Abstract
Plastid signals are among the most potent regulators of genes that encode proteins active in photosynthesis. Plastid signals help coordinate the expression of the nuclear and chloroplast genomes and the expression of genes with the functional state of the chloroplast. Here, we report the isolation of new cryptochrome1 (cry1) alleles from a screen for Arabidopsis thaliana genomes uncoupled mutants, which have defects in plastid-to-nucleus signaling. We also report genetic experiments showing that a previously unidentified plastid signal converts multiple light signaling pathways that perceive distinct qualities of light from positive to negative regulators of some but not all photosynthesis-associated nuclear genes (PhANGs) and change the fluence rate response of PhANGs. At least part of this remodeling of light signaling networks involves converting HY5, a positive regulator of PhANGs, into a negative regulator of PhANGs. We also observed that mutants with defects in both plastid-to-nucleus and cry1 signaling exhibited severe chlorophyll deficiencies. These data show that the remodeling of light signaling networks by plastid signals is a mechanism that plants use to integrate signals describing the functional and developmental state of plastids with signals describing particular light environments when regulating PhANG expression and performing chloroplast biogenesis.
INTRODUCTION
Proteins that perform functions related to photosynthesis are encoded by both nuclear and chloroplast genomes. The chloroplast contains a small genome that encodes <100 proteins; some 2100 nuclear genes are predicted to encode chloroplast proteins in Arabidopsis thaliana, and as many as 4800 are predicted in rice (Oryza sativa) (Richly and Leister, 2004). Thus, the majority of proteins active in photosynthesis are encoded by nuclear genes. Coordinating photosynthesis-associated nuclear gene (PhANG) expression with the expression of photosynthesis-associated plastidic genes is central to the establishment and maintenance of the photoautotrophic lifestyle of plants. The regulation of PhANGs has been studied for decades, but a number of significant gaps remain in our knowledge of their regulation (Nott et al., 2006; Rook et al., 2006; Jiao et al., 2007).
The regulation of PhANGs is complex, involving signals perceived by multiple signaling pathways, such as those triggered by light, the circadian clock, tissue-specific signals, carbohydrates, hormones, and plastids (Nott et al., 2006; Rook et al., 2006; Jiao et al., 2007). Plastid signals affect photosystem stoichiometry and stress responses and are thought to regulate PhANGs as proplastids develop into chloroplasts, a process that is coordinated with the development of leaf cells from the leaf primordia and with the transition of cotyledons from heterotrophic to photoautotrophic organs after germination (Mullet, 1988, 1993; Nott et al., 2006). To study plastid signals, laboratories often use inhibitors or mutations to block chloroplast development or perturb chloroplast function. PhANG expression is most potently repressed by plastid signals when chloroplast biogenesis is blocked (Nott et al., 2006). These repressive plastid signals are stronger than inductive signals, such as the robust inductive signals from extraplastidic photoreceptors, and can repress PhANG expression to lower levels than are observed in the dark (Sullivan and Gray, 1999). The plastid signals emitted when chloroplast biogenesis is blocked are thought to contribute to proper gene expression during the early stages of chloroplast biogenesis and to help coordinate the expression of genes that encode functions related to photosynthesis that reside in both nuclear and chloroplast genomes. Proper coordination of nuclear and chloroplast genome expression is thought to be critical for proper chloroplast biogenesis, because much of the photosynthetic machinery is composed of large multisubunit protein complexes composed of both plastid and nuclear gene products (Nott et al., 2006).
Retrograde signaling pathways analogous to these plastid-to-nucleus signaling pathways have been reported for the mitochondria and endoplasmic reticulum. These other retrograde signaling pathways, which are understood in much more detail, inform the nucleus of the status of their respective organelles, causing an adjustment in the anterograde flow of gene products from the nucleus (Liu and Butow, 2006; Ron and Walter, 2007). During chloroplast biogenesis, in an analogous fashion, retrograde plastid-to-nucleus signaling pathways are thought to regulate the anterograde flow of gene products from the nucleus to the plastid, which is driven by extraplastidic signaling pathways that sense endogenous and environmental cues.
Although the molecular nature of plastid-to-nucleus signaling pathways active during chloroplast biogenesis remains poorly understood, our understanding of the regulation of PhANG expression by plastid-to-nucleus signaling pathways has been enhanced by the genomes uncoupled (gun) mutants. gun mutants uncouple the expression of genes that encode proteins active in photosynthesis with chloroplast function. When chloroplast biogenesis is blocked in wild-type seedlings, genes that encode proteins active in photosynthesis are repressed, but when chloroplast biogenesis is blocked in gun mutants, PhANGs are partially derepressed. The derepression of PhANG expression in gun mutants is thought to be due to at least partial inactivation of a plastid-to-nucleus signaling pathway that represses nuclear gene expression in response to particular plastid signals (Nott et al., 2006; Koussevitzky et al., 2007). The expression of the nuclear and chloroplastic genomes has also been reported to be uncoupled in gun1 mutants when chloroplast biogenesis is blocked (Susek et al., 1993).
Previous work with gun mutants indicates that the accumulation of Mg-protoporphyrin IX, inhibiting the expression of the chloroplast genome, high levels of glucose, and exposure to high-intensity light all produce a second messenger that triggers a plastid-to-nucleus signaling pathway that represses PhANG expression. GUN1, a chloroplastic pentatricopeptide repeat protein, is required for the biosynthesis or transduction of this second messenger. ABI4, an Apetala2-type transcription factor, functions downstream of GUN1 by binding promoter elements found in PhANGs (Nott et al., 2006; Koussevitzky et al., 2007). The discoveries that GUN1 and ABI4 act downstream of multiple plastid signals are consistent with the master switch integrating diverse plastid signals proposed by Richly et al. (2003). Although these recent advances are very exciting, significant gaps in our understanding of this form of interorganellar communication remain. For example, the identity of plastid signals, the mechanism by which signals exit the plastid, the mechanism by which the plastid interacts with other cellular compartments, and the impact of plastid signals on growth and development remain open questions. Additionally, because strong gun1 alleles express lower levels of PhANGs, when treated with inhibitors of chloroplast biogenesis compared with wild-type seedlings that are not treated with inhibitors of chloroplast biogenesis (Koussevitzky et al., 2007), it is likely that one or more additional plastid-to-nucleus signaling pathways that do not utilize GUN1 help coordinate PhANG expression with chloroplast biogenesis and function.
Much more is known about the regulation of PhANG expression by light than by plastid signals. In Arabidopsis and rice, at least 20% of the transcriptome is regulated by light. A number of photoreceptors and downstream signaling components have been shown to function in light-regulated transcriptional networks, and multiple light-responsive promoter elements have been identified in PhANGs (Jiao et al., 2007). Well-studied PhANGs, such as the genes that encode the light-harvesting chlorophyll a/b binding protein of photosystem II (Lhcb or CAB; hereafter referred to as Lhcb) and the ribulose-1,5-bis-phosphate carboxylase/oxygenase small subunit (Rbcs), are light-induced via the phytochrome and cryptochrome signaling pathways (Gao and Kaufman, 1994; Reed et al., 1994; Folta and Kaufman, 1999; Mazzella et al., 2001; Martinez-Hernandez et al., 2002). These pathways transduce far-red, red, and blue light signals (Jiao et al., 2007). The regulation of transcription by light is complex, involving the regulation of activity, subcellular localization, and concentrations of particular photoreceptors and downstream signaling components (Jiao et al., 2007).
Although light and plastid signals trigger distinct signaling pathways (Sullivan and Gray, 1999), it is known that plastid signals and light signals can regulate PhANG expression using common or adjacent promoter elements (Nott et al., 2006; Koussevitzky et al., 2007). To test whether plastid-to-nucleus signaling pathways and light signaling pathways might interact at some point upstream of these common promoter elements, we isolated a group of new gun mutants and screened them for light signaling phenotypes. We discovered that four of these mutants were allelic to cry1 and that cry1 alleles isolated by other laboratories were also gun mutants. We found that along with plastid signals and cry1, phyB and likely another phytochrome can also contribute to the repression of Lhcb when chloroplast biogenesis is blocked. Moreover, we found that the mechanism by which cry1 represses Lhcb expression when chloroplast biogenesis is blocked involves the conversion of HY5, a well-studied basic domain/leucine zipper transcription factor that acts downstream of cry1 and other photoreceptors (Jiao et al., 2007), from a positive regulator to a negative regulator of Lhcb and Rbcs. This remodeling of light signaling pathways was independent of the plastid-to-nucleus signaling pathway defined in previously isolated gun mutants and affected the response of PhANGs to both light quality and quantity. We also observed that gun1-101, a null allele, exhibited chlorophyll deficiencies that increased strikingly in either cry1 or hy5 null mutant backgrounds, which indicates that GUN1 and cry1 signaling pathways contribute to efficient chloroplast biogenesis in a redundant manner.
RESULTS
Isolation of cry1 Mutants from a gun Mutant Screen
We performed a screen to isolate new gun mutants that might contain defects in one or more of the steps in plastid-to-nucleus signaling that are poorly understood (see Supplemental Figure 1 online). In this screen, following ethyl methanesulfonate (EMS) mutagenesis, new gun mutants were identified as M2 seedlings that exhibited derepression of an Lhcb1*1:luciferase+ (luc+) reporter gene, as judged by bioluminescence of seedlings grown on medium containing norflurazon. Norflurazon blocks carotenoid biosynthesis by inhibiting phytoene desaturase. Without carotenoids, plastids experience severe photooxidative stress in bright light, chloroplast biogenesis is arrested at an early stage that resembles the proplastid, and PhANGs are severely repressed (Oelmüller, 1989). We expected that the Lhcb:luc+ reporter gene would provide a more sensitive screen than the original gun screen, which utilized an Lhcb-driven reporter gene that conferred hygromycin resistance upon gun mutants (Susek et al., 1993). With greater sensitivity, we expected to identify mutants with subtle phenotypes that were missed in the original screen. Progeny that inherited the Lhcb:luc+ reporter gene–based gun phenotype were grown on medium that contained norflurazon and screened for derepression of the endogenous Lhcb genes. Mutants that exhibited derepression of endogenous Lhcb genes when grown on medium containing norflurazon were then grown on medium containing lincomycin and tested for derepression of endogenous Lhcb genes. Like norflurazon, lincomycin blocks chloroplast biogenesis and causes severe repression of PhANGs. Lincomycin inhibits chloroplast biogenesis by specifically inhibiting plastid translation, which is an entirely different mechanism than norflurazon (Mulo et al., 2003). Thus, the mutants obtained at the end of this process cannot be resistant to a particular inhibitor and likely have defects in signaling pathways that regulate endogenous Lhcb genes.
