphotransfer from the sensor kinase to the response regulator. Receiver domain phosphorylation induces conformational changes, which, in most response regulators, release repression of the output domain to allow the activation of downstream processes, often transcriptional regulation. In some response regulators, these conformational changes allow specific interactions with target proteins.
The cytokinin signaling pathway is the best-characterized system employing two-component elements in plants (Kakimoto, 2003; Ferreira and Kieber, 2005; Maxwell and Kieber, 2005; Muller and Sheen, 2007). Cytokinins were discovered by their ability to promote division in cultured cells (Miller et al., 1955) and have since been implicated in almost every aspect of plant growth and development and in the responses to various biotic and abiotic environmental cues (Mok and Mok, 2001; Sakakibara, 2006).
In Arabidopsis thaliana, the three cytokinin receptors (Arabidopsis Histidine Kinase2 [AHK2], AHK3, and AHK4) are hybrid His kinases that contain a fused receiver domain in addition to an input (a cytokinin binding CHASE domain) and a His kinase domain (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001a, 2001b; Yamada et al., 2001). In response to cytokinin binding, these receptors autophosphorylate on a conserved His residue and relay this phosphoryl group to Arabidopsis Response Regulators (ARRs) via an intermediate set of histidine phosphotransfer (Hpt) proteins called the Arabidopsis Hpt proteins (AHPs) (Suzuki et al., 1998; Hutchison et al., 2006). Similar cytokinin signaling components have been characterized in other plant species (Asakura et al., 2003; Ito and Kurata, 2006; Jain et al., 2006; Pareek et al., 2006; Du et al., 2007).
The Arabidopsis response regulators fall into four classes based on phylogenetic analysis and domain structure: type-A ARRs, type-B ARRs, type-C ARRs, and the Arabidopsis pseudoresponse regulators (APRRs) (Schaller et al., 2007). The 10 type-A ARRs are primary transcriptional targets of cytokinin signaling and contain short C-terminal extensions beyond the conserved receiver domain (Brandstatter and Kieber, 1998; Imamura et al., 1998; D'Agostino et al., 2000). The 11 type-B ARRs contain C-terminal output domains that have DNA binding and transactivating activity (Sakai et al., 1998, 2000). Type-B ARRs are positive regulators of cytokinin signaling that control the transcription of a subset of cytokinin-regulated targets, including the type-A ARRs (Hwang and Sheen, 2001; Sakai et al., 2001; Tajima et al., 2004; Mason et al., 2005; Taniguchi et al., 2007; Yokoyama et al., 2007). The type-C ARRs are more distantly related to type-A and type-B ARR receiver domain sequences. They do not contain the output domain of type-B ARRs and are not transcriptionally regulated by cytokinin, although their overexpression results in reduced sensitivity to cytokinin (Kiba et al., 2004). The ARRs all contain the conserved Asp required for receiver domain phosphorylation in bacterial response regulators, and phosphotransfer from an AHP to representative members of all three ARR groups has been demonstrated in vitro (Suzuki et al., 1998; Imamura et al., 2001, 2003; Kiba et al., 2004; Mahonen et al., 2006a). The APRRs lack the conserved Asp phosphorylation site, and some play a role in modulating circadian rhythms (McClung, 2006).
At least 8 of the 10 type-A ARRs act as partially redundant negative regulators of cytokinin signaling (Kiba et al., 2003; To et al., 2004; Lee et al., 2007b). ARR4 interacts directly with the red light receptor phytochrome B and, along with other type-A ARRs, modulates the response to red light (Sweere et al., 2001; To et al., 2004). A subset of type-A ARRs are direct targets of the transcription factor WUSCHEL and regulate shoot apical meristem function (Leibfried et al., 2005). ARR3 and ARR4 are involved in controlling the circadian clock, and this function is opposed by ARR8 and ARR9 (Salomé et al., 2005). While it is clear that type-A ARRs play a role in multiple signaling pathways, little is known with regard to their mechanism of action.
There are two general models by which type-A ARRs can act to negatively regulate cytokinin signaling (Figure 1A). In the first, the type-A ARRs may compete with positively acting type-B ARRs for phosphoryl transfer from the upstream AHPs, similar to the chemotaxis system in Sinorhizobium meliloti (Schmitt, 2002). A second model is that type-A ARRs regulate the pathway through direct or indirect interactions with pathway components, as observed in Escherichia coli chemotaxis (Bourret and Stock, 2002). These two models are not mutually exclusive.
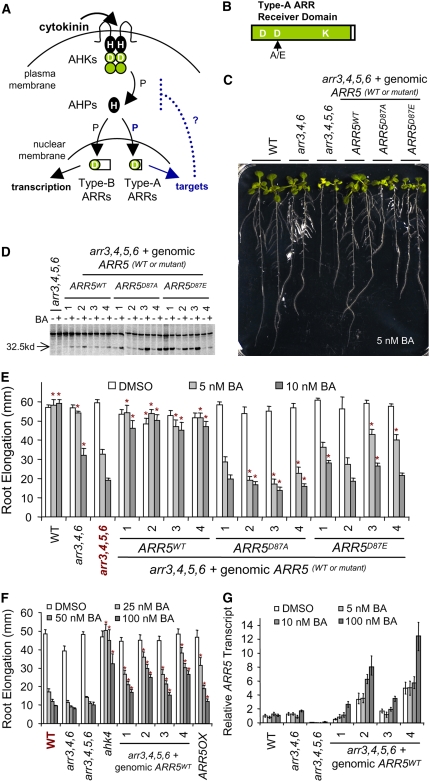
Figure 1.
ARR5 Function Is Dependent on the Phosphorylation of Its Receiver Domain.
(A) Model of type-A ARR function in cytokinin signaling. Cytokinin is perceived by AHKs, which autophosphorylate and transmit the signal via AHPs to ARRs in a His (H)-to-Asp (D) multistep phosphorelay. Type-A ARRs may compete for phosphotransfer with type-B ARRs or interact with targets to negatively regulate the pathway.
(B) Type-A ARR protein is shown with conserved Asp (D) and Lys (K) residues characteristic of receiver domains. The conserved phosphorylation target Asp (D) in the receiver domain is mutated to Ala (A) or Glu (E).
(C) to (G) Complementation of arr3,4,5,6 hypersensitivity to cytokinin inhibition of root elongation. Homozygous T3 seedlings were grown on vertical Murashige and Skoog (MS) plates supplemented with the specified concentrations of BA or 0.1% DMSO control under constant light for 9 d.
(C) Two representative seedlings grown on 5 nM BA per genotype are pictured. Note: line 3 of arr3,4,5,6+ genomicARR5WT, line 4 of arr3,4,5,6+ genomicARR5D87A, and line 1 of arr3,4,5,6+ genomicARR5D87E in (D) are shown.
(D) Transgenic seedlings express cytokinin-inducible myc-tagged wild-type and mutant ARR5 proteins. Full-length roots were harvested from 9-d-old light-grown seedlings and were treated with 1 μM BA or 0.1% DMSO control for 2 h. Total proteins were extracted from root tissues and separated by SDS-PAGE. ARR5-myc proteins were detected by protein gel blotting with anti-c-myc antibody. Note that a protein band of ∼35 to 40 kD that binds nonspecifically to anti-c-myc can be detected in all lanes, including untransformed arr3,4,5,6. The arrowhead indicates the position of a 32.5-kD protein marker.
(E) Root elongation of seedlings from four independent transgenic lines was quantified between days 4 and 9 at the indicated cytokinin concentrations. Error bars represent se (n > 30). Asterisks indicate statistically significant differences from arr3,4,5,6 (indicated in red) at the given concentrations of BA (Student's t test, P < 0.05).
(F) Root elongation of seedlings were analyzed as in (E). Error bars represent se (n > 30). Asterisks indicate statistically significant differences from the wild type (indicated in red) at the given concentrations of BA (Student's t test, P < 0.05). ARR5OX is as described for Figure 2.
(G) arr3,4,5,6+ genomicARR5WT expresses ARR5 transcript. RNA was extracted from seedlings grown under the same conditions as in (C) to (F) and used for real-time RT-PCR analysis. ARR5 relative expression was normalized to β-tubulin levels and to the wild-type DMSO control using REST 2005 version 1.9.12. Error bars represent the upper and lower limits of 95% confidence intervals.
