Abstract
NORK in legumes encodes a receptor-like kinase that is required for Nod factor signaling and root nodule development. Using Medicago truncatula NORK as bait in a yeast two-hybrid assay, we identified 3-hydroxy-3-methylglutaryl CoA reductase 1 (Mt HMGR1) as a NORK interacting partner. HMGR1 belongs to a multigene family in M. truncatula, and different HMGR isoforms are key enzymes in the mevalonate biosynthetic pathway leading to the production of a diverse array of isoprenoid compounds. Testing other HMGR members revealed a specific interaction between NORK and HMGR1. Mutagenesis and deletion analysis showed that this interaction requires the cytosolic active kinase domain of NORK and the cytosolic catalytic domain of HMGR1. NORK homologs from Lotus japonicus and Sesbania rostrata also interacted with Mt HMGR1, but homologous nonsymbiotic kinases of M. truncatula did not. Pharmacological inhibition of HMGR activities decreased nodule number and delayed nodulation, supporting the importance of the mevalonate pathway in symbiotic development. Decreasing HMGR1 expression in M. truncatula transgenic roots by RNA interference led to a dramatic decrease in nodulation, confirming that HMGR1 is essential for nodule development. Recruitment of HMGR1 by NORK could be required for production of specific isoprenoid compounds, such as cytokinins, phytosteroids, or isoprenoid moieties involved in modification of signaling proteins.
INTRODUCTION
The symbiosis between leguminous plants and bacteria collectively named rhizobia leads to the formation of nitrogen-fixing root nodules. Depending on the host plant, nodules can be of determinate or indeterminate type, for which Lotus japonicus and Medicago truncatula have been selected as models, respectively. The first interaction between rhizobia and the host plant occurs at the root hair level in a restricted root zone that is competent for nodulation. Rhizobia attach to the root hair tip that curls and entraps the bacteria, which then enter the root hairs through the formation of infection threads. Infection threads progress through epidermal cells and reach the root cortex. Meanwhile, cortical cells dedifferentiate and start to divide leading to the formation of a nodule primordium. During the differentiation of an indeterminate nodule primordium, an apical nodule meristem is established (nodule zone I). Postmitotic cells exiting from the meristem continually become infected by rhizobia via budding of the infection threads into organelle-like structures called symbiosomes. Infected cells differentiate along several cell layers of the so-called nodule zone II until they reach their fully differentiated and nitrogen-fixing state in zone III.
Initiation and development of nodules is mediated by signal exchanges between the host plant and its rhizobial partner. This molecular dialog controls the specificity of the interaction, nodule organogenesis, and the infection process. The earliest signals are flavonoid and isoflavonoid molecules produced by the host plant. Interaction of these plant signals with rhizobial NodD transcription factors activates the expression of rhizobial nodulation genes, which leads to the production of bacterial lipochitooligosaccharidic signals named Nod factors. Perception of Nod factors by the host plant induces many early events related to infection thread formation and primordium development (D'Haeze and Holsters, 2002).
In recent years, forward genetics and map-based cloning approaches have identified major components of the Nod factor signaling pathway in M. truncatula and L. japonicus (for review, see Stacey et al., 2006). Receptor-like kinases, such as Lj NFR1, Lj NFR5, or Mt NFP, with chitin binding LysM motifs in their extracellular domain are most likely the receptors for the chitin-like Nod factors since the corresponding mutants do not show any response to Nod factors (Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Smit et al., 2007). Immediately downstream of these Nod factor receptors, another receptor-like kinase known as NORK (also called Mt DMI2 or Lj SYMRK) is essential for transmitting the Nod factor signal (Endre et al., 2002; Stracke et al., 2002). Downstream of the Nod factor receptors and genetically unresolved from NORK, putative ion channels known as Mt DMI1, Lj POLLUX, and Lj CASTOR are required for the release of Ca2+ ions from internal stores (Ané et al., 2004; Imaizumi-Anraku et al., 2005). The Ca2+ fluxes take the form of spikes (periodic peaks and valleys of Ca2+ concentrations) in the perinuclear and nuclear regions (Ehrhardt et al., 1996). Ca2+ spikes are believed to be interpreted in the nucleus by a calcium/calmodulin-dependent protein kinase known as DMI3 (Lévy et al., 2004; Mitra et al., 2004; Tirichine et al., 2006). Finally, the activated calcium/calmodulin-dependent protein kinase stimulates NSP1, NSP2, and NIN transcription factors, which leads to changes in expression of symbiotic genes (Schauser et al., 1999; Kaló et al., 2005; Smit et al., 2005; Marsh et al., 2007).