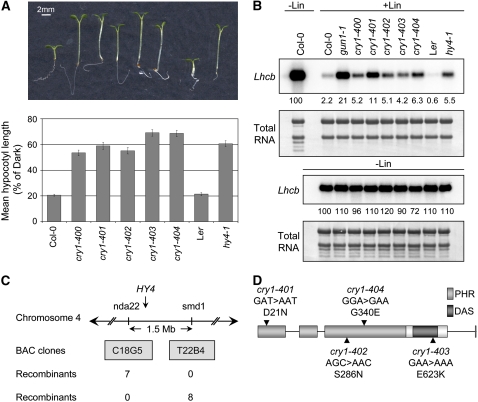
Because light and plastid-to-nucleus signaling pathways regulate Lhcb and a number of other PhANGs through common promoter elements, we tested whether some of our new gun mutants might also have light signaling defects. We found that four of our mutants exhibited long hypocotyls when grown in high-fluence-rate blue light (Figure 1A), which is consistent with these mutants having defects in cry1 signaling (Ahmad and Cashmore, 1993; Ahmad et al., 1995; Shalitin et al., 2003; Ohgishi et al., 2004). The gun phenotypes of these mutants were subtle compared with mutants isolated from the first gun screen, such as gun1-1 (Susek et al., 1993; Mochizuki et al., 2001). In fact, Lhcb mRNA accumulated to only twofold to threefold above wild-type levels in most of these new gun mutants. By contrast, we repeatedly observed that gun1-1 accumulated approximately 8- to 10-fold more Lhcb mRNA than the wild type when chloroplast biogenesis was blocked (Figure 1B). Like other gun mutants, Lhcb mRNA accumulated to similar levels as in the wild type when these mutants were not treated with inhibitors of chloroplast biogenesis (Figure 1B).
Figure 1.
Allelism of New gun Mutants and cry1 Mutants.
(A) Similar long-hypocotyl phenotypes of new gun mutants and cry1 mutants. Seedlings were grown in 25 μmol·m−2·s−1 blue light. Representative seedlings (top) and hypocotyl measurements (bottom) are shown. Hypocotyls were measured in blue light and in the dark for each line shown. Error bars indicate 95% confidence intervals (n > 36).
(B) Similar gun phenotypes of new gun mutants and cry1 mutants. Seedlings were grown in 125 μmol·m−2·s−1 white light on medium that either contained (+Lin) or lacked (−Lin) lincomycin. RNA was extracted and Lhcb mRNA levels were determined by RNA gel blotting using 3.0 μg of RNA. The levels of Lhcb transcripts were normalized to total RNA stained with methylene blue. Numbers below each lane indicate the amount of hybridized RNA as a percentage of hybridized RNA in untreated wild-type seedlings grown in the same light condition.
(C) Rough mapping of the long hypocotyl in blue light phenotype. The wild-type hypocotyl phenotype was rough-mapped based on an analysis of 118 chromosomes from F2 progeny that were obtained from a cry1-401 (Col-0) × Ler cross and exhibited wild-type hypocotyl lengths in 25 μmol·m−2·s−1 blue light. Chromosomes were analyzed using two simple sequence length polymorphism (SSLP) markers, nda22 and smd1 (see Supplemental Table 2 online). BAC clones that contain the SSLP marker sequences for nda22 and smd1 are indicated. The numbers of centromere proximal recombinants (top) and centromere distal recombinants (bottom) identified with each marker are indicated.
(D) CRY1 nucleotide and derived amino acid substitutions found in the new gun mutants. The altered codons and the resulting amino acid substitutions found in new cry1 mutants are indicated. Lines and boxes indicate introns and exons, respectively. The photolyase-related (PHR) domain and the DAS motifs have been reviewed by Lin and Shalitin (2003).
To test whether the gun phenotypes and the long-hypocotyl phenotypes might be linked, one of these new mutants was crossed to the parental line, Columbia-0 (Col-0), containing the Lhcb:luc+ reporter gene. In the F2 progeny, the long-hypocotyl phenotype segregated like a semidominant allele when seedlings were grown in high-fluence-rate blue light (S.M. DeMarco, M.E. Ruckle, and R.M. Larkin, unpublished data), as has been reported for cry1 mutants (Koornneef et al., 1980; Ahmad and Cashmore, 1993). Four F2 seedlings that exhibited long hypocotyls in blue light were propagated, and F3 progeny were found to be homozygous for the long hypocotyl in blue light and gun phenotypes (see Supplemental Table 1 and Figure 2 online), which indicates that these two phenotypes are linked. Because the wild-type hypocotyl phenotype could be scored unambiguously, we were able to determine that this phenotype mapped to a 1.5-Mb interval on chromosome 4 that contained CRY1 (Figure 1C). From these data, we hypothesized that this new mutant and possibly all four of our new gun mutants that exhibited long hypocotyls in blue light might be allelic to cry1. To test this idea, we sequenced CRY1 in all four mutants. We found G-to-A transitions in each mutant that caused substitutions in the derived amino acid sequence (Figure 1D). To further test the possibility that cry1 mutants were also gun mutants, we obtained a cry1 allele in which a T-DNA is inserted into the third exon of CRY1 (Salk_069292; Alonso et al., 2003). Because the last published set of cry1 alleles was numbered in the three hundreds (Shalitin et al., 2003), we refer to this T-DNA allele as cry1-400. We determined that cry1-400 does not accumulate CRY1 mRNA (see Supplemental Figure 3C online) and therefore must be a null allele. We observed that cry1-400 is a gun mutant and exhibits a similar gun phenotype compared with the aforementioned missense alleles. hy4-1, a cry1 mutant in the Landsberg erecta (Ler) ecotype (Ahmad and Cashmore, 1993), accumulated similar amounts of Lhcb mRNA compared with the other mutants, but because Lhcb is more severely repressed in Ler compared with Col-0, the gun phenotype of hy4-1 is actually more robust than a typical cry1 mutant in the Col-0 ecotype (Figure 1B). From these data, we concluded that cry1 contributes to the repression of Lhcb genes when chloroplast biogenesis is blocked and that our four new gun mutants are allelic to cry1. We named these new missense alleles cry1-401, cry1-402, cry1-403, and cry1-404. We have not yet determined whether the twofold stronger gun phenotype observed in cry1-401 is caused by an unlinked mutation or by the amino acid substitution caused by the cry1-401 allele.
Double Mutant Studies with cry1 and gun1 Mutants
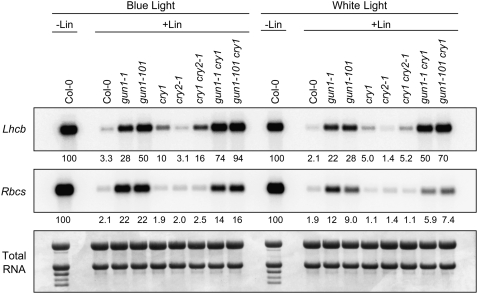
To learn more about the mechanism of PhANG regulation by GUN1 and cry1 when chloroplast biogenesis is blocked, we analyzed gun1 cry1 double mutants. Double mutants were constructed using cry1-400 and either gun1-1, a leaky missense allele (Susek et al., 1993; Koussevitzky et al., 2007), or a publicly available gun1 T-DNA allele (SAIL_33_D01; Sessions et al., 2002) that we refer to as gun1-101. gun1-101 expresses a partial GUN1 mRNA that encodes a truncated pentatricopeptide repeat domain but not the small mutS-related domain that is thought to be required for DNA binding activity (Koussevitzky et al., 2007) (see Supplemental Figures 3A and 3B online). According to the current model of GUN1 activity (Koussevitzky et al., 2007), gun1-101 should at least be a severe loss-of-function allele and possibly a null allele. Because cry1-400 was used in all subsequent experiments, for simplicity, we hereafter refer to cry1-400 as cry1. gun1-101 accumulated more Lhcb mRNA than gun1-1 when chloroplast biogenesis was blocked. This difference was always greater in blue light than in white light. In nine independent experiments with different inhibitors of chloroplast biogenesis and in either blue or white light, we observed 1.5- to 2.7-fold more Lhcb mRNA when chloroplast biogenesis was blocked in cry1 gun1-1 or cry1 gun1-101 double mutants than would be expected for additive increases caused by two pathways acting independently (Figures 2 to 4). These data indicate that GUN1 and cry1 are partially redundant in their repression of Lhcb gene expression when chloroplast biogenesis is blocked in these light conditions.
Figure 2.
Expression of Lhcb and Rbcs in gun1 and cry Mutants after Chloroplast Biogenesis Was Blocked.
Seedlings were grown in either 50 μmol·m−2·s−1 blue or 125 μmol·m−2·s−1 white light on medium that either contained (+Lin) or lacked (−Lin) lincomycin. The levels of Lhcb and Rbcs mRNA were quantitated as described for Figure 1B, except that 4.0 μg of RNA was used.
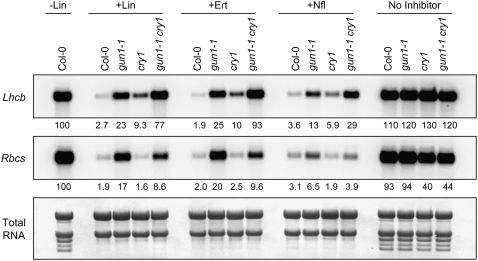
Figure 3.
Expression of Lhcb and Rbcs in gun1 and cry1 after Chloroplast Biogenesis Was Blocked with Various Inhibitors of Chloroplast Biogenesis.
Seedlings were grown in 50 μmol·m−2·s−1 blue light on medium that lacked any inhibitor of chloroplast biogenesis (−Lin) or contained lincomycin (+Lin), erythromycin (+Ert), or norflurazon (+Nfl). Lhcb and Rbcs mRNA levels were quantitated as described for Figure 1B, except that 4.0 μg of RNA was used.
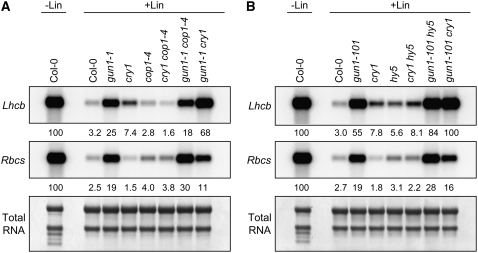
Figure 4.