Here, we explore the mechanism by which the type-A ARRs negatively regulate cytokinin signaling and the role of phosphorylation in this process. We show that type-A ARR function requires phosphorylation and that the type-A ARRs likely interact with other components in a phosphorylation-dependent manner to generate negative feedback on the signaling pathway. In addition, we show that a subset of the type-A ARR proteins are stabilized by cytokinin, revealing a novel level of control of these components.
RESULTS
To investigate the role of phosphorylation in type-A ARR function, we generated site-directed mutations in ARR5 that alter the conserved phosphorylation site in the receiver domain (Figure 1B). The analogous conserved Asp in ARR7 (Asp-85), a closely related type-A ARR, has been shown to be required for receiver domain phosphorylation (Lee et al., 2007a). We mutated the conserved phosphoryl-accepting Asp-87 in ARR5 to Ala (ARR5D87A) to test whether phosphorylation of the type-A ARRs is necessary for their function. An analogous D→A mutation in the bacterial response regulator CheY has been shown to disrupt gene function with negligible changes in protein structure compared with the unphosphorylated wild-type CheY protein (Bourret et al., 1993; Alon et al., 1998; Sola et al., 2000). The conserved Asp-87 residue in ARR5 was also mutated to Glu (ARR5D87E). Analogous D→E changes in some bacterial and yeast response regulators can partially mimic the phosphorylated and active protein form (Klose et al., 1993; Moore et al., 1993; Brown et al., 1994; Gupte et al., 1997; Lan and Igo, 1998). Previously, we had further shown that a similar D→E change in ARR7, another type-A ARR, acts as a gain-of-function mutation (Leibfried et al., 2005). Likewise, a similar D→E change in the receiver domain of a type-B ARR (ARR1) resulted in an activated form of this transcription factor, presumably mimicking phosphorylation (Sakai et al., 2001). All wild-type and mutant ARR5 proteins could interact with AHP2 in a yeast two-hybrid assay (see Supplemental Figure 1 online), indicating that the Asp-87 mutations do not strongly disrupt ARR5 protein folding and that the interaction between ARR5 and AHP2 is not dependent on ARR5 Asp-87 phosphorylation.
ARR5 Function Requires Receiver Domain Phosphorylation
To test whether ARR5WT, ARR5D87A, and ARR5D87E are functional in planta, an arr3,4,5,6 mutant, which is hypersensitive to cytokinin, was transformed with genomic constructs expressing myc-tagged wild-type and mutant ARR5 from the endogenous ARR5 promoter (Figure 1). We identified multiple independent transgenic lines, and four lines that represented a range of expression levels of the different transgenes (Figure 1D) were tested for cytokinin sensitivity by a seedling root elongation assay, as described previously (To et al., 2004).
Reintroduction of a wild-type genomic ARR5 gene was sufficient to restore wild-type–like cytokinin sensitivity to the arr3,4,5,6 mutant (Figures 1C and 1E). If the ARR5 transgene were expressed identically to the endogenous ARR5 gene, then the ARR5WT transgenic lines should closely resemble the arr3,4,6 mutant. However, in the four lines examined, cytokinin resistance was restored beyond that of arr3,4,6 to nearly wild-type levels and further increased resistance to higher levels of cytokinin (Figures 1C, 1E, and 1F). One explanation for this is that the roles of ARR3, ARR4, ARR5, and ARR6 are interchangeable in this cytokinin assay and that the transgenic copy of ARR5 in these lines is overexpressed. To test this, we isolated RNA from whole seedlings and seedling roots grown under assay conditions and analyzed the level of ARR5 transcripts by real-time PCR. In wild-type seedlings, no significant increase in the steady state level of ARR5 transcript was observed in response to the low levels of cytokinin used in this assay (Figure 1G). In three of the four arr3,4,5,6+ genomicARR5WT lines, the steady state level of ARR5 transcripts was significantly higher than in the wild type on 10 nM benzyladenine (BA) in both whole seedlings and seedling roots (Figure 1G; data not shown). Consistent with the model that cytokinin resistance correlates with the level of ARR5, all four transgenic lines showed increased resistance to cytokinin at 25 to 100 nM BA comparable to an ARR5-overexpressing line (Figures 1F and 2), which correlates with higher ARR5 expression at 100 nM BA than in the wild type (Figure 1G). The two lines displaying the highest level of ARR5 (lines 2 and 4) also showed the strongest cytokinin resistance at 25 to 100 nM BA (Figure 1F). Overexpression of ARR5 in these lines is most likely due to positional effects of the transgene and/or the insertion of multiple tandem copies of ARR5. Surprisingly, one line (line 1) displayed close to wild-type levels of ARR5, despite displaying nearly wild-type cytokinin sensitivity in root assays. One explanation could be that this line may overexpress ARR5 in a specific subset of root cells, which may not be detected in our analysis of RNA from whole seedlings or full-length roots. An alternative explanation is that the addition of the myc epitope tag may increase the translatability of the transgenic ARR5 transcript, or may increase the stability of the protein relative to endogenous ARR5, thus allowing higher levels of protein accumulation.
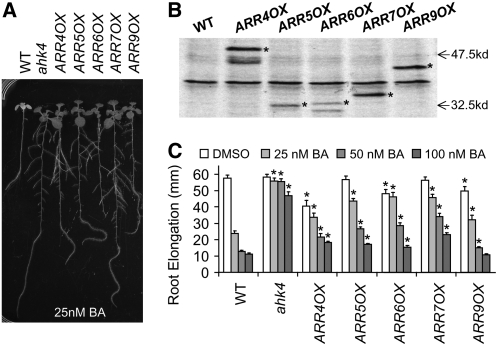
Figure 2.
Wild-Type Type-A ARR Overexpression Confers Cytokinin Resistance.
Overexpression of ARR4, ARR5, ARR6, ARR7, and ARR9 confers cytokinin resistance.
(A) Seedlings were grown as described for Figure 1E with the specified concentrations of BA or 0.1% DMSO control.
(B) Transgenic seedlings express myc-tagged ARR proteins, detected as in Figure 1E. Bands corresponding to the relevant protein products quantified in Figure 3 are noted with asterisks. Lower molecular mass bands in ARR4 and ARR6 may represent degradation products. Arrows indicate the positions of 47.5- and 32.5-kD protein markers.
(C) Root elongation was measured as described for Figure 1. Error bars represent se (n > 30). Asterisks indicate statistically significant differences from the wild type at the given concentrations of BA (Student's t test, P < 0.05).
If phosphorylation is required for ARR5 function, then introducing an ARR5D87A genomic fragment should not rescue the cytokinin-hypersensitive phenotype of arr3,4,5,6. We analyzed four independent transgenic lines that expressed ARR5D87A protein at levels comparable to the four arr3,4,5,6+ genomicARR5WT lines (Figure 1D). In all four lines, introduction of the ARR5D87A transgene into arr3,4,5,6 did not decrease the sensitivity to cytokinin, and in three lines, ARR5D87A expression further increased cytokinin sensitivity compared with the parental line (Figures 1C and 1E). Thus, phosphorylation of the receiver domain is required for ARR5 function. The increased sensitivity in some transgenic lines may be explained by ARR5D87A acting in a dominant negative manner.
ARR5D87E Phosphomimic Is Partially Active
In bacterial systems, altering the Asp phosphorylation target to a Glu can sometimes mimic the phosphorylated form, resulting in a partially activated response regulator (Klose et al., 1993; Moore et al., 1993; Brown et al., 1994; Gupte et al., 1997; Lan and Igo, 1998). This change can also block phosphorylation of the activated response regulator, thus preventing further activation (Klose et al., 1993; Moore et al., 1993). If type-A ARRs negatively regulate cytokinin signaling by acting as phosphate sinks and thus reducing the flow of phosphates to the type-B ARRs, then ARR5D87E should be completely nonfunctional. By contrast, if type-A ARRs act by interacting with other proteins in a phosphorylation-dependent manner, then a phosphomimic mutant may partially complement the arr5 loss-of-function mutation in the arr3,4,5,6 parental line. To test this, we introduced a genomic ARR5D87E transgene into arr3,4,5,6. Four independent transgenic lines showed transgenic protein expression comparable to that of arr3,4,5,6+ genomicARR5WT and arr3,4,5,6+ genomicARR5D87A (Figure 1D). In three of the four lines examined, ARR5D87E partially restored cytokinin resistance significantly above that of the arr3,4,5,6 parental line (Figures 1C and 1E). Importantly, in three arr3,4,5,6+ genomicARR5D87E lines, cytokinin responsiveness was restored significantly above that of the arr3,4,5,6+ genomicARR5D87A lines (Student's t test, P < 0.05 at 5 nM BA), indicating that the activation of the ARR5D87E protein is specific to phosphomimickry and is not due to nonspecific phosphorylation at other sites in the absence of the conserved Asp. The results indicate that mimicking the phosphorylated protein form is at least partially sufficient for ARR5 function. The effect of ARR5D87E is weaker than that of ARR5WT, which is consistent with a partial activation of the mutant receiver domain and the inability of ARR5D87E to be fully activated by phosphorylation (Figure 1E) (Moore et al., 1993). This partial complementation by ARR5D87E, which is unlikely to receive a phosphoryl group from the AHPs, indicates that ARR5 does not function entirely as a phosphate sink. Furthermore, it suggests that the conformational state of phosphorylated ARR5 is likely to be the active state for interactions with target proteins.