This cell-autonomous or autocrine pathway operates in root hair epidermal cells that are directly receiving the Nod factor signal. However, Nod factors also trigger long-distance responses in the cortex leading to cell divisions (paracrine signaling), which are likely generated by secondary signals after Nod factor perception in the root hairs. Physiological data have implied cytokinins in the induction of nodule-specific gene expression and cortical cell divisions (Cooper and Long, 1994; Bauer et al., 1996; Fang and Hirsch, 1998; Mathesius et al., 2000). Using a cytokinin-responsive promoter controlling a marker gene, it was shown that the endogenous cytokinin levels increase in Rhizobium-stimulated root hairs and nodule primordia (Lohar et al., 2004). Genetic evidence for the role of cytokinin was provided recently, demonstrating that a His kinase cytokinin receptor is required and sufficient to induce cortical cell divisions and the establishment of a nodule primordium (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007). These findings suggest that Nod factors generate a localized production of cytokinins that could act as secondary signals for nodule primordium formation. However, it remains unclear how Nod factors stimulate cytokinin production.
Forward genetics approaches have been fruitful in the study of Nod factor signaling in legumes. Nevertheless, the nodulation process (especially the Nod factor signal transduction pathway) probably interacts with other pathways necessary for general cellular functions and plant development. Such interactions may be difficult to identify through genetic screens since mutations affecting such processes may be lethal or lead to severe pleiotropic phenotypes. On the other hand, genes with redundant functions are rarely identified by such approaches.
In this study, we aimed to identify novel elements of the Nod factor signal transduction pathway, focusing on the receptor-like kinase NORK (Endre et al., 2002; Stracke et al., 2002). NORK possesses an extracellular sensor domain with three Leu-rich repeats generally involved in protein–protein interactions and an NSL domain (NORK extracellular-sequence-like) with unknown function, a transmembrane domain, and an intracellular kinase domain. No ligands of NORK have been identified so far. In response to purified Nod factors or rhizobia, NORK null mutants are unable to elicit calcium spikes, nodulin gene expression, or cortical cell divisions (Catoira et al., 2000; Wais et al., 2000; Esseling et al., 2004). Knocking down NORK expression by RNA interference (RNAi) in M. truncatula and Sesbania rostrata revealed that NORK is essential not only for epidermal functions but also for the release of bacteria from infection threads and symbiosome formation (Capoen et al., 2005; Limpens et al., 2005). In line with these observations, NORK expression was detected in the infection zone of M. truncatula nodules (Bersoult et al., 2005; Limpens et al., 2005). In this region, the NORK protein was localized to the plasma membrane and to the infection thread membrane (Limpens et al., 2005).
To gain insight into NORK function, we searched for proteins that interact with it. Using yeast two-hybrid (Y2H) screenings and immunoprecipitations, we identified 3-hydroxy-3-methylglutaryl CoA reductase 1 (Mt HMGR1) as a specific interactor of the NORK kinase domain. We show that this enzyme is crucial for nodule development. Our data couple the production of isoprenoid compounds (such as cytokinins and steroids) to Nod factor signaling and thus provide critical information toward understanding the function of NORK in symbiotic signaling and nodule development.
RESULTS
The M. truncatula NORK Protein Interacts with an HMGR
To identify interacting partners of NORK, Y2H screenings were performed with the extracellular 540 amino acids from the N terminus and the intracellular 383 amino acids from the C terminus domains of NORK using a Y2H cDNA library made of young nodules and roots of M. truncatula line R108 (Györgyey et al., 2000). Interactions were tested in repeated experiments under stringent selective conditions. Screenings with the extracellular domain of NORK did not result in the identification of interacting clones. By contrast, screenings with the intracellular domain (NORK-IC) revealed three independent positive clones. Each of them corresponded to the same HMGR gene that we named Mt HMGR1. These three clones were partial but overlapping and provided similar yeast growth under stringent selective conditions.