Expression of Lhcb and Rbcs in cop1-4 and hy5 Mutants after Chloroplast Biogenesis Was Blocked.
(A) gun phenotypes of cop1-4. Seedlings were grown in 50 μmol·m−2·s−1 blue light on medium that contained (+Lin) or lacked (−Lin) lincomycin. Lhcb and Rbcs mRNA levels were quantitated as described for Figure 1B.
(B) gun phenotypes of hy5. The growth of seedlings and the quantitation of the levels of Lhcb and Rbcs mRNA were as described for (A).
In blue light, Lhcb mRNA accumulated to the same level in wild-type seedlings that were not treated with inhibitors of chloroplast biogenesis (i.e., green seedlings) and lincomycin-treated gun1-101 cry1 double mutants (Figures 2 and 4). These data suggest that when chloroplast biogenesis is blocked in blue light, most, if not all, of the repression of Lhcb is mediated by both GUN1 and cry1 and that gun1-101 is likely null. In white light, the gun1-101 cry1 double mutant accumulated 70% of Lhcb mRNA found in untreated controls, which suggests that perception of at least one additional light quality besides blue light or the presence of higher fluence rates might be important for maximal repression of Lhcb under these conditions.
We observed that although Lhcb was derepressed in cry1 when chloroplast biogenesis is blocked in either blue light or white light, Lhcb mRNA accumulated to similar levels in cry1 and the wild type when chloroplast biogenesis was blocked in red light (i.e., cry1 is not a gun mutant in red light) (see Supplemental Figure 4 online). We conclude that when chloroplast biogenesis is blocked, maximum repression of Lhcb is dependent on photoactivated cry1. The long hypocotyl in blue light phenotypes of all of the cry1 alleles described in this report (Figure 1A) further supports our conclusion that photoactivated cry1 represses Lhcb when chloroplast biogenesis is blocked.
Because redundancies have been observed between cry1 and cry2 in the regulation of some blue light–responsive processes (Casal, 2006), we tested whether a cry2 mutant might also be a gun mutant. We observed that cry2-1, a null allele (Guo et al., 1998), is not a gun mutant. However, the cry1 cry2-1 double knockout repeatedly exhibited a slightly stronger gun phenotype than cry1, but only in blue light (Figure 2). These results indicate that cry2 can partially compensate for cry1 in the cry1 background under these conditions. We also tested whether other blue light photoreceptor T-DNA insertion mutants (see Supplemental Figure 3 online) might also be gun mutants. We found that phot1, phot2, phyA, phyB, nph3, and cry3 T-DNA insertion mutants are not gun mutants in blue light (see Supplemental Figure 5 online).
Like Lhcb, Rbcs is repressed in the wild type and derepressed in gun1 mutants when chloroplast biogenesis is blocked (Figure 2) (Susek et al., 1993). In contrast with Lhcb, however, Rbcs is not derepressed in cry1 or gun1 cry1 when chloroplast biogenesis is blocked. In fact, Rbcs mRNA usually accumulated to slightly lower levels in cry1 than in the wild type and always accumulated to lower levels in gun1 cry1 double mutants than in gun1 mutants (Figures 2 to 4). These results indicate that cry1 induces Rbcs gene expression under these conditions. Because in gun1 mutants Rbcs mRNA accumulated to only ∼10 to 20% of that in untreated wild-type controls (Figure 2), we conclude that at least one additional pathway besides the GUN1 pathway likely contributes to the repression of Rbcs under these conditions. The more severe repression of Rbcs in white light than in blue light (Figure 2) suggests that, as with Lhcb, perception of multiple qualities of light or higher fluence rates might be required for the maximal repression of Rbcs when chloroplast biogenesis is blocked.
We observed the same patterns of Lhcb and Rbcs expression in gun1, cry1, and gun1-1 cry1 double mutants regardless of whether chloroplast biogenesis was blocked by treatments using lincomycin, erythromycin, or norflurazon (Figure 3). Both lincomycin and erythromycin specifically inhibit plastid translation, but they utilize different mechanisms (Mulo et al., 2003), and, as mentioned above, norflurazon is a phytoene desaturase inhibitor. Each of these inhibitors was shown previously to arrest chloroplast biogenesis at a stage resembling a proplastid and to severely repress PhANGs in Arabidopsis (Nott et al., 2006). Therefore, we conclude that the repression of Lhcb and Rbcs expression is likely caused by a reduction in the activities of particular signaling pathways and cannot be caused by resistance to particular inhibitors. Moreover, when seedlings were not treated with inhibitors of chloroplast biogenesis (i.e., in green seedlings), Lhcb mRNA accumulated to similar levels in gun1-1, cry1, gun1-1 cry1, and the wild type (Figure 3). When seedlings were not treated with inhibitors of chloroplast biogenesis, Rbcs mRNA accumulated to essentially the same levels in gun1-1 and the wild type, but Rbcs mRNA accumulated to lower levels in cry1 backgrounds regardless of whether chloroplast biogenesis was blocked (Figures 2 to 4). These data indicate that cry1 does not play an essential role in regulating Lhcb in green seedlings under these light conditions and that cry1 induces Rbcs regardless of the developmental state of the plastid.
Genetic Analyses of Downstream Signaling Components
cry1 has been shown to promote photomorphogenesis by inhibiting COP1, an E3 ubiquitin ligase that targets positive regulators of photomorphogenesis for degradation via the proteasome (Yi and Deng, 2005; Jiao et al., 2007). We tested whether cry1 utilized a COP1-dependent mechanism to repress Lhcb and simultaneously induce Rbcs expression when chloroplast biogenesis was blocked and whether GUN1 utilizes a COP1-dependent mechanism to repress PhANGs. When we analyzed Lhcb expression, we observed that cop1-4, a weak allele (Deng and Quail, 1992; McNellis et al., 1994), was not a gun mutant and that cop1-4 was epistatic to cry1 but had a minor impact on gun1-1 when chloroplast biogenesis was blocked (Figure 4A). These data indicate that cry1 functions through a COP1-dependent mechanism to repress Lhcb and suggest that GUN1 does not likely utilize a COP1-dependent mechanism to repress Lhcb under these conditions.
A different pattern of regulation was observed when we monitored Rbcs expression in cop1-4 single and double mutants. Because cry1 induces Rbcs when chloroplast biogenesis is blocked, we expected that lincomycin-treated cop1-4 would express higher levels of Rbcs than the wild type. cop1-4 and cry1 cop1-4 repeatedly accumulated slightly higher levels of Rbcs than the wild type under these conditions. Moreover, we did observe an enhanced derepression of Rbcs in cop1-4 gun1-1 compared with the cop1-4 and gun1-1 single mutants (Figure 4A). These data indicate that cop1-4 is epistatic to cry1 but not gun1-1, suggesting that cry1 and COP1 function in the same pathway and that GUN1 and cry1 function in different pathways.
Because cry1 contributes to the repression of Lhcb when chloroplast biogenesis is blocked, we speculated that other positive regulators of PhANG expression might also inhibit Lhcb expression when chloroplast biogenesis is blocked. We chose to test whether HY5, a positive regulator of PhANG expression in vivo (Lee et al., 2007) that acts downstream of multiple photoreceptors (Jiao et al., 2007), might contribute to the repression of PhANGs when chloroplast biogenesis is blocked. Indeed, we observed that a T-DNA insertion in HY5 (Salk_096651; Alonso et al., 2003) that we determined to be a null allele (see Supplemental Figure 3 online) and refer to as hy5 was a subtle gun mutant with an Lhcb expression phenotype similar to that of cry1 (Figure 4B). We also observed that Lhcb was expressed at similar levels in the cry1 hy5 double mutant as in the cry1 and hy5 single mutants. Additionally, the gun1-101 hy5 double mutant resembled the gun1 cry1 double mutants in that it exhibited enhanced Lhcb expression compared with the gun1-101 and hy5 single mutants. These data indicate that HY5 functions in the same pathway as cry1 and is responsible for much of the cry1-mediated repression of Lhcb when chloroplast biogenesis is blocked. These data also indicate that HY5 functions in a pathway that is distinct from GUN1 that represses Lhcb when chloroplast biogenesis is blocked. When we monitored Rbcs expression in the same mutants, we observed that hy5 and the wild type contained similar levels of Rbcs mRNA and that more Rbcs mRNA accumulated in the gun1-101 hy5 double mutant than in gun1-101 (Figure 4B). These data indicate that HY5 does not contribute to the cry1-mediated induction of Rbcs under these conditions. HY5 may contribute to the repression of Rbcs as it does for Lhcb, but these repressive effects can be observed only when GUN1 is not active.
Analysis of gun Phenotypes in Different Light Qualities and in phy Mutants
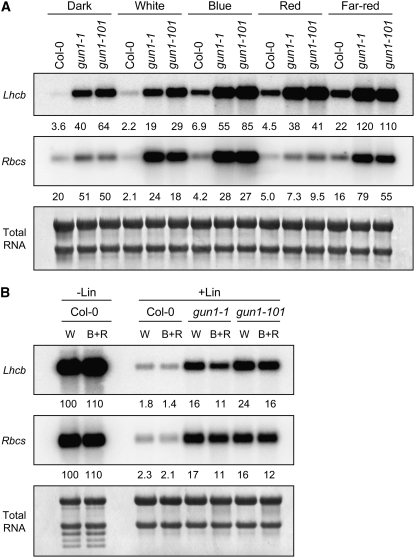
Because we repeatedly observed that regardless of genetic background, Lhcb and Rbcs mRNAs accumulated to higher levels when chloroplast biogenesis was blocked in blue light compared with white light, we hypothesized that the crosstalk between plastid-to-nucleus signaling pathways and light signaling pathways may be more complex than the interactions between the GUN1 and cry1 pathways described above. To test this idea, we compared PhANG expression in lincomycin-treated and untreated wild-type and gun1 seedlings in darkness, white light, or a fluence rate of blue, red, and far-red light that was equivalent to the fluence rate of each of these light qualities in our 125 μmol·m−2·s−1 white light. In these experiments, we blocked etioplast and chloroplast biogenesis by treating seedlings with lincomycin, which prevents plastid development beyond a proplastid stage and severely represses PhANG expression in either dark- or light-grown seedlings (Sullivan and Gray, 1999). We observed that in lincomycin-treated wild type, gun1-1, and gun1-101, Lhcb mRNA accumulated to the lowest levels in darkness but accumulated to only slightly higher levels in white light within a genetic background (Figure 5A). By contrast, Lhcb was expressed at approximately threefold or higher levels in blue, red, or far-red light than in white light and darkness within a genetic background (Figure 5A).