Overexpression of Type-A ARRs Confers Cytokinin Resistance
To test whether increasing the levels of other type-A ARRs can confer cytokinin resistance, we expressed ARR4, ARR5, ARR6, ARR7, and ARR9 in wild-type Arabidopsis as myc-epitope–tagged fusion proteins from the constitutive cauliflower mosaic virus (CaMV) 35S promoter. One representative line that expressed a detectable level of myc-ARR fusion protein was selected and analyzed for cytokinin responsiveness (Figures 2A and 2B). All transgenic lines tested were significantly more resistant to 25 nM BA than the wild type in root elongation assays (Figure 2C) (Student's t test, P < 0.05) but less resistant than the loss-of-function cytokinin receptor mutant ahk4.
A Subset of Type-A ARR Proteins Are Stabilized by Cytokinin
The regulation of protein turnover plays an important role in controlling several phytohormone signaling and biosynthetic pathways (reviewed in Dreher and Callis, 2007). We analyzed ARR5 protein turnover using a dexamethasome (DEX)-inducible myc-tagged ARR5 line (DMA5). Continuous growth of DMA5 seedlings on 10 nM DEX results in reduced sensitivity to cytokinin, indicating that the ARR5 myc fusion protein in DMA5 is functional (see Supplemental Figure 2 online).
The myc-ARR5 protein is rapidly degraded following the inhibition of de novo protein synthesis by cycloheximide (CHX). To test whether ARR5 protein turnover is regulated by cytokinin, we compared ARR5 protein steady state levels and degradation rates in the presence and absence of cytokinin. ARR5 protein accumulated to higher steady state levels in the presence of cytokinin, and this is the result of a decreased rate of protein degradation (Figure 3A). Stabilization of ARR5 was effective within 30 min of cytokinin application and was sensitive to concentrations of BA as low as 10 nM (Figures 3A and 3B). Cytokinin increased ARR5 protein stability when added simultaneously with the CHX treatment, indicating that the stabilization of ARR5 protein by cytokinin does not require de novo protein synthesis (Figure 3C).
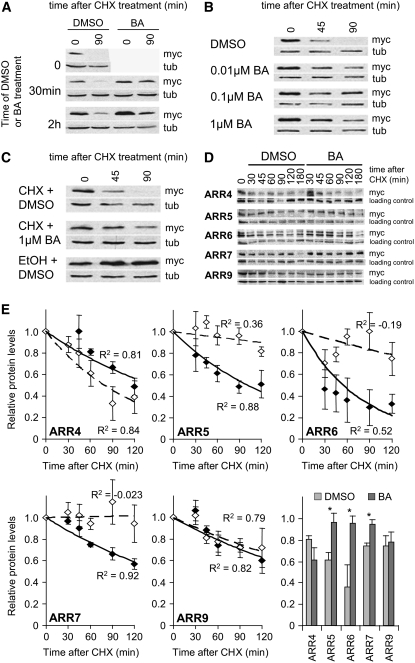
Figure 3.
A Subset of Type-A ARR Proteins Are Stabilized by Exogenous Cytokinin Application.
(A) to (D) myc-ARR5 protein is stabilized by exogenous cytokinin. myc-ARR5 protein was generated in 7-d-old light-grown seedlings in a DEX-inducible myc-ARR5 (DMA5) line by a 2-h 1 μM DEX treatment.
(A) Cytokinin stabilization of myc-ARR5 protein occurs within 30 min and is effective at 2 h. After DEX induction of myc-ARR5 protein production, 1 μM BA or 0.1% DMSO control was added for the times indicated at left before CHX treatment.
(B) Cytokinin stabilization of ARR5 is sensitive to low concentrations of BA. Seedlings were treated with the indicated concentrations of BA or DMSO control during DEX treatment, followed by CHX application.
(C) Cytokinin stabilization of myc-ARR5 does not require new protein synthesis. After DEX treatment, 200 μM CHX (or ethanol [EtOH] control) and 1 μM BA (or 0.1% DMSO control) were applied simultaneously and myc-ARR5 protein turnover was analyzed as in (A).
(D) A subset of type-A ARRs are stabilized by exogenous cytokinin application. Seven-day-old light-grown ARR4OX, ARR5OX, ARR6OX, ARR7OX, and ARR9OX seedlings were treated simultaneously with 200 μM CHX and 1 μM BA or 0.1% DMSO control. Three independent experiments were conducted with consistent results, and one representative blot is shown.
(E) Relative myc-ARR protein levels were normalized to loading control and to myc-ARR protein levels at time 0. The results from three independent experiments were averaged and shown with error bars indicating se. Note that the upper band for ARR6 was quantified. An exponential best-fit curve was fitted through the data points to estimate protein half-life. Correlation coefficient (R2) values are indicated as a measure of curve fit. Closed symbols and solid lines represent DMSO control. Open symbols and broken lines represent BA treatment. The bottom right panel shows relative protein levels at 60 min after CHX treatment. Asterisks indicate statistical differences between BA treatment and DMSO control within each transgenic line (Student's t test, P < 0.05).
To test whether other type-A ARR proteins are stabilized by cytokinin, we analyzed the turnover of their respective myc fusion proteins expressed from the CaMV 35S promoter. The five type-A ARR proteins that we examined exhibited different rates of protein turnover (Figures 3D and 3E). The half-lives of the myc-ARR5 and myc-ARR6 fusion proteins were estimated to be 100 and 60 min, respectively. myc-ARR4, myc-ARR7, and myc-ARR9 proteins exhibited longer protein half-lives, ∼140, 160, and 180 min, respectively. In the presence of exogenous cytokinin, the myc-ARR5, myc-ARR6, and myc-ARR7 fusion proteins were stabilized, with protein half-lives estimated to be >300 min. The turnover of the myc-ARR4 and myc-ARR9 fusion proteins was not significantly affected by cytokinin (Figures 3D and 3E).
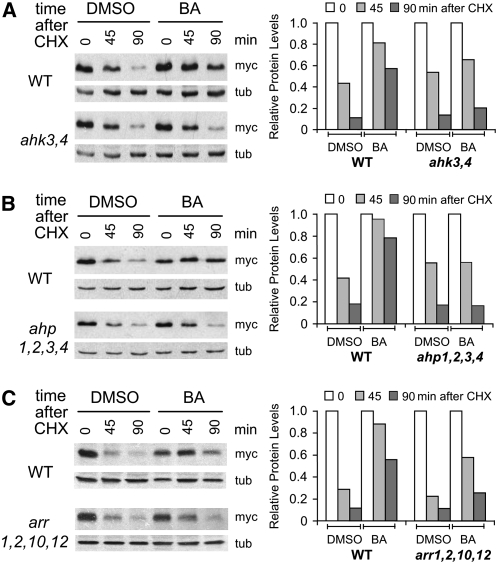
Cytokinin-Mediated Stabilization of ARR5 Involves Two-Component Phosphorelay
To test whether the stabilization of ARR5 by cytokinin is mediated by the two-component signaling pathway, we expressed myc-ARR5 in the background of two-component element mutants. In the ahk3,4 and ahp1,2,3,4 mutants, cytokinin treatment failed to stabilize myc-ARR5 (Figures 4A and 4B). These data indicate that an intact AHK-AHP phosphorelay is required for cytokinin to delay the turnover of type-A ARR proteins. Interestingly, cytokinin-mediated stabilization of myc-ARR5 was also reduced in a multiple type-B ARR loss-of-function mutant (arr1,2,10,12) (Figure 4C). As de novo protein synthesis is not required for the stabilization of ARR5 by cytokinin, this result suggests that type-B ARRs are required for the transcription of an element involved in the stabilization of ARR5 that is expressed prior to cytokinin application in this assay. However, arr1,2,10,12 mutants still retain some response to cytokinin stabilization of myc-ARR5, supporting the model that phosphorelay plays a role in regulating myc-ARR5 turnover.