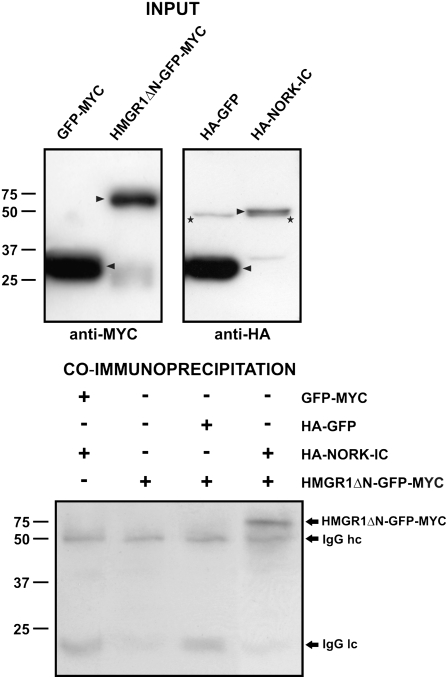
HMGRs are conserved enzymes of the mevalonate pathway and control the biosynthesis of diverse isoprenoid compounds in eukaryotes. The C terminus of HMGRs contains a catalytic domain, which allows a NADPH-dependent reduction of 3-hydroxy-3-methyl-glutaryl-CoA into mevalonic acid. This catalytic region was present in the three yeast clones that were identified. The longest clone (HMGR1ΔN), encoding 379 amino acids, was used to verify the NORK-IC–HMGR1 interaction. It was confirmed both in yeast by pairwise two-hybrid interactions and also by coimmunoprecipitations of the proteins from extracts of Arabidopsis thaliana protoplasts. In the latter case, the Mt HMGR1ΔN and NORK-IC sequences were cloned under the control of a cauliflower mosaic virus 35S promoter and tagged with different epitopes. An N-terminal 3HA tag was added to NORK-IC, while HMGR1ΔN was fused to the green fluorescent protein (GFP) and a c-Myc epitope (GFP-MYC tag) in the C terminus. HA-NORK-IC, HMGR1ΔN-GFP-MYC, and the GFP-MYC and HA-GFP control constructs were expressed transiently in Arabidopsis protoplasts. The presence of tagged proteins in the transfected protoplasts was verified by protein gel blot analysis. GFP was produced in high, HMGR1ΔN-GFP-MYC in medium, and HA-NORK-IC in lower amounts (Figure 1, top panel). To test the interactions, coexpressed or mixed protein extracts were immunoprecipitated with a mouse anti-HA antibody and separated by SDS-PAGE. The presence of c-Myc–tagged proteins was revealed with an anti-c-Myc antibody by protein gel blot analysis (Figure 1, bottom panel). In all samples, the heavy and light chains of the anti-HA antibody used for immunoprecipitation were detectable as background signals due to their cross-reactivity with the secondary antibody. Nevertheless, the interaction between HA-NORK-IC and HMGR1ΔN-GFP-MYC was clear and specific, as no signal was observed between HA-NORK-IC and GFP-MYC or between HA-GFP and HMGR1ΔN-GFP-MYC. Consequently, we demonstrated in two independent experimental systems that NORK-IC interacts with the catalytic domain of HMGR1.
Figure 1.
Mt HMGR1ΔN Coimmunoprecipitates with Mt NORK-IC.
Top panel: Arrowheads indicate c-Myc–tagged (GFP-MYC and HMGR1ΔN-GFP-MYC) and HA-tagged (HA-GFP and HA-NORK-IC) proteins produced in Arabidopsis protoplasts by protein gel blot analysis using anti-MYC and anti-HA antibodies, respectively. In the HA-tagged protein extracts, asterisks indicate an aspecific band that runs slightly faster but was clearly different from HA-NORK-IC. Bottom panel: Detection of c-Myc–tagged proteins in the anti-HA immunoprecipitates. Immunoglobulin G heavy chain (IgG hc) and light chain (IgG lc) of rat anti-HA antibody used for the immunoprecipitation are present in all samples and detected by weak cross-reaction with the sheep anti-mouse antibody. + and − indicate the presence or the absence, respectively, of given proteins in the reactions.
HMGRs Are Encoded by a Multigenic Family in M. truncatula
The number of genes encoding HMGRs varies significantly among plant species. The NORK-IC interacting partner isolated by Y2H derives from the R108 ecotype of M. truncatula. However, the ongoing genome-sequencing program uses the Jemalong A17 reference line. Homology searches in EST and genomic databases of M. truncatula allowed the identification of seven distinct homologs of Mt HMGR1 in Jemalong A17. Five of them are full-length sequences (Figure 2), and two of them are partial ones (data not shown). Since the genome sequence has not been completed, M. truncatula may contain even more HMGR genes. Although the catalytic domains are well conserved in the five complete HMGR sequences, significant variations in enzymatic function are possible due to the presence of nonhomologous amino acid replacements in the catalytic domains (Figure 2). Unlike mammalian HMGRs, which have eight N-terminal membrane-spanning domains, M. truncatula and other plant HMGRs only have two transmembrane domains. These two domains and the linker between them are well conserved among Mt HMGR isoforms, but the N terminus and the linker between the second transmembrane domain and catalytic domain are highly variable. Based on homology scores and phylogenetic analysis, a single sequence of Jemalong A17 (TC106633) corresponded to Mt HMGR1 of R108. This Jemalong A17 sequence was used for subsequent studies.