Figure 5.
Expression of Lhcb and Rbcs in the Dark or in Various Qualities of Light.
(A) Lhcb and Rbcs expression levels in darkness, white light, blue light, red light, and far-red light. Seedlings were grown on medium containing lincomycin in the dark or in 125 μmol·m−2·s−1 white light, 25 μmol·m−2·s−1 blue light, 35 μmol·m−2·s−1 red light, or 2 μmol·m−2·s−1 far-red light. The levels of Lhcb and Rbcs mRNA were quantitated as described for Figure 1B, except that 2.5 μg of RNA was used.
(B) Expression of Lhcb and Rbcs in white light and in a combination of blue and red light. Seedlings were grown on medium containing (+Lin) or lacking (−Lin) lincomycin in either white light (W) or a combination of blue and red light (B+R). Fluence rates were as described for (A). The levels of Lhcb and Rbcs mRNA were quantitated as described for (A). The numbers below the B+R (−Lin) lanes indicate the amount of hybridized RNA as a percentage of mRNA in the untreated control grown in white light (−Lin).
These data indicate that the perception of multiple qualities of light might be necessary for maximum repression of Lhcb or that maximum repression of Lhcb might require high fluence rates of light when chloroplast biogenesis is blocked. Moreover, these data show that different light qualities do not have a major impact on GUN1 activity and that light stimulates Lhcb expression compared with darkness under these conditions. When Lhcb expression levels were analyzed as a percentage of untreated wild-type seedlings in the same light conditions, we observed that Lhcb mRNA accumulated to lower levels in treated gun1 mutants compared with untreated wild-type seedlings in all light conditions except far-red. In far-red light, Lhcb mRNA accumulated to similar levels in treated gun1 mutants and untreated wild-type seedlings (Figure 5A). These data indicate that, in addition to GUN1, at least one other pathway is required to repress Lhcb expression in the dark, blue light, and red light when etioplast or chloroplast biogenesis is blocked and that only the GUN1 pathway is necessary to repress Lhcb in far-red light.
To test whether the simultaneous perception of blue and red light might be sufficient to produce the strong repression of Lhcb observed in white light, we compared Lhcb mRNA levels in lincomycin-treated and untreated wild-type and gun1 seedlings irradiated with white light and seedlings irradiated with a combination of blue and red light that were equivalent to the fluence rates of each of these light qualities in our 125 μmol·m−2·s−1 white light. Although seedlings exposed to white light received a larger quantity of light than seedlings exposed to a combination of blue and red light, we observed that seedlings accumulated lower levels of Lhcb mRNA when chloroplast biogenesis was blocked in a combination of blue and red light than in white light (Figure 5B). From these data, we conclude that most if not all of the Lhcb repression observed when chloroplast biogenesis is blocked in white light is caused by GUN1 and a combination of blue and red light and that a component of white light that is distinct from blue and red light may have a slight stimulatory effect on Lhcb when chloroplast biogenesis is blocked.
We obtained different results from a similar analysis of Rbcs expression. Like Lhcb, Rbcs was expressed at higher levels in blue light than in white light when chloroplast biogenesis was blocked (Figure 5A), as observed previously (Figure 2). In contrast with Lhcb, however, Rbcs was expressed at very low levels in red light compared with white light. In contrast with untreated seedlings (see Supplemental Figure 6 online), treated seedlings accumulated similar levels of Rbcs mRNA when chloroplast biogenesis was blocked in either red light or darkness. GUN1 had a minor effect on Rbcs expression in red light compared with white and blue light. An additional difference between Lhcb and Rbcs was that Rbcs mRNA did not accumulate to the same level as untreated controls in far-red light (Figure 5A). These data suggest that GUN1 and at least one other pathway are required to repress Rbcs when chloroplast biogenesis is blocked in far-red light. From these and previous results, it would appear that cryptochromes and phytochromes regulate Rbcs and Lhcb very differently when chloroplast biogenesis is blocked, in contrast with the similar regulation that we observed in these same light conditions when seedlings were not treated with inhibitors of chloroplast biogenesis (see Supplemental Figure 6 online) and the similar regulation that was reported previously for both Lhcb and Rbcs by cryptochromes and phytochromes when seedlings were not treated with inhibitors of chloroplast biogenesis (Mazzella et al., 2001; Martinez-Hernandez et al., 2002).
We observed that the gun1 mutants accumulate more Lhcb and Rbcs mRNA than the wild type when etioplast biogenesis is blocked in the dark and that in gun1 mutants neither Lhcb nor Rbcs mRNA accumulates to the levels observed in untreated wild-type seedlings grown in the dark (Figure 5A). These data are consistent with a previous report showing that lincomycin treatments can block etioplast biogenesis and that plastid-to-nucleus signaling does not depend on light (Sullivan and Gray, 1999). These data also show that GUN1 and at least one additional light-independent pathway repress these PhANGs when etioplast biogenesis is blocked. In the dark, as in all of the different light conditions that we tested, untreated gun1 mutants and untreated wild-type seedlings accumulated similar levels of Lhcb and Rbcs mRNA (see Supplemental Figures 6 and 7 online). Therefore, gun1 is distinct from the det/cop/fus mutants, which can express higher levels of PhANGs than the wild type, regardless of whether they are treated with inhibitors of etioplast biogenesis (Sullivan and Gray, 1999) (see Supplemental Figure 7 online).
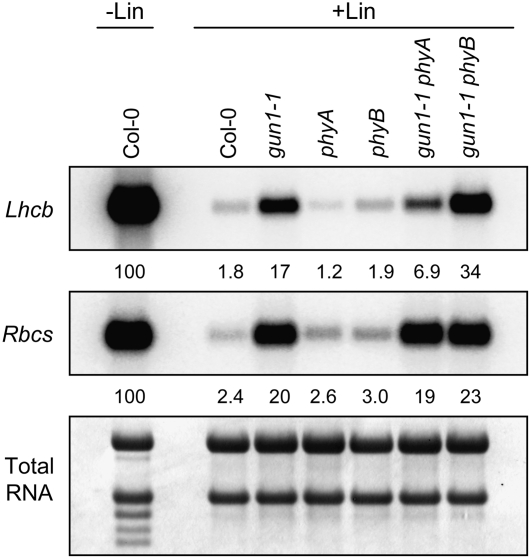
Because our data indicate that perception of red light is required for maximal repression of PhANGs when chloroplast biogenesis is blocked, we tested whether phyA, phyB, phyA gun1-1, and phyB gun1-1 exhibit gun phenotypes. In these experiments, we used T-DNA insertion alleles of phyA and phyB (Salk_ 014575 and Salk_ 022035; Alonso et al., 2003), which we determined to be null alleles (see Supplemental Figure 3 online). When Lhcb was monitored, we observed that neither phyA nor phyB was a gun mutant. We observed that phyA gun1-1 accumulated less and that phyB gun1-1 accumulated more Lhcb mRNA than gun1-1 (Figure 6). These data indicate that, like GUN1 and cry1, phyB can contribute to the repression of Lhcb when chloroplast biogenesis is blocked but that phyB is only a gun mutant when GUN1 is not active. By contrast, phyA only induces Lhcb when GUN1 is inactive. Rbcs mRNA accumulated to similar levels in phyA and phyB mutants, regardless of whether these alleles were in a wild-type or a gun1-1 background (Figure 6). These data indicate that, by themselves, neither phyA nor phyB is critical for either inducing or repressing Rbcs in wild-type seedlings when chloroplast biogenesis is blocked.
Figure 6.
Expression of Lhcb and Rbcs in gun1, phyA, and phyB Mutants after Chloroplast Biogenesis Was Blocked.
Seedlings were grown in 125 μmol·m−2·s−1 white light on medium that lacked (−Lin) or contained (+Lin) lincomycin. The levels of Lhcb and Rbcs mRNA were quantitated as described for Figure 1B.
Analysis of gun Phenotypes in Different Fluence Rates of White Light
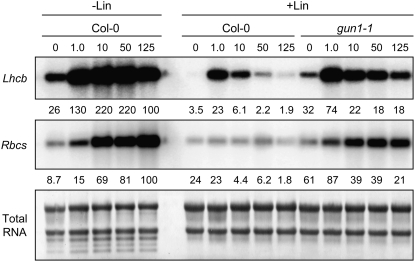
In addition to light quality, fluence rate can have a major impact on PhANG expression (Terzaghi and Cashmore, 1995). Because our data indicate that plastid signals can change the nature of PhANG regulation in response to light quality, we tested whether plastid signals might also influence the response of Lhcb and Rbcs to different quantities of light. The Lhcb and Rbcs mRNA levels were measured in wild-type and gun1 seedlings that were grown in increasing fluence rates of white light and either treated or not treated with lincomycin. In untreated seedlings, we found that the fluence rate response of Lhcb and Rbcs expression differed: Lhcb expression was inhibited at the highest fluence rate, but Rbcs was stimulated only by increasing the fluence rate (Figure 7). By contrast, when either the wild type or gun1-1 was treated with lincomycin, the peak of Lhcb expression was shifted to a lower fluence rate relative to the untreated control and Rbcs expression was inhibited by higher fluence rates, which was not observed in untreated seedlings. When seedlings were treated with lincomycin, the expression of each gene was most strongly inhibited by fluence rates >1 μmol·m−2·s−1 relative to the untreated control grown at the same fluence rate (Figure 7). These data indicate that the inhibition of Lhcb and Rbcs expression by increasing fluence rates of white light is enhanced when chloroplast biogenesis is blocked and that GUN1 does not affect this response to fluence rate. These data also show that under these conditions, GUN1 plays a major role in repressing Lhcb at low fluence rates but plays a less important role at higher fluence rates.
Figure 7.
Effects of Plastid Development on the Fluence Rate Response of Lhcb and Rbcs.
Seedlings were grown in white light fluence rates of 0, 1.0, 10, 50, or 125 μmol·m−2·s−1 in either the presence (+Lin) or absence (−Lin) of lincomycin. The levels of Lhcb and Rbcs mRNA were quantitated as described for Figure 1B. For the untreated wild type, the number below each lane indicates the amount of hybridized RNA as a percentage of hybridized RNA in the untreated wild type grown in 125 μmol·m−2·s−1 white light.