Figure 4.
Cytokinin Stabilization of ARR5 Requires Upstream Cytokinin Signaling Genes.
Protein turnover of DEX-inducible myc-ARR5 was examined in the background of the cytokinin signaling mutants indicated. Seedlings were treated and analyzed as described for Figure 3B. Relative protein levels were normalized to tubulin and to myc-ARR5 levels at 0 min after CHX treatment.
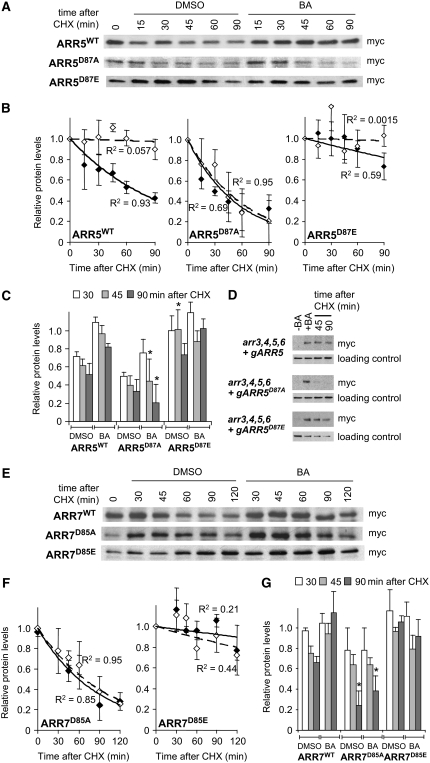
We tested the hypothesis that type-A ARR proteins are stabilized by phosphorylation by analyzing the turnover of ARR proteins mutated in the conserved Asp phosphorylation target. We expressed ARR5D87A, ARR5D87E, ARR7D85A, and ARR7D85E from the CaMV 35S promoter and compared their kinetics of protein turnover with those of their respective wild-type proteins. The myc-ARR5D87A protein was degraded more rapidly than myc-ARR5WT in the absence of exogenous cytokinin; 15 min after CHX treatment, myc-ARR5WT levels decreased 20%, whereas myc-ARR5D87A levels decreased ∼40%, compared with the initial protein levels (Figures 5A to 5C). Cytokinin treatment resulted in a strong stabilization of myc-ARR5WT protein, but this was not observed with the myc-ARR5D87A protein (Figures 5A to 5C). Consistent results were observed in wild-type and mutant ARR5 proteins expressed from genomic constructs used for the complementation of arr3,4,5,6 (Figure 5D). Similarly, in the absence of cytokinin, myc-ARR7D85A protein was turned over more rapidly than myc-ARR7WT (Figures 5E to 5G), and cytokinin treatment resulted in a stabilization of ARR7WT but not ARR7D85A (Figures 5E to 5G). The rapid turnover of ARR5D87A and ARR7D85A both in the presence and absence of cytokinin suggests that the phosphorylation of Asp-85/87 plays a role in regulating the turnover of these ARR proteins.
Figure 5.
ARR5 and ARR7 Protein Stability Is Dependent on the Conserved Phosphorylation Target Asp.
Seedlings expressing the proteins indicated were grown, treated, and analyzed as described for Figures 3D and 3E. In (C) and (G), asterisks indicate significant differences in relative protein levels from ARR5WT or ARR7WT after the same treatment (Student's t test, P < 0.05). The data for triplicate analysis of ARR7WT protein degradation is presented in Figure 3E. In (D), genomic versions of myc-ARR5 were first induced by cytokinin to elevate myc-ARR5 protein levels before treatment with CHX.
To further test the role of phosphorylation in type-A ARR protein stability, we analyzed the protein turnover of myc-ARR5D87E and myc-ARR7D85E phosphomimic mutants. When expressed from the CaMV 35S promoter, basal myc-ARR5D87E protein turnover was slower than that of myc-ARR5WT. At 60 min after CHX addition, myc-ARR5WT proteins decreased by >40%, whereas myc-ARR5D87E proteins only decreased by 10% (Figures 5A to 5C). In the presence of cytokinin, myc-ARR5D87E may be weakly stabilized (Figures 5A to 5C), but the response is greatly muted relative to that of myc-ARR5WT. Degradation of the myc-ARR7D85E protein was also reduced in the absence of cytokinin and was not altered significantly by cytokinin application (Figures 5E to 5G). The delayed protein turnover of myc-ARR5D87E and myc-ARR7D85E in the absence of cytokinin suggests that the protein conformation induced by phosphorylation of the conserved Asp contributes to protein stability.
DISCUSSION
Type-A ARRs Are Likely to Negatively Regulate Cytokinin Signaling by Phospho-Dependent Interactions
We investigated the mechanism by which type-A ARRs regulate cytokinin signaling. Two distinct, but not mutually exclusive, models for this mechanism are proposed: one invokes phosphocompetition between the type-A and type-B ARRs, and the other involves phospho-dependent interactions of the type-A ARRs with target proteins (Figure 1A). To test these models, we generated two site-directed mutants targeting the Asp-87 residue of ARR5, ARR5D87A, and ARR5D87E. This Asp residue is conserved among response regulator family proteins and has been shown to be the target of bacterial two-component phosphorelay (Bourret et al., 1990; Stock et al., 2000; West and Stock, 2001). Substitutions at this conserved Asp have also been shown to abolish receiver domain phosphorylation in other ARRs (Kim et al., 2006; Lee et al., 2007a). In bacterial response regulators, this invariant Asp resides in the conserved active site of the receiver domain that actively catalyzes phosphotransfer from histidine kinases, Hpts, and small molecular phosphodonors (Lukat et al., 1992; Stock et al., 2000; West and Stock, 2001). Analogous D→A and D→E substitutions in bacterial response regulators have been shown to eliminate receiver domain phosphorylation (Bourret et al., 1990, 1993; Drake et al., 1993; Klose et al., 1993; Moore et al., 1993). The D→A change has been shown to disrupt protein function (Bourret et al., 1993; Klose et al., 1993; Moore et al., 1993; Alon et al., 1998), and this defect is not due to perturbations in protein structure (Sola et al., 2000). In some bacterial response regulators, the D→E change has been shown to result in constitutive but partial activation that is independent of two-component phosphorelay (Klose et al., 1993; Moore et al., 1993; Gupte et al., 1997; Lan and Igo, 1998). Response regulator activation by the D→E mutation has also been demonstrated for Skn7 in yeast and a type-B ARR (Brown et al., 1994; Sakai et al., 2001).
The ARR5 D87A substitution did not disrupt protein interactions with AHP2 in our yeast two-hybrid analysis, indicating that the D→A change does not significantly alter ARR5 protein folding. When we expressed ARR5D87A under the control of the native ARR5 promoter in a multiple loss-of-function type-A arr mutant background, ARR5D87A failed to complement the cytokinin hypersensitivity defect of the arr mutant. In addition, the ARR5D87A protein further increased the cytokinin sensitivity of the arr3,4,5,6 mutant. One explanation for the dominant negative effect of ARR5D87A is that the protein is unable to be activated by phosphorylation but retains its ability to interact with the AHPs, thus reducing the activation of other type-A ARRs. In addition, cytokinin-inducible stabilization of the ARR5 protein, which is dependent on two-component phosphorelay, is abolished in the ARR5D87A protein. Together, these results indicate that the ARR5 D87A substitution eliminates phosphoryl Asp-57–dependent activation, which is required for type-A ARR function.
The ARR5D87E protein also retained its ability to interact with AHPs in yeast, indicating that the protein folding is relatively conserved. Expression of ARR5D87E in a multiple type-A arr mutant resulted in the partial rescue of cytokinin hypersensitivity, which is the opposite effect compared with ARR5D87A. In addition, basal protein stability of the ARR5D87E protein was elevated in both the absence and presence of exogenous cytokinin, consistent with the ARR5D87E protein being a phosphomimic. Together, these results indicate that the ARR5 D87E substitution renders a partially activated type-A ARR, which is functional in negatively regulating cytokinin signaling despite its inability to be phosphorylated on Asp-57.