Figure 2.
Alignment of the Full-Length M. truncatula HMGR Proteins.
TM1 and TM2 are the hydrophobic transmembrane domains. Lines above the sequence alignments indicate HMGR1ΔN (broken line), HMGR1ΔNΔC1 (dotted line), and HMGR1ΔNΔC2 (full line). Asterisks mark positions that are variable in the NORK interaction domain among the five Mt HMGR proteins.
NORK Interacts Specifically with the HMGR1 Isoform of the Multigenic HMGR Family
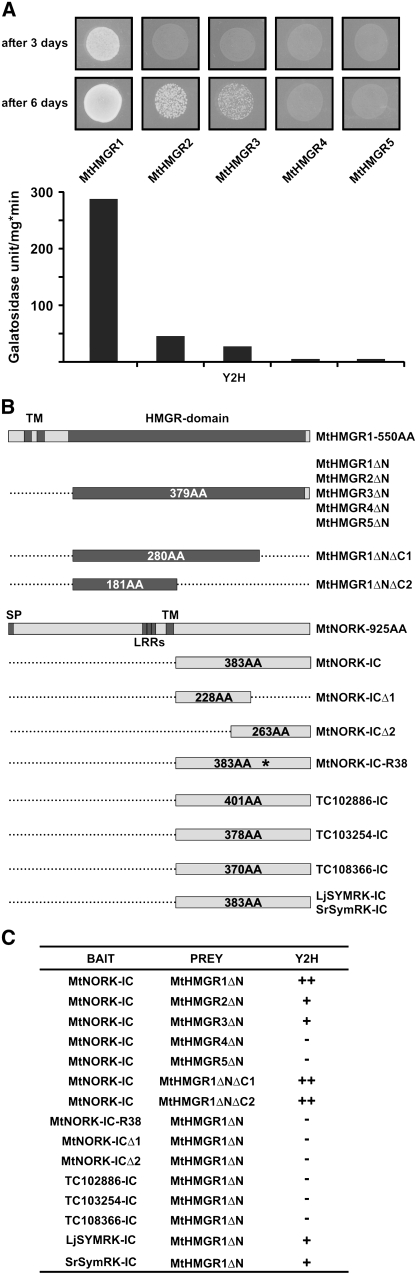
Since the highly conserved catalytic region of HMGR1 interacted with NORK-IC, we tested whether the binding is specific for HMGR1 or occurs with other members of the Mt HMGR family. Interaction of NORK-IC with the catalytic domain of the five full-length Jemalong HMGR proteins was tested by pairwise Y2H assays (Figure 3A). Despite the strong conservation of HMGR catalytic domains, the strength of interactions was remarkably different with the HMGR isoforms. Similarly to HMGR1 from R108, HMGR1 from Jemalong showed strong interaction with NORK-IC as yeast growth was confluent within 3 d, while no yeast growth was observed in the other pairwise interactions. After 6 d, however, yeast growth was also observed in the presence of HMGR2 (represented by EST contigs TC106634 and TC106637) and in a lesser extent in the presence of HMGR3 (TC106632 and TC106636), indicating that these Mt HMGR isoforms may bind weakly to NORK-IC. By contrast, HMGR4 (TC106631) and HMGR5 (TC106635) did not show any interaction with NORK-IC. In our Y2H system, interaction of tested proteins also activated the expression of a lacZ reporter gene. Therefore, the strength of interactions was measured through the β-galactosidase activity of yeast colonies (Figure 3A). Consistent with the yeast growth, the activity was high in the presence of HMGR1 and low in the case of HMGR2 or HMGR3. No β-galactosidase activity was measured in yeasts expressing HMGR4 or HMGR5. Thus, these results point to a specific interaction between Mt NORK-IC and HMGR1 in the Y2H system.
Figure 3.
Specificity of the NORK–HMGR Interactions.