Although Lhcb genes have been reported to be strongly light-responsive (Terzaghi and Cashmore, 1995), we observed only a threefold to fourfold increase in Lhcb mRNA levels in white light–grown compared with dark-grown seedlings (Figure 7; see Supplemental Figure 6 online). Differences in experimental conditions likely account for these conflicting results. For example, most reports on the light regulation of Lhcb genes analyze the transient induction of Lhcb in etiolated seedlings grown on medium that lacks sucrose (Terzaghi and Cashmore, 1995). By contrast, we compared the steady state levels of Lhcb after several days of growth in either the dark or the light, and we included sucrose in the growth medium, which has been shown to inhibit Lhcb expression (Rook et al., 2006). Additionally, we found that the 125 μmol·m−2·s−1 used in these experiments can have an inhibitory effect on Lhcb expression (Figure 7).
Analysis of Chlorophyll Deficiencies in gun1 and Light Signaling Mutants
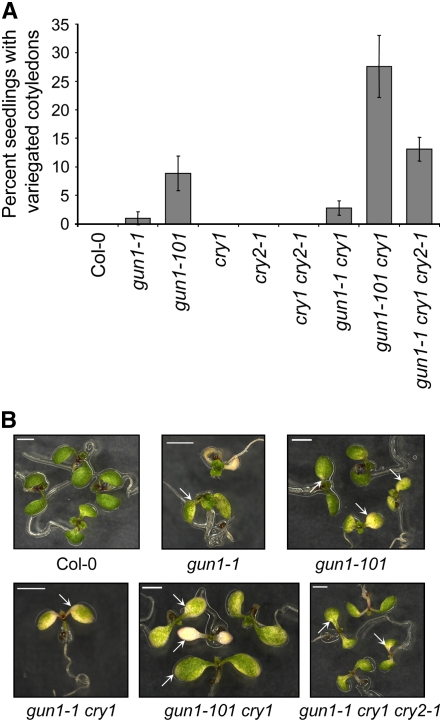
If plastid-to-nucleus signaling contributes to efficient chloroplast biogenesis, we would expect that chloroplast biogenesis would be less efficient in gun mutants than in the wild type and even more inefficient in gun1 cry1 double mutants compared with the wild type. Indeed, we observed that under growth conditions in which wild-type seedlings greened normally, a small percentage of gun1 mutants developed chlorophyll-deficient cotyledons (Figure 8). Consistent with our earlier experiments indicating that gun1-101 was a stronger allele than gun1-1 (Figures 2, 4, and 5), chlorophyll-deficient cotyledons were observed more frequently in gun1-101 than in gun1-1 (Figure 8). By contrast, neither cry1 nor cry1 cry2 developed chlorophyll-deficient cotyledons under these same conditions (Figure 8). Consistent with the idea that GUN1 and cry1 are at least partially redundant in the regulation of genes required for proper chloroplast biogenesis, the frequency of seedlings with chlorophyll-deficient cotyledons increased by approximately threefold in the gun1-101 cry1 double mutant compared with gun1-101. A similar increase was observed when gun1-1 cry1 cry2 was compared with gun1-1 cry1, which indicates that cry2 also contributes to efficient chloroplast biogenesis (Figure 8). Chlorophyll-deficient cotyledons varied from partially green organs that contained chlorophyll-deficient areas to uniformly albino cotyledons in both single and higher order mutants. Regardless of genetic background, seedlings with chlorophyll-deficient cotyledons produced mostly green primary leaves, but a minority produced primary leaves that only partially greened and in some cases appeared variegated (see Supplemental Figure 8 online).
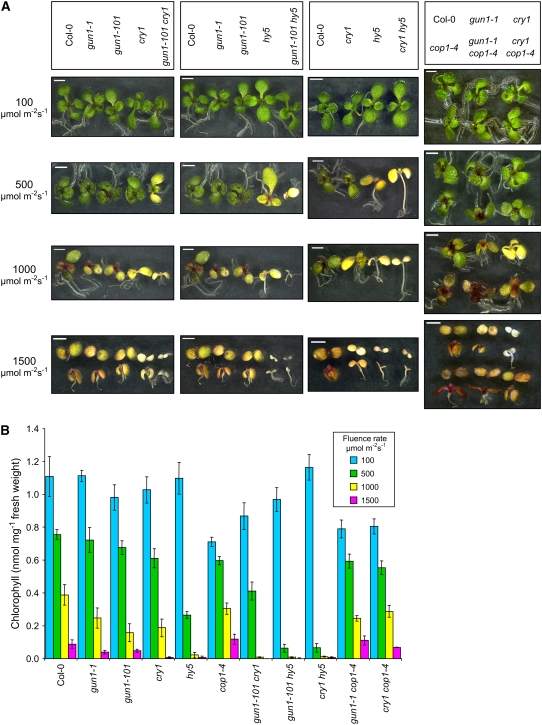
Figure 8.
Chlorophyll-Deficient Cotyledons in gun1 and gun1 cry Mutants.
(A) Percentage of seedlings exhibiting chlorophyll-deficient phenotypes. Seedlings were grown on medium without an inhibitor of chloroplast biogenesis. The total number of seedlings and the seedlings that were visibly chlorophyll-deficient were counted after 6 d of growth in 125 μmol·m−2·s−1 white light. Four independent experiments were performed, and each experiment contained a total of ∼50 seedlings. Error bars represent 95% confidence intervals between independent experiments.
(B) Chlorophyll-deficient seedlings. Representative wild-type (Col-0) and chlorophyll-deficient mutant seedlings are shown after 6 d (Col-0, gun1-101, gun1-1 cry1, and gun1-1 cry1 cry2) or 7 d (gun1-1 and gun1-1 cry1) of growth in white light. Arrows indicate chlorophyll-deficient areas. Bars = 2 mm.
If chloroplast biogenesis were inefficient in these mutants, we would expect them to be more susceptible than the wild type to photooxidative stress and albinism when chloroplast biogenesis proceeds in continuous high-intensity white light (HL). To test these mutants for HL sensitivity during chloroplast biogenesis, we allowed these seedlings to germinate in the dark for 23 h and then transferred them to various fluence rates of continuous white light. Smaller seedlings that contained less chlorophyll were judged to be more sensitive to HL. With the exception of gun1-101 cry1, cop1-4, and all cop1-4 double mutants, all mutants contained essentially the same level of chlorophyll as the wild type when seedlings were grown in 100 μmol·m−2·s−1 white light. At the higher fluence rates of white light, chlorophyll deficiency was uniformly distributed throughout a population of seedlings and not restricted to the cotyledons. We found that as the fluence rate of continuous white light was increased, the severity of chlorophyll deficiency increased more in the gun1, cry1, and hy5 single and double mutants compared with the wild type and that chlorophyll deficiency increased more in the double mutants made from combinations of gun1, cry1, and hy5 compared with the corresponding single mutants. At 500 μmol·m−2·s−1, chlorophyll levels were reduced synergistically in gun1 cry1 and gun1 hy5 double mutants compared with the corresponding single mutants. At higher fluence rates, the differences between these single and double mutants were less pronounced (Figure 9). We observed that gun1 mutants and cry1 mutants were similarly sensitive to HL under most fluence rates but that cry1 was more sensitive to HL than gun1 mutants at 1500 μmol·m−2·s−1. hy5 appeared to be more sensitive to HL than either gun1 or cry1 mutants. Accordingly, gun1-101 hy5 was more sensitive than gun1-101 cry1. When seedling size was considered, gun1-101 hy5 appeared to be more sensitive to HL than cry1 hy5 (Figure 9). These data are consistent with previous studies indicating that HY5 is a downstream component of multiple signaling pathways (Jiao et al., 2007) and with our gene expression results indicating that cry1 and GUN1 trigger distinct pathways.
Figure 9.
HL Sensitivity of gun1 and Light Signaling Mutants.
(A) gun1 and light signaling mutant seedlings grown in the indicated fluence rates of continuous white light. One-day-old etiolated seedlings were irradiated with the indicated fluence rates of continuous white light for 6 d. Representative seedlings are shown. Bars = 2 mm.
(B) Comparisons of total chlorophyll levels in gun1 and light signaling mutants in various fluence rates of continuous white light. Seedlings were grown as described for (A). Chlorophyll was extracted from at least three samples for each line in each condition. Error bars represent 95% confidence intervals.
We found that cop1-4 contained significantly less chlorophyll than the wild type at 100 and 500 μmol·m−2·s−1. Similar results have been reported previously (Deng and Quail, 1992). However, we observed that cop1-4, gun1-1 cop1-4, cry1 cop1-4, and the wild type contained similar levels of chlorophyll at 1000 and 1500 μmol·m−2·s−1. These data indicate that inhibition of COP1 is a major component of HL stress protection and are consistent with cry1 using a COP1-dependent mechanism to protect plants from HL. Because our gene expression studies indicate that GUN1 probably does not utilize a COP1-dependent mechanism to regulate PhANG expression, we suggest that the suppression of HL sensitivity in gun1-1 cop1-4 is likely caused by indirect effects.
To test whether mature chloroplasts in these single and double mutants are simply more sensitive to HL than those in the wild type, we grew wild-type seedlings and all of these mutants at 125 μmol·m−2·s−1 for 6 d and then transferred green seedlings to HL for 3 d. All of these mutants were green before they were transferred to continuous HL. We did not observe any consistent and striking differences in the pigmentation of mutants and the wild type except that the hy5 single and double mutants were always noticeably paler than wild-type plants at the highest fluence rate, but only in the youngest leaves (see Supplemental Figure 9 online). Similar results were obtained in an experiment with 3-week-old plants grown in soil (M.E. Ruckle and R.M. Larkin, unpublished data). Altogether, our analysis of greening in these mutants indicates that the photoprotective functions provided by GUN1, cry1, and HY5 are more important during chloroplast biogenesis than in seedlings that contain mature chloroplasts.