Our results from both the ARR5D87A and ARR5D87E mutants have demonstrated that type-A ARRs require phosphorylation on the conserved Asp for function in vivo and that a nonphosphorylatable, partially activated form of the type-A ARR protein can partially rescue a loss-of-function mutant. Together, our data provide evidence that the phosphorylated type-A ARR protein can negatively regulate cytokinin response independently of its ability to compete for phosphoryl groups with the type-B ARRs and suggest that this negative regulation may be mediated through phosphospecific interactions with target proteins.
It is interesting that the expression of ARR5 wild type, ARR5D87A, and ARR5D87E produced distinct effects on cytokinin sensitivity in planta and that the three proteins also displayed different levels of basal protein stability. In fact, the population of the unphosphorylated bacterial response regulator NtrC has been reported to consist of a mix of both active and inactive receiver domain conformations, and the population is shifted to predominantly active forms upon phosphorylation (Volkman et al., 2001). This finding is consistent with the residual activity found in the unphosphorylated bacterial response regulator CheY (Barak and Eisenbach, 1992). Our data suggest that the ARR5 D→A and D→E mutations shift the ARR5 protein population toward the inactive and active receiver domain conformations, respectively, which may exhibit distinct properties.
Although our results suggest that type-A ARRs function in cytokinin signaling through phospho-dependent interactions, they do not rule out a role for type-A ARRs in phosphocompetition. A recent study indicates that the cytokinin receptor AHK4 determines phosphate flux through the system by regulating a bidirectional phosphorelay to and from the AHPs (Mahonen et al., 2006b). A bidirectional phosphorelay is also used by the bacterial Arc two-component system to mediate signal decay: the phosphoryl group from the ArcB response regulator is transferred back to the receiver domain of the ArcA tripartite His kinase via its His transmitter domain (Georgellis et al., 1998; Pena-Sandoval et al., 2005). It is possible that type-A ARR function may act by a similar mechanism of reverse phosphotransfer from type-B ARRs to type-A ARRs via AHPs, because phosphotransfer from AHPs to both type-A and type-B ARRs has been demonstrated in vitro (Suzuki et al., 1998; Imamura et al., 2001, 2003) and, in the presence of cytokinins, type-A ARRs, type-B ARRs, and AHPs mostly localize to the same subcellular compartments (Hwang and Sheen, 2001; Imamura et al., 2001, 2003; Kiba et al., 2003). One argument against ARR5D87E acting as a phosphomimic is that the protein may be nonspecifically and less efficiently phosphorylated at an alternative site in the absence of the conserved Asp. However, the catalytic nature of the conserved active site surrounding the Asp phosphorylation target suggests that nonspecific phosphorylation on the receiver domain is unlikely (Stock et al., 2000; West and Stock, 2001). More importantly, the lack of activation of the ARR5D87A control indicates that the activation of ARR5D87E must be a property specific to the D87E substitution.
A previous study examined shoot formation from cultured Arabidopsis roots overexpressing ARR4 and ARR8 and reported that overexpression of ARR4 resulted in cytokinin hypersensitivity, whereas overexpression of ARR8 resulted in cytokinin insensitivity (Osakabe et al., 2002). While we have not examined the effect of ARR8 overexpression, our analysis of ARR4 (as well as ARR5, ARR6, ARR7, and ARR9) overexpression in this study, combined with other overexpression reports (Hwang and Sheen, 2001; Kiba et al., 2003; Lee et al., 2007b), as well as loss-of-function mutants from our previous work (To et al., 2004), are consistent with ARR4, as well as the other type-A ARRs, acting as negative regulators of cytokinin signaling. One explanation for this discrepancy is that ARR4 may act as a positive element in a subset of cytokinin responses, such as shoot initiation. Indeed, we have found antagonistic interactions among type-A ARRs in other physiological roles, such as in controlling rosette size, petiole length, and circadian rhythms (To et al., 2004; Salomé et al., 2005).
Previous overexpression studies have also produced conflicting data on the role of phosphorylation on type-A RR function. Cytokinin resistance conferred by overexpression of a rice (Oryza sativa) type-A RR was disrupted by mutating the conserved phosphorylation target to either an unphosphorylatable residue or a phosphomimic (Hirose et al., 2007). Similar results have also been reported for overexpression of ARR22, which is a type-C ARR (Kiba et al., 2004). One explanation is that the cytokinin-insensitive phenotype conferred by overexpression of wild-type type-A and type-C RRs may reflect an inappropriate diversion of phosphate flow from the Hpts to the abnormally high levels of type-A and type-C RR proteins, which would decrease the activation of the type-B RRs. Whether this proposed phosphocompetition is an artifact of overexpression or accurately reflects the role of endogenous type-A RR proteins is an open question. By contrast, disruption of the conserved phosphorylation site did not significantly alter the ability of type-A ARRs to reduce or enhance a cytokinin-responsive reporter when overexpressed in protoplasts (Hwang and Sheen, 2001). These differences may reflect differences between the assay systems in these overexpression studies. Our study expressing ARR5 under its endogenous promoter in a loss-of-function mutant background clearly shows that phosphorylated type-A ARRs can negatively regulate cytokinin response in vivo, independently of phosphocompetition with the type-B ARRs, and that this negative feedback regulation may be mediated via interactions between the type-A ARR in its phosphorylated protein conformation and target proteins.
Cytokinin Regulates Type-A ARR Function in Part by Protein Stabilization
Control of protein stability through the proteasome degradation machinery is a common mechanism for the regulation of plant hormone responses (reviewed in Dreher and Callis, 2007). Indeed, mutants of RPN12 and COP9/CIN4/ FUS10, which are subunits of proteasome regulatory structures (reviewed in Dreher and Callis, 2007), are cytokinin-insensitive (Vogel et al., 1998; Smalle et al., 2002), suggesting that cytokinin signaling may also be regulated by the proteasome. One possible explanation is that these mutants have higher levels of type-A ARR protein due to decreased degradation. However, ARR5 protein stability is not altered in rpn12a-1 or cin4/cop9/fus10 (see Supplemental Figure 3 online), indicating that cytokinin insensitivity in these mutants is probably due to a distinct mechanism.
In this study, we have shown that cytokinin regulates the turnover of a subset of type-A ARR proteins and that this occurs in the absence of de novo protein synthesis. Cytokinin-mediated stabilization of ARR5 is disrupted in mutants of upstream phosphorelay components, suggesting that phosphorylation of type-A ARRs by two-component elements is required for protein stabilization by cytokinin. In addition, the unphosphorylatable ARR5D87A and ARR7D85A mutant proteins are less stable and their stability is not altered by cytokinin treatment, whereas the partial phosphomimics, ARR5D87E and ARR7D85E, exhibit reduced protein turnover compared with the wild-type proteins, consistent with the idea that type-A ARR protein turnover is determined by the phosphorylation state of the receiver domain. Furthermore, stabilization by cytokinin is compromised in the arr1,2,10,12 mutant, which is disrupted in cytokinin-activated transcription factors and thus should have no direct effect on the phosphorylation state of the type-A ARRs. These results suggest that the mechanism for the stabilization of ARR5/ARR7 is dependent on type-B ARR basal transcription, because de novo protein synthesis is not required for type-A ARR stabilization.
A model in which ARR5 and ARR7 turnover is regulated by the phosphorylation status of their receiver domains is consistent with the finding that the yeast response regulator, SSK1, is degraded by the 26S proteasome pathway and the degradation of SSK1 is inhibited by the upstream phosphotransfer protein YPD1 (Sato et al., 2003). In our yeast two-hybrid analysis, the steady state protein levels of ARR5D87E prey fusion proteins are higher than those of ARR5 or ARR5D87A fusion proteins, suggesting that the ARR5 protein may also be subject to phosphorylation-dependent proteasome degradation in yeast.
Why Are a Subset of Type-A ARRs Stabilized?
The finding that cytokinin stabilizes a subset of type-A ARRs, apparent negative regulators of cytokinin signaling, appears distinct from other known phytohormone signaling pathways involving proteasome degradation machinery, such as auxin, gibberellin, and ethylene, which generally function to activate or stabilize positively acting transcription factors (reviewed in Moon et al., 2004; Fleet and Sun, 2005; Dreher and Callis, 2007). However, in this study, we have shown that expression of the phosphomimic ARR5D87E can partially complement a multiple type-A ARR loss-of-function mutant; furthermore, our previous results showed that ARR7D85E overexpression can further induce meristem arrest at a low frequency (Leibfried et al., 2005). Together, these results indicate that these proteins can function without direct phosphotransfer from the AHPs and suggest that phosphorylated, activated, and stabilized type-A ARR proteins may interact with other targets, possibly to regulate output beyond the cytokinin signaling circuitry.