(A) Interaction of the NORK-IC with the different members of the Mt HMGR family in the Y2H system was evaluated by the growth (after 3 and 6 d) and β-galactosidase activity of yeasts.
(B) The different constructs of HMGRs and NORK or NORK homologs used for pairwise Y2H interactions. The size of the different derivates and the special motifs in the proteins are indicated. TM, transmembrane domain; SP, signal peptide; LRRs, Leu-rich repeats; AA, amino acids. The asterisk indicates the mutation in NORK-R38.
(C) Results of the pairwise Y2H interactions. ++ indicates yeast growth at 3 d (strong interaction), + indicates yeast growth at 6 d (weak interaction), and − indicates no yeast growth (no interaction).
The N-Terminal Half of the Mt HMGR1 Catalytic Region Is Sufficient and Specific for Binding to Mt NORK and Its Putative Orthologs from Other Legumes
To delimit the protein regions responsible for the interaction between NORK and HMGR1, deletion derivatives of both proteins were created (Figure 3B) and tested in the Y2H system in pairwise interactions (Figure 3C). The 3′ deletions in HMGR1ΔNΔC1, reducing the 379–amino acid region from the C-terminal part to 280 or 181 amino acids, had no effect on binding, demonstrating that the NORK binding site resides within the N-terminal part of the catalytic domain.
The NORK-IC contains 383 amino acids from the C terminus of NORK. Separating this region into two overlapping halves, one containing 228 N-terminal amino acids and the other 266 amino acids from the C terminus, abolished the interaction with HMGR1. Thus, either a longer or the entire intracellular domain of NORK is needed for this interaction or for correct folding of the binding domain. To test whether the binding requires the NORK kinase activity, the R38 Nod− mutant allele of NORK (dmi2-4) was tested by Y2H. This mutation, a Gly-to-Glu amino acid replacement (Endre et al., 2002) converted the protein to an inactive kinase as in vitro autophosphorylation activity of the wild-type NORK kinase domain was abolished in the R38 mutant (data not shown). Using this clone, no interaction was detected with HMGR1ΔN, indicating that the NORK kinase activity is probably necessary for the interaction. This could either be required for substrate phosphorylation or self-phosphorylation of the NORK protein. Subsequently, we tested whether other kinases can substitute NORK in the interaction with HMGR1ΔN. We chose three M. truncatula kinases (TC102886, TC103254, and TC108366) that showed the highest homologies with NORK in their kinase region at the current status of available genomic sequence. In pairwise combinations with HMGR1ΔN, the kinase domain of these proteins resulted in no yeast growth, indicating that HMGR1ΔN binds specifically to NORK-IC.
This raised the possibility that Mt HMGR1 may also bind putative orthologs of NORK from other legumes. Therefore, we cloned the intracellular regions (383 amino acids), equivalent to that of Mt NORK-IC, from L. japonicus (Lj SYMRK-IC) and S. rostrata (Sr SYMRK-IC). Although weaker than Mt NORK, both putative NORK orthologs interacted with Mt HMGR1ΔN in the Y2H system (Figures 3B and 3C), providing support for the notion that the NORK/SYMRK–HMGR1 interaction is conserved among legumes.
Expression and Subcellular Localization of Mt HMGR1
The possible interaction of NORK and HMGR1 in vivo presumes their overlapping expression patterns during nodulation. Previous works demonstrated that the Mt NORK/DMI2 gene is expressed in the root, in the nodulation competent zone, in nodule primordia, and in nitrogen fixing nodules in a few cell layers of zone II adjacent to the meristem (Bersoult et al., 2005; Limpens et al., 2005; our unpublished data). At the subcellular level, NORK:GFP (DMI2:GFP) proteins have been localized to the plasma membrane and to the infection thread membrane (Limpens et al., 2005).