DISCUSSION
New cry1 Alleles Isolated from a gun Mutant Screen
The cryptochromes are composed of N-terminal DNA photolyase-related domains and C termini that are not related to DNA photolyases. The N termini bind the flavin adenine dinucleotide and methenyltetrahydrofolate chromophores and are necessary for dimerization. Both of these activities are necessary for light-dependent activation of the C-terminal domains (Lin and Shalitin, 2003; Sang et al., 2005). The blue light signal perceived by the photolyase-related domain is transduced by stimulating the C-terminal domains, which inhibit COP1 (Yi and Deng, 2005; Jiao et al., 2007). A large number of missense alleles that cause amino acid substitutions throughout cry1 were isolated previously. Like the new cry1 mutants described here, all of the previously isolated missense alleles exhibit long hypocotyls in blue light and all are loss-of-function alleles (Ahmad and Cashmore, 1993; Ahmad et al., 1995; Shalitin et al., 2003). cry1-401, cry1-402, and cry1-404 cause the amino acid substitutions D21N, S286N, and G340E, respectively, and likely render cry1 defective in one or more of the activities attributed to the N terminus. A missense allele that, like cry1-404, causes a G340E substitution in a photolyase signature sequence was isolated previously (Ahmad and Cashmore, 1993; Ahmad et al., 1995), but missense alleles that cause D21N and S286N substitutions have not been reported previously. The S286N substitution in cry1-402 is interesting because the mechanism by which cry1 transduces the blue light signal involves the phosphorylation of Ser residue(s) (Bouly et al., 2003; Shalitin et al., 2003) and because missense alleles that cause substitutions at Ser residues have not been reported previously. Although it is possible that the S286N substitution in cry1-402 removes an important phosphorylation site, it is also possible that replacing a Ser with an Asn residue at position 286 is simply disruptive to folding.
cry1-403 was the only allele we isolated that caused an amino acid substitution in the C terminus of cry1. C termini are poorly conserved among cryptochromes, but the C termini of most cryptochromes contain three well-conserved motifs referred to as DAS. The three motifs that make up the DAS motif are DQXVP (D), an acidic region (A), and STAES followed by GGXVP (S) (Lin and Shalitin, 2003). Other missense alleles have previously been reported to cause amino acid substitutions in and around the D and A motifs (Ahmad et al., 1995). cry1-403 is the only missense allele reported to alter the S motif, changing the highly conserved STAES motif to STAKS.
Plastid Signals Change the Nature of Lhcb Regulation by HY5
The cryptochromes have been shown to regulate gene expression in a blue light–dependent manner by binding and inhibiting COP1, an E3 ubiquitin ligase that targets photoreceptors and transcription factors that positively regulate photomorphogenesis (e.g., HY5) for degradation in the dark via the proteasome. Consistent with this mechanism, the short-hypocotyl phenotype of cop1 has been reported to be epistatic to the long-hypocotyl phenotype of cry1 (Ang and Deng, 1994). The results from our analysis of PhANG expression in double mutants resembles the analysis of hypocotyl length reported by Ang and Deng (1994) and are consistent (1) with cry1 utilizing a COP1-based mechanism to regulate PhANG expression regardless of whether cry1 is functioning as a positive or a negative regulator of PhANG expression and (2) with GUN1 not using a COP1-dependent mechanism to repress PhANGs when both etioplast and chloroplast biogenesis are blocked. From our double mutant studies, we also conclude that plastid signals convert cry1 signaling pathways from positive to negative regulators of Lhcb by converting HY5 from a positive to a negative regulator of Lhcb. Previously, HY5 has been reported to function only as a positive regulator of PhANGs like Lhcb and Rbcs in vivo, but HY5 has been reported to negatively regulate a number of other genes (Lee et al., 2007). The mechanism by which HY5 is converted from a positive regulator to a negative regulator of Lhcb expression is an open question whose answer may include posttranslational modifications, heterodimerization with distinct transcription factors, changes in the concentrations of coactivators and corepressors, or some combination of these mechanisms.
Koussevitzky et al. (2007) showed that GUN1 prevents light signaling pathways from inducing PhANGs by promoting the binding of ABI4 adjacent to promoter elements that contain G-boxes, which are important for the light induction of PhANGs. Our finding that HY5 only functions as a negative regulator of Rbcs expression when the GUN1 pathway was inactivated is consistent with the GUN1 pathway preventing G-box binding factors such as HY5 (Lee et al., 2007) from regulating Rbcs expression, as proposed by Koussevitzky et al. (2007). By contrast, we found that full repression of Lhcb genes requires not only GUN1 but also cry1 and HY5. Thus, our findings indicate that Lhcb and Rbcs are repressed by distinct mechanisms when chloroplast biogenesis is blocked.
Crosstalk between Plastids and Light Signaling Networks
Cryptochromes have previously been shown to regulate blue light–inducible genes, especially genes that encode proteins with functions related to photosynthesis (Ohgishi et al., 2004). cry1 induces Rbcs expression (Martinez-Hernandez et al., 2002). cry1 can induce Lhcb expression but has been suggested to repress Lhcb during high light stress (Mazzella et al., 2001). Consistent with these reports, we observed that Lhcb mRNA accumulated to lower levels in green seedlings when white light fluence rates were increased to >50 μmol·m−2·s−1. Little is known about the mechanisms that plants use to sense different quantities of light (Jiao et al., 2007). Our finding that Lhcb was repressed at much lower fluence rates when chloroplast biogenesis was blocked than in the untreated green seedlings indicates that the functional and developmental state of chloroplasts has a major impact on the response of PhANGs to fluence rates of light.
In the light, cry1 induces Lhcb, but it is not possible to observe these inductive effects in a cry1 mutant because of redundant induction by phyA and phyB (Mazzella et al., 2001). Moreover, phyA, phyB, and cry1 are all important for the light induction of Rbcs (Martinez-Hernandez et al., 2002). Interactions between cry1 and phytochromes that affect photomorphogenesis have also been reported previously (Casal, 2006). Therefore, we expected that the crosstalk between plastid and light signaling pathways that affect PhANG expression when chloroplast biogenesis is blocked might also involve one or more phytochromes in addition to GUN1 and cry1. Indeed, our results showing (1) that GUN1 and cry1 are necessary and sufficient for most if not all repression of Lhcb in high-fluence-rate blue light but not in white light and (2) that perception of both blue light and red light is essential for maximal repression of Lhcb and Rbcs when chloroplast biogenesis is blocked suggest that phy activity is probably also required for the repression of Lhcb under these conditions.
Both phyA and phyB have been shown to induce Lhcb and Rbcs expression in seedlings that are not treated with inhibitors of chloroplast biogenesis (Reed et al., 1994; Martinez-Hernandez et al., 2002). However, we found that phyA and phyB regulate Lhcb differently when chloroplast biogenesis is blocked. phyA remains a positive regulator of Lhcb in seedlings treated with inhibitors of chloroplast biogenesis, which is apparent when GUN1 is inactive. In contrast with phyA, and like cry1, phyB acts as a negative regulator of Lhcb when chloroplast biogenesis is blocked but only in the gun1-1 background. Although these data indicate that phyB contributes to the repression of Lhcb in white light when chloroplast biogenesis is blocked, they also suggest that the repression of Lhcb by phytochromes is likely complex. Our analysis of Rbcs expression in different light conditions also suggests that the perception of both blue and red light is critical for maximum repression when chloroplast biogenesis is blocked and that this repression likely requires multiple phytochromes. Consistent with this idea, Arabidopsis contains five phytochromes (phyA to phyE), and functional redundancies have been reported among some of these phytochromes (Casal, 2006). A comprehensive analysis of phy mutants will likely be required to understand the interactions between plastids and red light that affect PhANG expression.
Interactions between light and plastid signaling have already been suggested, because light and plastid signals utilize common or adjacent promoter elements (Nott et al., 2006; Koussevitzky et al., 2007) and phytochrome regulation of PhANGs was reported to be impaired in mutants with defective chloroplasts (Vinti et al., 2005). One interpretation of these data would be that PhANG expression is controlled by a balance between inductive light signaling pathways and repressive plastid signaling pathways acting independently. In this model, the gun phenotypes of cry1, hy5, and phyB could be explained if cry1, HY5, and phyB enhance plastid stress (e.g., photooxidative stress), thereby enhancing the activity of inhibitors that block chloroplast biogenesis. Although this model is difficult to rule out completely, it is inconsistent with our data and other published results. First, some of these light signaling proteins have been reported to protect chloroplasts from stress. For example, cry1 was reported previously to protect plants from chloroplast stress induced by high-intensity light (Kleine et al., 2007). We observed that cry1, HY5, and GUN1 also protect plants from albinism induced by HL (Figure 9) and that cry1 and GUN1 protect plants from albinism in 125 μmol·m−2·s−1 white light (Figure 8). Second, if cry1, HY5, and phyB promoted plastid stress in lincomycin-treated seedlings, plastids in dark-grown lincomycin-treated seedlings would be less stressed than those in light-grown lincomycin-treated seedlings. Such differences in plastid stress might affect plastid size and ultrastructure. However, plastid development has been reported to be similar in light-grown and dark-grown lincomycin-treated seedlings in both wild-type and COP1-deficient backgrounds (Sullivan and Gray, 1999, 2000). Analysis of plastid ultrastructure indicates that GUN1 also does not enhance plastid stress in norflurazon-treated seedlings (Susek et al., 1993). Nonetheless, analysis of plastid ultrastructure in the wild type and gun1 cry1 double mutants would help test this model.
Our analysis of PhANG expression in seedlings treated with inhibitors of chloroplast biogenesis also suggests that these light signaling proteins most likely do not induce plastid stress. For example, the GUN1 pathway appears to function as a master switch that integrates multiple plastid signals (Koussevitzky et al., 2007). If cry1, HY5, and phyB promote stress that triggers the GUN1 pathway, a seedling would not have a more robust gun phenotype than that observed in gun1-101. In other words, gun1-101 would be epistatic to cry1, hy5, and phyB; however, gun1 mutants are not epistatic to any of these mutants. If these light signaling proteins induce plastid stress that triggers a GUN1-independent plastid-to-nucleus signaling pathway, we would expect enhanced derepression of PhANG expression in cry1, hy5, and phyB mutants treated with inhibitors of chloroplast biogenesis. However, our analysis of Lhcb expression is consistent with both cry1 and HY5 inducing plastid stress in lincomycin-treated seedlings, but our analysis of Rbcs expression is consistent with HY5 inducing plastid stress and cry protecting plastids from stress under these conditions (Figure 4). It is difficult to imagine a simple mechanism in which HY5 could promote plastid stress that simultaneously represses Lhcb and Rbcs while cry1 concurrently promotes plastid stress that represses Lhcb and in parallel reduces plastid stress that represses Rbcs. Moreover, because our analysis of Lhcb and Rbcs expression indicates that these PhANGs are similarly repressed when chloroplast biogenesis is blocked, these data are also not likely explained by Rbcs being more sensitive than Lhcb to chloroplast stress. Although a mechanism in which cry1, HY5, and phyB promote plastid stress when seedlings are treated with inhibitors of chloroplast biogenesis would need to be complex to be consistent with the available data, more work will be required to completely rule out this possibility.