Type-A ARRs, as a group, have been shown to be transcriptionally upregulated by cytokinin and to function as redundant negative regulators of cytokinin signaling (Brandstatter and Kieber, 1998; Imamura et al., 1998; D'Agostino et al., 2000; Hwang and Sheen, 2001; Kiba et al., 2003; To et al., 2004; Lee et al., 2007b). Phenotypic analyses of loss-of-function and gain-of-function mutants have indicated that subsets of type-A ARRs may play distinct physiological roles (Sweere et al., 2001; To et al., 2004; Leibfried et al., 2005; Salomé et al., 2005; Lee et al., 2007b). In this study, we have shown that individual type-A ARRs differ in both their intrinsic protein stabilities and the effect of cytokinin on their protein turnover, which may suggest a mechanistic basis for functional specificity among type-A ARRs. Interestingly, the effect of cytokinin on type-A ARR protein turnover appears to correlate with their phylogenetic and functional relationships. The type-A ARR proteins that are stabilized by cytokinin, ARR5, ARR6, and ARR7, fall into a subset of ARRs that are more similar in receiver domain sequence and contain shorter C-terminal sequences (D'Agostino et al., 2000). ARR5, ARR6, and ARR7 transcription are also highly induced by cytokinin (D'Agostino et al., 2000) and are regulated by WUSCHEL, likely to mediate interaction between cytokinin signaling and meristem activity (Leibfried et al., 2005). The type-A ARR proteins that are not stabilized by cytokinin, ARR4 and ARR9, are less similar to ARR5, ARR6, and ARR7 in receiver domain sequence and contain longer C-terminal regions (D'Agostino et al., 2000). In addition, ARR4 and ARR9 are also less transcriptionally upregulated by cytokinin (D'Agostino et al., 2000) and play a cytokinin-independent role in modulating the circadian clock (Salomé et al., 2005). The C-terminal regions of type-A ARR proteins may impart specificity in protein regulation. Cytokinin regulation of protein turnover of a subset of type-A ARRs may reflect another mechanism for modulating their function in specific processes, such as meristem activity.
In summary, we have shown that cytokinin regulates type-A ARR activity by two-component phosphorelay, in part through the control of protein stability. Targets of phosphorylated and activated type-A ARRs may modulate cytokinin signaling or other functions and remain to be determined.
METHODS
Plasmid Constructs
A genomic ARR5 DNA fragment (from 1.6 kb upstream of ATG through the entire length of cDNA excluding the stop codon) (D'Agostino et al., 2000) was amplified by PCR from genomic DNA isolated from wild-type Arabidopsis thaliana ecotype Columbia seedlings and inserted into the pENTR/D-TOPO vector (Invitrogen) to generate Gateway entry clone pAR5g. Full-length cDNAs of ARR4, ARR5, ARR6, ARR7, ARR9, and AHP2 were amplified by PCR and inserted into the pENTR-D Gateway entry clone vector (Invitrogen) to generate Gateway entry clones pAR4cs, pAR5cs, pAR6cs, pAR7cs, pAR9cs, and pAP2cs. In the coding region for ARR5 in pAR5g and pAR5cs, the 87th codon GAT encoding Asp-87 of ARR5 cDNA, was changed to GCT, encoding Ala, by site-directed mutagenesis to generate pAR5gDA and pAR5DAcs, respectively. The same codon for Asp-87 was changed to GAG, encoding Glu, to generate pAR5DEs and pAR5gDE. In pAR7s, Asp-85 was changed to Ala and Glu by site-directed mutagenesis to generate pAR7DAcs and pAR7DEcs, respectively. All entry clones were sequence-verified.
For ARR5 complementation constructs, a genomic ARR5 fragment was transferred from pAR5g, pAR5gDA, and pAR5gDE into Gateway-compatible binary vector pGWB16 (a gift from Tsuyoshi Nakagawa, Shimane University) to generate pB16-5gw, pB16-5gDA, and pB16-5gDE, respectively. Each of the resulting constructs carried the endogenous ARR5 promoter driving the expression of wild-type or mutant ARR5 with a 4× C-terminal myc tag.
For ARR overexpression constructs, full-length ARR cDNAs were transferred from Gateway entry vectors pAR4cs, pAR5cs, pAR6cs, pAR7cs, pAR9cs, pAR5DAcs, pAR5DEcs, pAR7DAcs, and pAR7DEcs into the Gateway-compatible binary vector pGWB18 (a gift from Tsuyoshi Nakagawa) by LR recombination (Invitrogen) to generate pB18-4w, pB18-5w, pB18-6w, pB18-7w, pB18-9w, pB18-5DA, pB18-5DE, pB18-7DA, and pB18-7DE, respectively. In each of the resulting constructs, expression of an ARR cDNA carrying a 4× N-terminal myc tag was driven by the constitutive CaMV 35S promoter.
To generate a DEX-inducible 6× N-terminal myc-tagged ARR5 construct, a full-length ARR5 cDNA fragment was introduced into a 6× myc vector via EcoRI sites and subcloned into pTA7002 (Aoyama and Chua, 1997) to generate pDMA5.
Plant Materials and Transgenic Lines
Arabidopsis plants of the Columbia ecotype were used in all experiments as the wild-type control unless stated otherwise. Mutant lines arr3,4,5,6 (To et al., 2004), ahk3,4 (Rashotte et al., 2006), arr1,2,10,12 (Rashotte et al., 2006), and ahp1,2,3,4 (Hutchison et al., 2006) have been described previously.
All transgenic plant lines described here were generated in the Columbia ecotype background by introducing binary plasmid constructs via Agrobacterium tumefaciens–mediated floral dip (Clough and Bent, 1998). pB16-5gw, pB16-5gDA, and pB16-5gDE were introduced into arr3,4,5,6 to generate arr3,4,5,6+ genomicARR5WT, arr3,4,5,6+ genomicARR5D87A, and arr3,4,5,6+ genomicARR5D87E, respectively. At least eight independent single-locus insertion lines were analyzed in the T2 generation and taken to T3 homozygosity. Detailed characterization of homozygous T3 progeny from four independent lines is presented in this article.
pB18-4w, pB18-5w, pB18-6w, pB18-7w, pB18-9w, pB18-5DA, pB18-5DE, pB18-7DA, pB18-7DE, and pDMA5 were introduced into wild-type Columbia to generate ARR4OX, ARR5OX, ARR6OX, ARR7OX, ARR9OX, ARR5D87AOX, ARR5D87EOX, ARR7D85EOX, ARR7D85EOX, and DMA5, respectively. Transgenic T1 seedlings were selected on MS agar plates (see below) supplemented with 30 μg/mL hygromycin and 50 μg/mL carbenicillin. Transgene expression was confirmed in homozygous hygromycin-resistant T3 seedlings by protein gel blotting of whole seedling protein extracts and detection with anti-c-myc POD antibody (Roche Applied Science). For each construct, the results from one representative line are presented.
To generate DEX-inducible myc-ARR5 lines in the various genetic backgrounds, ahk3,4 was crossed to DMA5. pDMA5 was introduced into ahp1,2,3,4 and arr1,2,10,12 and selected as described above.
Plant Growth Conditions
Seeds were surface-sterilized and cold-treated at 4°C for 3 d in the dark and grown at 23°C under constant white light (∼100 μE). Seedlings were grown on MS medium containing 1× MS salts, 0.05% MES buffer, and 1% sucrose, pH 5.7. For cytokinin response assays, seedlings were grown on vertical MS plates with 0.6% phytagel (Sigma-Aldrich) supplemented with a dose range of N6-BA or 0.1% (v/v) DMSO carrier control as described previously (To et al., 2004). For protein assays and transgenic seedling selection, seedlings were grown on horizontal MS plates with 0.8% bactoagar.