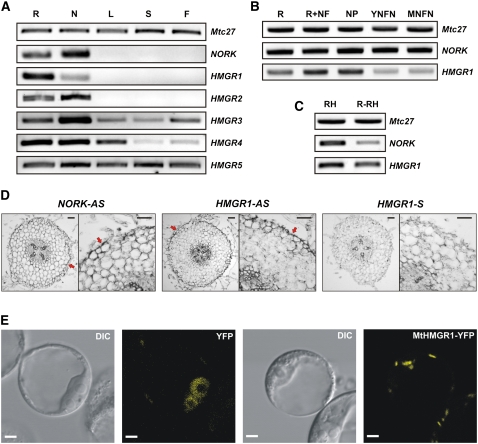
To obtain a global view on the HMGR genes, their expression was first tested in different M. truncatula organs by RT-PCR experiments (Figure 4A). All five HMGR genes were expressed in roots and nodules. Mt HMGR3, HMGR4, and HMGR5 were also transcribed in the aerial organs, such as leaves, stems, and flowers at varying levels. Like NORK, expression of Mt HMGR1 and HMGR2 was restricted to the root system, where HMGR1 transcript levels were more abundant in the root than in the nodules. This prompted us to test and compare expression patterns of HMGR1 and NORK during nodulation (Figure 4B). NORK, similarly to the constitutive Mtc27 gene, was expressed at a constant level in roots, Nod factor-treated roots, and nodule primordia as well as in young and mature nitrogen fixing nodules. By contrast, expression of HMGR1 increased in Nod factor–treated roots and in the nodule primordium and decayed in the nitrogen fixing nodules. NORK is expressed in the root hairs where Nod factor perception occurs and signaling events begin. Therefore, we studied expression of HMGR1 compared with NORK in isolated root hairs and in roots devoid of root hairs (Figure 4C). Both genes were expressed in root hairs and in roots lacking root hairs, in line with the postulated function of HMGR1 in the NORK signaling pathway. In situ hybridizations on root sections in the nodulation competent root zone confirmed colocalization of HMGR1 and NORK transcripts in the epidermal cell layer where Nod factor signaling takes place (Figure 4D).
Figure 4.
Expression Pattern and Subcellular Localization of Mt HMGR1.
(A) Expression pattern of NORK and the HMGR genes in different plant organs by RT-PCR analysis. The constitutively expressed Mtc27 was used as control for RNA loading in (A) to (C). R, roots; N, nodules; L, leaves; S, stems; F, flowers.
(B) Expression pattern of NORK and HMGR1 in roots, Nod factor–treated roots (R+NF), nodule primordia (NP), young nitrogen fixing nodules (YNFN), and mature nitrogen fixing nodules (MNFN).
(C) Expression of NORK and HMGR1 in root hairs (RH) and roots devoid of root hairs (R-RH).
(D) Localization of NORK (NORK-AS) and HMGR1 (HMGR1-AS) transcripts by in situ hybridization in the nodulation-competent root zone. Dark color (arrows) corresponds to the hybridization signal, which is absent in the control section hybridized with the sense HMGR1 (HMGR1-S) probe. Bars = 50 μm.
(E) Subcellular localization of the YFP control and MtHMGR1-YFP in Arabidopsis protoplasts. DIC, differential interference contrast. Bars = 10 μm.
To determine the subcellular localization of HMGR1, the protein was fused to yellow fluorescent protein (YFP) and expressed from the cauliflower mosaic virus 35S promoter in Arabidopsis protoplasts. The HMGR1-YFP signals were found predominantly on vesicle-like structures in this system (Figure 4E). This localization is in agreement with the immunolocalization of At HMGR1 in Arabidopsis cells where the protein was present on uncharacterized vesicles that most probably originate from the endoplasmic reticulum (ER) (Leivar et al., 2005).
Pharmacological Inhibition of HMGR Activity Reduces the Nodulation Efficiency
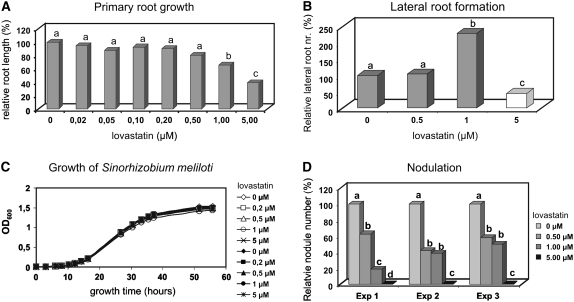
Lovastatin (mevinolin) is a potent and widely used inhibitor of HMGR enzymatic activity (Alberts et al., 1980). To confirm the role of HMGR in nodulation, the effect of lovastatin treatment on M. truncatula nodulation was investigated. To select a maximal lovastatin concentration with minimal nonspecific effect on plant development, the growth capacity of the main root (Figure 5A) and formation of lateral roots (Figure 5B) were tested at different concentrations of lovastatin (0 to 5 μM). Lovastatin up to 0.5 μM concentrations had a negligible-to-weak effect on primary and lateral root growth. Lovastatin at 1 μM strongly modified root architecture, reducing primary root growth and stimulating lateral root formation. Lovastatin at 5 μM strongly inhibited the primary root growth, and although it induced the formation of numerous lateral root primordia, these primordia were arrested and never developed into lateral roots (Figure 5B).