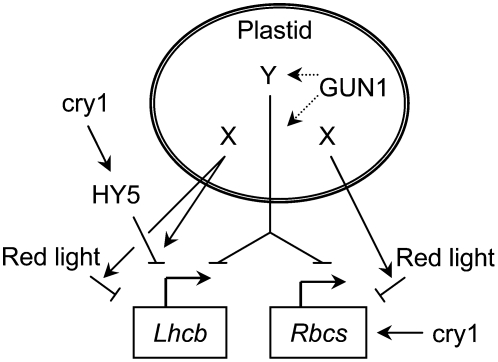
A model in which the functional and developmental state of the plastid controls the nature of PhANG regulation by light signaling pathways is consistent with all available data (Figure 10). In this model, PhANG expression (1) is repressed by the GUN1-dependent plastid-to-nucleus signaling pathway and (2) may (e.g., Lhcb) or may not (e.g., Rbcs) be repressed by a plastid signal that converts HY5 from a positive to a negative regulator of PhANGs and is distinct from the plastid signal that is either produced or transduced by a GUN1-dependent pathway. In this model, a plastid signal determines whether cry1 is a positive or a negative regulator of Lhcb and the fluence rate of blue light determines the amount of pathway activity. The conversion of light signaling pathways from positive to negative regulators of Lhcb allows plants to repress Lhcb more severely than if light signaling pathways remained inductive and simply competed with repressive plastid-to-nucleus signaling pathways. The additional flexibility afforded by integrating light and plastid signals in this manner might facilitate chloroplast biogenesis and repair in diverse light environments.
Figure 10.
Model for PhANG Regulation by a Network of Plastid and Light Signaling Pathways.
The current model for the GUN1-dependent plastid-to-nucleus signaling pathway was adapted from Koussevitzky et al. (2007). In this model, a second messenger (indicated with Y) that requires GUN1 for either its production or transduction (dotted arrows) triggers a plastid-to-nucleus signaling pathway that represses PhANGs. A plastid signal(s) that is independent of GUN1 (indicated with X) represses both Lhcb and Rbcs in the dark (data not shown) and also converts cry1 and one or more photoreceptors that perceive red light into negative regulators of Lhcb and Rbcs. cry1 becomes a negative regulator of Lhcb when X converts HY5 from a positive to a negative regulator of Lhcb. Under these same conditions, Rbcs is induced by cry1 and simultaneously repressed by a combination of blue and red light.
Because cry1 remains a positive regulator of Rbcs regardless of whether seedlings are treated with inhibitors of chloroplast biogenesis, we might expect that Rbcs would be expressed at higher levels than Lhcb when chloroplast biogenesis is blocked. However, we found that Lhcb and Rbcs are similarly repressed when chloroplast biogenesis is blocked in white light. These data, our analysis of Lhcb and Rbcs expression in darkness and in particular light qualities, and our analysis of phy mutants argue that the plastid signals have a broad impact on the nature of PhANG regulation by light signaling pathways and that multiple pathways repress PhANGs in response to chloroplast function and developmental state. Thus, the mechanisms by which plastid signals inhibit PhANG expression appear to be more complex than suggested by Koussevitzky et al. (2007).
Plastid Signals Are Required for Efficient Chloroplast Biogenesis
gun1 mutants were shown to have much greater difficulty greening than wild-type seedlings after prolonged periods of growth in the dark, which is consistent with GUN1 performing an important function during chloroplast biogenesis (Mochizuki et al., 1996). Aside from this phenotype, gun1 mutants have not been reported to have other morphological or pigmentation defects (Nott et al., 2006; Koussevitzky et al., 2007). However, the triggering of the GUN1 pathway by HL implicates this pathway in HL resistance (Koussevitzky et al., 2007). Indeed, we observed that gun1 mutants are more sensitive to HL than are wild-type plants. Moreover, the striking increase in chlorophyll deficiencies that we observed in gun1 cry mutants in 125 μmol·m−2·s−1 white light and the synergistic decrease in chlorophyll levels that we observed in gun1 cry and gun1 hy5 double mutants in 500 μmol·m−2·s−1 white light lead us to several conclusions: (1) these two pathways are required for efficient chloroplast biogenesis; (2) efficient chloroplast biogenesis likely requires two plastid signals, one transduced by the GUN1-dependent pathway and the other by the GUN1-independent pathway that is partially transduced by cry1, phyB, and HY5; and (3) the full impact of plastid-to-nucleus signaling on chloroplast biogenesis has been underappreciated because of redundancies between these two plastid-to-nucleus signaling pathways.
In 125 μmol·m−2·s−1 white light, we observed that GUN1 and cryptochromes are more important for chloroplast biogenesis in cotyledons than in primary leaves, which is consistent with previous reports indicating that chloroplast biogenesis in cotyledons and true leaves requires different genes (Yamamoto et al., 2000; Albrecht et al., 2006). However, the impact of plastid-to-nucleus signaling on chloroplast biogenesis in both cotyledons and primary leaves may be far greater than suggested by these data. Because we found that the plastid-to-nucleus signaling pathway that controls the nature of PhANG regulation affects both blue and red light signaling and that GUN1 and at least one other GUN1-independent pathway repress PhANGs when etioplast biogenesis is blocked in the dark, we conclude that plastid-to-nucleus signaling is likely complex. To determine the full impact of plastid-to-nucleus signaling on chloroplast biogenesis, it will be necessary to analyze chloroplast biogenesis in mutants in which all plastid-to-nucleus signaling pathways are inactivated.
METHODS
Plant Materials and Growth Conditions
hy4-1 was in the Ler ecotype of Arabidopsis thaliana (Ahmad and Cashmore, 1993). All other mutants were in the Col-0 ecotype. cry2-1, hy4-1, and all of the T-DNA alleles were obtained from the ABRC (Ohio State University). Seeds were surface-sterilized by mixing them in 70% ethanol, 0.5% Triton X-100 solution for 10 min on a tube mixer, incubating them in 95% ethanol for 10 min on a tube mixer, followed by air drying on filter paper soaked in 95% ethanol in a laminar flow hood. Seeds were plated on Linsmaier and Skoog medium containing 2.0% sucrose and 0.5% Phytoblend (Caisson Laboratories). Five micromolar norflurazon, 0.5 mM lincomycin, or 0.5 mM erythromycin was included in the growth medium to block chloroplast biogenesis (Nott et al., 2006); all were purchased from Sigma-Aldrich. Seeds were stratified for 4 d at 4°C, irradiated with 125 μmol·m−2·s−1 red light for 1 h at 21°C, incubated in the dark for 23 h at 21°C as recommended by Fankhauser and Casal (2004), and grown for 6 d at 21°C in the specified light conditions in environmentally controlled chambers (Percival Scientific). For experiments in white light, other than HL, light was provided by broad-spectrum fluorescent tube lamps at 125 μmol·m−2·s−1. To measure the spectral quality of our white light, we used a StellarNet EPP2000 spectroradiometer (Apogee Instruments). For HL experiments, a combination of high-pressure sodium and metal halide lamps was used. For single light quality experiments and experiments with combinations of blue and red light, seedlings were grown in controlled-environment chambers containing light-emitting diodes (Percival Scientific). In these chambers, the blue light peak was at 470 nm with a spectral bandwidth of 25 nm, the red light peak was at 669 nm with a spectral bandwidth of 25 nm, and the far-red light peak was at 739 nm with a spectral bandwidth of 31 nm. Far-red light was passed through one filter (No. 116; Lee Filters) to remove wavelengths that were <700 nm. During fluence rate response experiments, white light was filtered through neutral density filters (Roscolux 397; Rosco Laboratories). For far-red light, fluence rates were measured with a StellarNet EPP2000 spectroradiometer (Apogee Instruments). All other fluence rates were measured with an LI-250A photometer using a PAR sensor (LI-COR Biosciences).
Genetic Methods
A Col-0 line harboring an Lhcb:luc+ reporter gene was mutagenized using EMS as recommended by Weigel and Glazebrook (2002). Pools of M2 seeds representing ∼20 to 30 M2 families were surface-sterilized and plated in medium that contained 5 μM norflurazon as described above. Seeds were stratified as described above and then incubated for 8 to 9 d in constant 125 μmol·m−2·s−1 white light at 21°C. Photobleached seedlings were screened for derepression of Lhcb:luc+ by imaging bioluminescence as recommended by Chinnusamy et al. (2002) using a low-light imaging camera from EG&G Berthold. The bioluminescence of putative mutants was compared with that of the Col-0 Lhcb:luc+ parental line and the gun1-1 and gun5 lines (Mochizuki et al., 2001) in which the Lhcb:luc+ reporter gene was introduced from the Col-0 Lhcb:luc+ parental line by crossing.
For mapping, gun mutants were crossed to a Ler line in which the Lhcb:luc+ reporter gene was introgressed by 12 crosses to Ler. F2 progeny that exhibited a wild-type hypocotyl phenotype in 25 μmol·m−2·s−1 blue light were used to map cry1-401 with SSLP markers (Bell and Ecker, 1994) using the Cereon Genomics Indel database (Jander et al., 2002) (see Supplemental Table 2 online) and procedures described by Weigel and Glazebrook (2002). To sequence CRY1 in the mutants isolated from the gun mutant screen, the CRY1 coding sequence was amplified by means of Platinum Pfx DNA polymerase (Invitrogen) using CRY1-specific oligonucleotides in at least 10 aliquots that were subsequently pooled, purified from agarose gels using the QIAquick gel extraction kit (Qiagen), and sequenced with gene-specific oligonucleotides by the Research Technology Support Facility (Michigan State University).
Oligonucleotides for identifying T-DNA alleles (see Supplemental Table 3 online) were designed using the recommendations of the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/). Double mutants were identified among progeny of appropriate crosses using SSLP, cleaved-amplified polymorphic sequence (CAPS) (Konieczny and Ausubel, 1993; Bell and Ecker, 1994), or derived CAPS (Neff et al., 1998) markers (see Supplemental Table 4 online).
Hypocotyl Measurements
Seedlings were grown as described above but on medium lacking sucrose. Hypocotyl measurements were performed as recommended by Fankhauser and Casal (2004).