Quantitative Real-Time RT-PCR
Ten-day-old light-grown seedlings in cytokinin response assays were harvested, and total RNA was extracted from both whole seedlings and full-length roots using an RNeasy kit according to the manufacturer's instructions (Qiagen). cDNA was generated from the RNA with Moloney murine leukemia virus reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed with Taq DNA Polymerase Hot-Start Version, buffer, and deoxynucleotide triphosphates according to the manufacturer's instructions (Takara Mirus Bio) supplemented with 0.3× SYBR Green (Molecular Probes) and ARR5 primers ARR5F3 (5′-TCTGAAGATTAATTTGATAATGACGG-3′) and ARR5R2 (5′-TCACAGGCTTCAATAAGAAATCTTCA-3′) or β-tubulin primers TUB4s (5′-AGAGGTTGACGAGCAAGATGA-3′) and TUB4a (5′-AACAATGAAAGTAGACGCCA-3′). Real-time PCR was performed in an Opticon2 PCR machine (MJ Research) using the following thermocycler program: (1) 2 min at 95°C; (2) 15 s at 95°C; (3) 15s at 60°C; (4) 15 s at 72°C; (5) optical read, repeat 34 cycles of steps 2 through 5, followed by a final analysis of product melting temperature to confirm the PCR product. Each biological sample was analyzed at least twice in triplicate. The relative expression for ARR5 (normalized to β-tubulin as a reference gene and to the wild type grown on DMSO as a control sample) and 95% confidence intervals were determined using REST 2005 version 1.9.12 (Pfaffl et al., 2002; http://rest-2005.gene-quantification.info). Two independent experiments were performed with consistent results. The data from one triplicate analysis are presented.
Analysis of Protein Stability
For DEX-inducible myc-tagged ARR5, myc-ARR5 protein expression was induced by incubating 7-d-old light-grown seedlings in liquid MS medium with 1 μM DEX supplemented with 1 μM BA or 0.1% (v/v) DMSO carrier control for 2 h. Protein synthesis was inhibited by 200 μM CHX. Seedlings were harvested by flash freezing in liquid nitrogen at the time points indicated.
For lines constitutively overexpressing ARRs, 7-d-old light-grown seedlings were incubated in liquid MS medium with 200 μM CHX supplemented with 1 μM BA or DMSO carrier control. Seedlings were harvested at the time points indicated.
Protein extracts were prepared in 250 mM Tris, pH 8, 150 mM NaCl, 5 mM EDTA, 1× Complete protease inhibitors (Roche Applied Science), and 0.5% β-mercaptoethanol. Protein extracts were separated by SDS-PAGE and transferred to Nitropure membranes (GE). myc-tagged proteins were detected with anti-c-myc POD (Roche Applied Science), and tubulin was detected by rabbit polyclonal anti-tubulin and secondary goat anti-rabbit POD antibodies (Chemicon) and visualized by chemiluminescent detection (Perkin-Elmer) by autoradiography. Films were quantified using ImageQuant software (Molecular Dynamics). myc-ARR protein levels were normalized to signal from β-tubulin or from nonspecific anti-c-myc hybridization to a 35- to 40-kD protein. Three independent ARR protein degradation time course experiments were conducted for each line, and the results were averaged. Protein half-lives of myc-ARRs were estimated by plotting an exponential best-fit curve to the averaged data from three independent experiments.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes characterized in this study are as follows: ARR4 (At1g10470), ARR5 (At3g48100), ARR6 (At5g62960), ARR7 (At1g19050), and ARR9 (At3g57040).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mutations Targeting the Conserved Phosphorylation Target Asp Do Not Disrupt ARR5 Protein Interaction with AHP2 in the Yeast Two-Hybrid Assay.
Supplemental Figure 2. Exogenous DEX Application Enhances Cytokinin Resistance in DMA5 Seedlings.
Supplemental Figure 3. Mutations in RPN12a and COP9/CIN4/FUS10 Do Not Alter myc-ARR5 Protein Stability.
Supplementary Material
Acknowledgments
We thank T. Nakagawa for supplying pGWB16 and pGWB18 plasmids and H. Kaminaka and J. Dangl for providing pEG202gw and pjg4-5gw plasmids. We also thank J. Smalle and R. Vierstra for sharing rpn12a-1 seeds. We thank Kieber laboratory members for helpful discussions and for critiquing the manuscript. This work was supported by funding from the National Institutes of Health and the National Science Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Joseph J. Kieber (jkieber@unc.edu).
Online version contains Web-only data.
References
- Alon, U., Camarena, L., Surette, M.G., Aguera y Arcas, B., Liu, Y., Leibler, S., and Stock, J.B. (1998). Response regulator output in bacterial chemotaxis. EMBO J. 17 4238–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11 605–612. [DOI] [PubMed] [Google Scholar]
- Asakura, Y., Hagino, T., Ohta, Y., Aoki, K., Yonekura-Sakakibara, K., Deji, A., Yamaya, T., Sugiyama, T., and Sakakibara, H. (2003). Molecular characterization of His-Asp phosphorelay signaling factors in maize leaves: Implications of the signal divergence by cytokinin-inducible response regulators in the cytosol and the nuclei. Plant Mol. Biol. 52 331–341. [DOI] [PubMed] [Google Scholar]
- Barak, R., and Eisenbach, M. (1992). Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry 31 1821–1826. [DOI] [PubMed] [Google Scholar]
- Bourret, R.B., Drake, S.K., Chervitz, S.A., Simon, M.I., and Falke, J.J. (1993). Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J. Biol. Chem. 268 13089–13096. [PMC free article] [PubMed] [Google Scholar]
- Bourret, R.B., Hess, J.F., and Simon, M.I. (1990). Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc. Natl. Acad. Sci. USA 87 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret, R.B., and Stock, A.M. (2002). Molecular information processing: Lessons from bacterial chemotaxis. J. Biol. Chem. 277 9625–9628. [DOI] [PubMed] [Google Scholar]
- Brandstatter, I., and Kieber, J.J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.L., Bussey, H., and Stewart, R.C. (1994). Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13 5186–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- D'Agostino, I., Deruère, J., and Kieber, J.J. (2000). Characterization of the response of the Arabidopsis ARR gene family to cytokinin. Plant Physiol. 124 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, S.K., Bourret, R.B., Luck, L.A., Simon, M.I., and Falke, J.J. (1993). Activation of the phosphosignaling protein CheY. I. Analysis of the phosphorylated conformation by 19F NMR and protein engineering. J. Biol. Chem. 268 13081–13088. [PMC free article] [PubMed] [Google Scholar]
- Dreher, K., and Callis, J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L., Jiao, F., Chu, J., Chen, M., and Wu, P. (2007). The two-component signal system in rice (Oryza sativa L.): A genome-wide study of cytokinin signal perception and transduction. Genomics, 89 697–707. [DOI] [PubMed] [Google Scholar]
- Ferreira, F.J., and Kieber, J.J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8 518–525. [DOI] [PubMed] [Google Scholar]
- Fleet, C.M., and Sun, T.-p. (2005). A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85. [DOI] [PubMed] [Google Scholar]
- Georgellis, D., Kwon, O., De Wulf, P., and Lin, E.C.C. (1998). Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J. Biol. Chem. 273 32864–32869. [DOI] [PubMed] [Google Scholar]
- Gupte, G., Woodward, C., and Stout, V. (1997). Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J. Bacteriol. 179 4328–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, N., Makita, N., Kojima, M., Kamada-Nobusada, T., and Sakakibara, H. (2007). Over-expression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 48 523–539. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.E., Li, J., Argueso, C., Gonzalez, M., Lee, E., Lewis, M.W., Maxwell, B.B., Perdue, T.D., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18 3073–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis signal transduction. Nature 413 383–389. [DOI] [PubMed] [Google Scholar]
- Imamura, A., Hanaki, N., Umeda, H., Nakamura, A., Suzuki, T., Ueguchi, C., and Mizuno, T. (1998). Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 2691–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, A., Kiba, T., Tajima, Y., Yamashino, T., and Mizuno, T. (2003). In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44 122–131. [DOI] [PubMed] [Google Scholar]
- Imamura, A., Yoshino, Y., and Mizuno, T. (2001). Cellular localization of the signaling components of Arabidopsis His-to-Asp phosphorelay. Biosci. Biotechnol. Biochem. 65 2113–2117. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409 1060–1063. [DOI] [PubMed] [Google Scholar]
- Ito, Y., and Kurata, N. (2006). Identification and characterization of cytokinin-signalling gene families in rice. Gene 382 57–65. [DOI] [PubMed] [Google Scholar]
- Jain, M., Tyagi, A.K., and Khurana, J.P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 6 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto, T. (2003). Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 54 605–627. [DOI] [PubMed] [Google Scholar]