Figure 5.
Effect of the HMGR Inhibitor Lovastatin on Medicago Root Growth and Nodule Development.
(A) Primary root growth at different concentrations of lovastatin. Small letters (a to d) in (A), (B), and (D) represent significantly distinct categories by Student's t test at 95% confidence.
(B) Lateral root formation at different concentrations of lovastatin. At 5 μM lovastatin, only lateral root primordia were formed that did not develop further.
(C) Growth of S. meliloti is unaffected by lovastatin.
(D) Nodulation efficiency in the presence of different lovastatin concentrations in three independent experiments.
Sinorhizobium meliloti (Sm1021), the symbiotic partner of M. truncatula, has no HMGR gene; therefore, it was unlikely that lovastatin affects its growth. Nevertheless, to exclude the formal possibility of a nonspecific effect, the growth rate of S. meliloti was measured and was found to be unaffected by lovastatin up to 5 μM (Figure 5C).
Nodulation of M. truncatula by S. meliloti (Sm1021) was performed in the absence and presence of lovastatin at 0.5, 1, and 5 μM concentrations. As expected, the toxic 5 μM concentration of lovastatin inhibited root growth and nodulation. Three independent experiments showed that the presence of lovastatin at 0.5 or 1 μM significantly reduced the nodule numbers (Figure 5D). Since 0.5 μM lovastatin did not significantly affect the root growth, this negative effect on nodulation indicates that HMGR activity is indeed required for nodule development.
RNAi of Mt HMGR1 Inhibits Nodulation
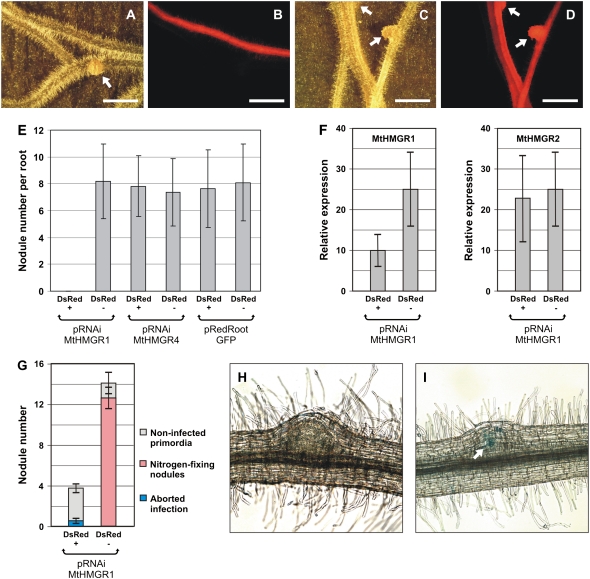
Since lovastatin is a general inhibitor of HMGR enzymes, our pharmacological studies did not specify which isoform is required for nodulation. Given the specific interaction observed between NORK and HMGR1 in the Y2H system, we studied the involvement of HMGR1 in nodulation by RNAi in transgenic hairy roots of M. truncatula (Figure 6). We cloned a 226-bp 5′ untranslated region of Mt HMGR1 in a pK7GWIWG2(II)-derived binary plasmid vector carrying the DsRED1 fluorescent marker resulting in the pRNAiMtHMGR1 construct. Similarly, pRNAiMtHMGR4 was created by cloning a 158-bp 5′ untranslated region of the noninteracting HMGR4 gene. These plasmids, as well as the pRedRoot:GFP control plasmid (Limpens et al., 2004; Riely et al., 2007), were introduced into Agrobacterium rhizogenes MSU440. By the transfer of the A. rhizogenes Ri T-DNA, these strains provoked the formation of transgenic hairy roots on M. truncatula seedlings. The DsRED1 fluorescent marker indicated the cotransformed roots. In two sets of experiments, >120 wild-type (Jemalong A17) plants were transformed with each construct. Hairy roots were scored for nodule formation 3 weeks after the inoculation with S. meliloti. In the first set of experiments, seven to eight nodules were formed in average on the DsRED1− and the DsRED1+ pRedRoot:GFP and pRNAiMtHMGR4 hairy roots (Figure 6E). By contrast, no nodules appeared on the cotransformed transgenic roots expressing pRNAiMtHMGR1 (DsRED+, Figure 6E), while on the same plant, roots that were not cotransformed with pRNAiMtHMGR1 developed a comparable number of nodules as the controls (DsRED−, Figure 6E; compare Figures 6A and 6B). In the pRNAiMtHMGR1 roots the expression of HMGR1 was reduced on average to 40% of the wild type level (Figure 6F). Due to the high degree of similarity of the Mt HMGR genes, we have also tested the specificity of this RNAi strategy for Mt HMGR1. The Mt HMGR1 sequence used for RNAi was the most similar to Mt HMGR2; therefore, we measured the expression level of this gene in the pRNAiMtHMGR1 background and in the DsRED1− roots (Figure 6F). Since the