Analysis of RNA
For RT-PCR, RNA was isolated from 7-d-old seedlings using the RNeasy Plant Miniprep kit (Qiagen), including the on-column DNase treatment. First-strand cDNA was synthesized from 2 μg of RNA using the Omniscript RT kit (Qiagen). PCR was programmed with Taq polymerase (Invitrogen) and gene-specific oligonucleotides (see Supplemental Table 3 online). PCR products were analyzed after 20, 25, and 30 cycles. UBQ10 expression was analyzed to test whether the same amounts of cDNA were used for each PCR, as recommended by Weigel and Glazebrook (2002).
For RNA gel blotting, RNA was extracted as described for RT-PCR without the on-column DNase treatment. RNA gel blotting was performed as recommended by Chory et al. (1991) with the indicated quantities of RNA. The Lhcb probe was prepared by amplifying the entire open reading frame from cDNA clone U13603, which encodes Lhcb1*1 (At1g29920), using 5′-CCGGAATTCATGGCCTCAACAAT-3′, 5′-TCCCCGCGGTCACTTTCCGGGAACAA-3′, and Taq DNA polymerase (Invitrogen). The Rbcs probe was prepared by amplifying part of the Rbcs open reading frame from cDNA clone U15710, which encodes Rbcs-1A (At1g67090), essentially as described for Lhcb but using 5′-TATGGTCGCTCCTTTCAACG-3′ and 5′-TGATGCACTGGACTTGACGG-3′. Both cDNA clones were obtained from the ABRC. To prepare each probe, PCR products were purified by agarose gel electrophoresis and extracted from gel slices using the QIAquick gel extraction kit (Qiagen). Purified PCR products were labeled using the Random Primers DNA Labeling system (Invitrogen). Hybridized RNA gel blots were imaged using Imaging Screen K (Bio-Rad) and analyzed using the Molecular Imager FX (Bio-Rad). Lhcb and Rbcs mRNAs were quantitated using Quantity One one-dimensional analysis software (Bio-Rad) and normalized to methylene blue–stained 18S rRNA that was quantitated using the same software. Normalization by this method was found to be in the linear range of detection from 2 to 6 μg of total RNA (R2 = 0.97). The same relative levels of Lhcb and Rbcs mRNA accumulation among genetic backgrounds were observed repeatedly and were consistent within a particular light condition.
Chlorophyll Measurements
Chlorophyll was extracted and quantitated as recommended by Porra et al. (1989).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The gun Mutant Screen Procedure.
Supplemental Figure 2. Cosegregation Analysis of the Long Hypocotyl in Blue Light and gun Phenotypes.
Supplemental Figure 3. Analysis of T-DNA Alleles.
Supplemental Figure 4. Expression of Lhcb in cry1 after Chloroplast Biogenesis Was Blocked in Red Light.
Supplemental Figure 5. Lhcb and Rbcs Expression in gun1-1 and Various Photoreceptor Mutants after Chloroplast Biogenesis Was Blocked in Blue Light.
Supplemental Figure 6. Lhcb and Rbcs mRNA Levels in the Wild Type and gun1 Mutants Grown in Darkness and Various Light Qualities without Inhibitors of Chloroplast Biogenesis.
Supplemental Figure 7. Analysis of gun Phenotypes in gun1-1 and cop1-4.
Supplemental Figure 8. Chlorophyll-Deficient Leaves in gun1 and gun1 cry1 Double Mutants.
Supplemental Figure 9. Phenotypes of gun1 and Light Signaling Mutants following HL Incubations.
Supplemental Table 1. Segregation of the Long-Hypocotyl Phenotype in 25 μmol·m2·s−1 Blue Light in F3 Seedlings.
Supplemental Table 2. SSLP Markers Used for Rough Mapping of cry1-401.
Supplemental Table 3. Primers Used for T-DNA Genotyping and RT-PCR.
Supplemental Table 4. CAPS and dCAPS Markers for gun1-1 and cop1-4.
Supplementary Material
Acknowledgments
We are grateful to Steve Kay (Scripps Research Institute) for providing the Arabidopsis line containing the Lhcb:luc+ reporter gene, Joanne Chory (Salk Institute for Biological Studies) for providing gun1-1 and gun5 seeds, Xing Wang Deng (Yale University) for providing cop1-4 seeds, and Neil Adhikari (Michigan State University) for providing the SSLP marker nda22. We thank Abby Lott, Todd Lydic, Stephanie Buck, and Jackson Gehan (Michigan State University) for providing helpful assistance during the EMS mutant screen and Beronda Montgomery-Kaguri (Michigan State University) for many helpful discussions during the course of this work and helpful comments on the manuscript. We thank Gregg Howe (Michigan State University) for helpful comments on the manuscript. This work was supported by Department of Energy Grant DE-FG02-91ER20021 and National Science Foundation Grant IOB 0517841 to R.M.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Robert M. Larkin (larkinr@msu.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Lin, C., and Cashmore, A.R. (1995). Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J. 8 653–658. [DOI] [PubMed] [Google Scholar]
- Albrecht, V., Ingenfeld, A., and Apel, K. (2006). Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60 507–518. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Ang, L.H., and Deng, X.-W. (1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bouly, J.P., Giovani, B., Djamei, A., Mueller, M., Zeugner, A., Dudkin, E.A., Batschauer, A., and Ahmad, M. (2003). Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur. J. Biochem. 270 2921–2928. [DOI] [PubMed] [Google Scholar]
- Casal, J.J. (2006). The photoreceptor interaction network. In Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms, E. Schaefer and F. Nagy, eds (Dordrecht, The Netherlands: Springer), pp. 407–437.
- Chinnusamy, V., Stevenson, B., Lee, B.-H., and Zhu, J.K. (2002). Screening for gene regulation mutants by bioluminescence imaging. Science's STKE, http://www.stke.org/cgi/content/full/sigtrans;2002/140/pl10. [DOI] [PubMed]
- Chory, J., Nagpal, P., and Peto, C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X.-W., and Quail, P.H. (1992). Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 2 83–95. [Google Scholar]
- Fankhauser, C., and Casal, J.J. (2004). Phenotypic characterization of a photomorphogenic mutant. Plant J. 39 747–760. [DOI] [PubMed] [Google Scholar]
- Folta, K.M., and Kaufman, L.S. (1999). Regions of the pea Lhcb1*4 promoter necessary for blue-light regulation in transgenic Arabidopsis. Plant Physiol. 120 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J., and Kaufman, L.S. (1994). Blue-light regulation of the Arabidopsis thaliana Cab1 gene. Plant Physiol. 104 1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279 1360–1363. [DOI] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y., Lau, O.S., and Deng, X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8 217–230. [DOI] [PubMed] [Google Scholar]
- Kleine, T., Kindgren, P., Benedict, C., Hendrickson, L., and Strand, Å. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. Bd. 100 147–160. [Google Scholar]
- Koussevitzky, S., Nott, A., Mockler, T.C., Hong, F., Sachetto-Martins, G., Surpin, M., Lim, J., Mittler, R., and Chory, J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316 715–719. [PubMed] [Google Scholar]
- Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., Tongprasit, W., Zhao, H., Lee, I., and Deng, X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., and Shalitin, D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54 469–496. [DOI] [PubMed] [Google Scholar]
- Liu, Z., and Butow, R.A. (2006). Mitochondrial retrograde signaling. Annu. Rev. Genet. 40 159–185. [DOI] [PubMed] [Google Scholar]
- Martinez-Hernandez, A., Lopez-Ochoa, L., Arguello-Astorga, G., and Herrera-Estrella, L. (2002). Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol. 128 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzella, M.A., Cerdan, P.D., Staneloni, R.J., and Casal, J.J. (2001). Hierarchical coupling of phytochromes and cryptochromes reconciles stability and light modulation of Arabidopsis development. Development 128 2291–2299. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.-W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, N., Brusslan, J.A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (gun5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, N., Susek, R., and Chory, J. (1996). An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol. 112 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet, J.E. (1988). Chloroplast development and gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39 475–502. [Google Scholar]
- Mullet, J.E. (1993). Dynamic regulation of chloroplast transcription. Plant Physiol. 103 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo, P., Pursiheimo, S., Hou, C.-X., Tyystjärvi, T., and Aro, E.-M. (2003). Multiple effects of antibiotics on chloroplast and nuclear gene expression. Funct. Plant Biol. 30 1097–1103. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14 387–392. [DOI] [PubMed] [Google Scholar]
- Nott, A., Jung, H.S., Koussevitzky, S., and Chory, J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57 739–759. [DOI] [PubMed] [Google Scholar]
- Oelmüller, R. (1989). Photooxidative destruction of chloroplasts and its effect on nuclear gene expression and extraplastidic enzyme levels. Photochem. Photobiol. 49 229–239. [Google Scholar]
- Ohgishi, M., Saji, K., Okada, K., and Sakai, T. (2004). Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 957 384–394. [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, E., Dietzmann, A., Biehl, A., Kurth, J., Laloi, C., Apel, K., Salamini, F., and Leister, D. (2003). Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep. 4 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, E., and Leister, D. (2004). An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329 11–16. [DOI] [PubMed] [Google Scholar]
- Ron, D., and Walter, P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8 519–529. [DOI] [PubMed] [Google Scholar]
- Rook, F., Hadingham, S.A., Li, Y., and Bevan, M.W. (2006). Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ. 29 426–434. [DOI] [PubMed] [Google Scholar]
- Sang, Y., Li, Q.H., Rubio, V., Zhang, Y.C., Mao, J., Deng, X.W., and Yang, H.Q. (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin, D., Yu, X., Maymon, M., Mockler, T., and Lin, C. (2003). Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15 2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., and Gray, J.C. (1999). Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., and Gray, J.C. (2000). The pea light-independent photomorphogenesis1 mutant results from partial duplication of COP1 generating an internal promoter and producing two distinct transcripts. Plant Cell 12 1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek, R.E., Ausubel, F.M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74 787–799. [DOI] [PubMed] [Google Scholar]
- Terzaghi, W.B., and Cashmore, A.R. (1995). Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46 445–474. [Google Scholar]
- Vinti, G., Fourrier, N., Bowyer, J.R., and López-Juez, E. (2005). Arabidopsis cue mutants with defective plastids are impaired primarily in the photocontrol of expression of photosynthesis-associated nuclear genes. Plant Mol. Biol. 57 343–357. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Yamamoto, Y.Y., Puente, P., and Deng, X.W. (2000). An Arabidopsis cotyledon-specific albino locus: A possible role in 16S rRNA maturation. Plant Cell Physiol. 41 68–76. [DOI] [PubMed] [Google Scholar]
- Yi, C., and Deng, X.W. (2005). COP1—From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15 618–625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.