- Kiba, T., Aoki, K., Sakakibara, H., and Mizuno, T. (2004). Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 45 1063–1077. [DOI] [PubMed] [Google Scholar]
- Kiba, T., Yamada, H., Sato, S., Kato, T., Tabata, S., Yamashino, T., and Mizuno, T. (2003). The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44 868–874. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., Ryu, H., Hong, S.H., Woo, H.R., Lim, P.O., Lee, I.C., Sheen, J., Nam, H.G., and Hwang, I. (2006). Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, K.E., Weiss, D.S., and Kustu, S. (1993). Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J. Mol. Biol. 232 67–78. [DOI] [PubMed] [Google Scholar]
- Lan, C.Y., and Igo, M.M. (1998). Differential expression of the OmpF and OmpC porin proteins in Escherichia coli K-12 depends upon the level of active OmpR. J. Bacteriol. 180 171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D., Kim, S., Ha, Y.-M., and Kim, J. (2007. a). Phosphorylation of Arabidopsis response regulator 7 (ARR7) at the putative phospho-accepting site is required for ARR7 to act as a negative regulator of cytokinin signaling. Planta, in press. [DOI] [PubMed]
- Lee, D.J., Park, J.-Y., Ku, S.-J., Ha, Y.-M., Kim, S., Kim, M.D., Oh, M.-H., and Kim, J. (2007. b). Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7(ARR7) overexpression in cytokinin response. Mol. Genet. Genomics 277 115–137. [DOI] [PubMed] [Google Scholar]
- Leibfried, A., To, J.P.C., Busch, W., Stehling, S.K., Kehle, A., Demar, M., Kieber, J.J., and Lohmann, J.U. (2005). WUSCHEL controls meristem size by direct transcriptional regulation of cytokinin inducible response regulators. Nature 438 1172–1175. [DOI] [PubMed] [Google Scholar]
- Lukat, G.S., McCleary, W.R., Stock, A.M., and Stock, J.B. (1992). Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89 718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen, A.P., Bishopp, A., Higuchi, M., Nieminen, K.M., Kinoshita, K., Tormakangas, K., Ikeda, Y., Oka, A., Kakimoto, T., and Helariutta, Y. (2006. a). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311 94–98. [DOI] [PubMed] [Google Scholar]
- Mahonen, A.P., Higuchi, M., Tormakangas, K., Miyawaki, K., Pischke, M.S., Sussman, M.R., Helariutta, Y., and Kakimoto, T. (2006. b). Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 16 1116–1122. [DOI] [PubMed] [Google Scholar]
- Mason, M.G., Mathews, D.E., Argyros, D.A., Maxwell, B.B., Kieber, J.J., Alonso, J.M., Ecker, J.R.S., and Shaller, G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, B.B., and Kieber, J.J. (2005). Cytokinin signal transduction. In Plant Hormones: Biosynthesis, Signal Transduction, Action! P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 321–349.
- McClung, C.R. (2006). Plant circadian rhythms. Plant Cell 18 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C.O., Skoog, F., Von Saltza, M.H., and Strong, F. (1955). Kinetin, a cell division factor from deoxyribonucleic acid. J. Am. Chem. Soc. 77 1392. [Google Scholar]
- Mok, D.W., and Mok, M.C. (2001). Cytokinins: Chemistry, Activity and Function. (Boca Raton, FL: CRC Press).
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J.B., Shiau, S.P., and Reitzer, L.J. (1993). Alterations of highly conserved residues in the regulatory domain of Nitrogen Regulator-I (NTRC) of Escherichia coli. J. Bacteriol. 175 2692–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, B., and Sheen, J. (2007). Advances in cytokinin signaling. Science 318 68–69. [DOI] [PubMed] [Google Scholar]
- Osakabe, Y., Miyata, S., Urao, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem. Biophys. Res. Commun. 293 806–815. [DOI] [PubMed] [Google Scholar]
- Pareek, A., Singh, A., Kumar, M., Kushwaha, H.R., Lynn, A.M., and Singla-Pareek, S.L. (2006). Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 142 380–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Sandoval, G.R., Kwon, O., and Georgellis, D. (2005). Requirement of the receiver and phosphotransfer domains of ArcB for efficient dephosphorylation of phosphorylated ArcA in vivo. J. Bacteriol. 187 3267–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W., Horgan, G.W., and Dempfle, L. (2002). Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., Mason, M.G., Hutchison, C.E., Ferreira, F.J., Schaller, G.E., and Kieber, J.J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103 11081–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., Aoyama, T., Bono, H., and Oka, A. (1998). Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol. 39 1232–1239. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Aoyama, T., and Oka, A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24 703–711. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., and Oka, A. (2001). Arabidopsis ARR1 is a transcription factor for genes immediately responsive to cytokinins. Science 294 1519–1521. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H. (2006). CYTOKININS: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57 431–449. [DOI] [PubMed] [Google Scholar]
- Salomé, P.A., To, J.P.C., Kieber, J.J., and McClung, C.R. (2005). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, N., Kawahara, H., Toh-e, A., and Maeda, T. (2003). Phosphorelay-regulated degradation of the yeast Ssk1p response regulator by the ubiquitin-proteasome system. Mol. Cell. Biol. 23 6662–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, G.E., Doi, K., Hwang, I., Kieber, J.J., Khurana, J.P., Kurata, N., Mizuno, T., Pareek, A., Shiu, S.-H., Wu, P., and Yip, W.K. (2007). Nomenclature for two-component signaling elements of rice. Plant Physiol. 143 555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, R. (2002). Sinorhizobial chemotaxis: A departure from the enterobacterial paradigm. Microbiology 148 627–631. [DOI] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Babiychuk, E., Kushnir, S., Durski, A., and Vierstra, R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola, M., Lopez-Hernandez, E., Cronet, P., Lacroix, E., Serrano, L., Coll, M., and Parraga, A. (2000). Towards understanding a molecular switch mechanism: Thermodynamic and crystallographic studies of the signal transduction protein CheY. J. Mol. Biol. 303 213–225. [DOI] [PubMed] [Google Scholar]
- Stock, A.M., Robinson, V.L., and Goudreau, P.N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69 183–215. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Imamura, A., Ueguchi, C., and Mizuno, T. (1998). Histidine-containing phosphotransfer (HPt) signal transducers implicated in His-to-Asp phosphorelay in Arabidopsis. Plant Cell Physiol. 39 1258–1268. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H., and Mizuno, T. (2001). The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42 107–113. [DOI] [PubMed] [Google Scholar]
- Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Bäurle, I., Kudla, J., Nagy, F., Schäfer, E., and Harter, K. (2001). Interaction of the response regulator ARR4 with the photoreceptor phytochrome B in modulating red light signaling. Science 294 1108–1111. [DOI] [PubMed] [Google Scholar]
- Tajima, Y., Imamura, A., Kiba, T., Amano, Y., Yamashino, T., and Mizuno, T. (2004). Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol. 45 28–39. [DOI] [PubMed] [Google Scholar]
- Taniguchi, M., Sasaki, N., Tsuge, T., Aoyama, T., and Oka, A. (2007). ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 48 263–277. [DOI] [PubMed] [Google Scholar]
- To, J.P.C., Haberer, G., Ferreira, F.J., Deruère, J., Mason, M.G., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2004). Type-A ARRs are partially redundant negative regulators of cytokinin signaling in Arabidopsis. Plant Cell 16 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi, C., Koizumi, H., Suzuki, T., and Mizuno, T. (2001. a). Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 42 231–235. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Sato, S., Kato, T., and Tabata, S. (2001. b). The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 42 751–755. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Schuerman, P., Woeste, K.W., Brandstatter, I., and Kieber, J.J. (1998). Isolation and characterization of Arabidopsis mutants defective in induction of ethylene biosynthesis by cytokinin. Genetics 149 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman, B.F., Lipson, D., Wemmer, D.E., and Kern, D. (2001). Two-state allosteric behavior in a single-domain signaling protein. Science 291 2429–2433. [DOI] [PubMed] [Google Scholar]
- West, A.H., and Stock, A.M. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26 369–376. [DOI] [PubMed] [Google Scholar]
- Yamada, H., Suzuki, T., Terada, K., Takei, K., Ishikawa, K., Miwa, K., Yamashino, T., and Mizuno, T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42 1017–1023. [DOI] [PubMed] [Google Scholar]
- Yokoyama, A., Yamashino, T., Amano, Y., Tajima, Y., Imamura, A., Sakakibara, H., and Mizuno, T. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 48 84–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.