expression level of HMGR2 was not affected by pRNAiMtHMGR1, we could conclude that the RNAi silencing was specific for Mt HMGR1 and that HMGR1 function is necessary for nodulation. In this set of experiments, the complete absence of nodulation was surprising, as one would expect with RNAi also milder effects on nodulation. By changing light intensity and humidity, we optimized the nodulation on hairy roots. Nodulation rates on wild-type or DsRED1− roots raised from 8.1 ± 2.9 nodules per root to 12.8 ± 2.3 (compare Figures 6E and 6G). Under these favorable conditions, a few nodules appeared also on the pRNAiMtHMGR1 roots (4 ± 1). However, these nodules only reached the primordium state. On very rare occasions, infection threads developed. They were usually arrested early in the root hairs or in the outer cortical cells and never lead to formation of nitrogen fixing nodules. These results thus confirmed an essential role for HMGR1 in nodulation and more particularly in the infection process that was consistent with the specific NORK–HMGR1 interaction.
Figure 6.
Downregulation of HMGR1 in Hairy Roots Inhibits Nodulation.
(A) and (B) Transgenic M. truncatula roots expressing pRNAiMtHMGR1.
(C) and (D) Transgenic M. truncatula roots expressing pRNAiMtHMGR4.
Roots were observed under bright field in (A) and (C) and fluorescence microscope in (B) and (D) to identify hairy roots that were DsRED1 positive (DsRED1+) and distinguish them from the DsRED1-negative (DsRED1−; non-cotransformed) roots. Arrows indicate nodules. Bars = 0.5 cm.
(E) Nodule number at 3 weeks after inoculation on DsRED1+ versus DsRED1− roots of pRNAiMtHMGR1, pRNAiMtHMGR4, or pRedRoot:GFP transformation. Nodulation was similar in all plants except the DsRED1+ pRNAiMtHMGR1 roots, where no nodules developed.
(F) Relative expression levels of HMGR1 and its closest homolog HMGR2 measured by quantitative RT-PCR in DsRED1+ roots expressing pRNAiMtHMGR1 compared with DsRED1− ones.
(G) Optimized hairy root nodulation allows nodule initiation on DsRED1+ pRNAiMtHMGR1 roots but in reduced number and with no or aborted infection. Error bars in (E) to (G) represent sd.
(H) Nodules on DsRED1+ pRNAiMtHMGR1 roots were mainly uninfected and halted in the primordium state.
(I) Occasional aborted surface infections marked by the blue color (arrow) of S. meliloti lacZ activity in a DsRED1+ pRNAiMtHMGR1 nodule.
DISCUSSION
The NORK–HMGR1 Interaction Is Specific and Conserved in Legumes
Our work demonstrates a specific interaction between Mt NORK and HMGR1 protein. The HMGR1 binding site in NORK resides within the intracellular kinase domain and probably requires the active kinase domain since a putative inactive kinase mutant, NORK-R38 (dmi2-4) (Endre et al., 2002), did not bind HMGR1 in the Y2H system. Because the M. truncatula dmi2-4 mutant is unable to form nodules, the inability of NORK-R38 to interact with HMGR1 identifies HMGR1 as a potential downstream element of NORK signaling. The kinase region of Mt NORK could be substituted by putative NORK orthologs from other legumes, such as Sr SymRK or Lj SYMRK, which revealed the conservation of this interaction and supported further the role of HMGR1 in nodulation. Consistent with this hypothesis, kinase domains of NORK homologous, but nonsymbiotic receptor-like kinases, from M. truncatula did not interact with HMGR1. Thus, these data collectively show a specific interaction of the NORK kinase domain with HMGR1 and the conservation of this interaction in other legumes.
In M. truncatula, the HMGR family is composed of at least seven members. A strong and specific interaction of NORK was detected only with HMGR1. The NORK binding domain within HMGR1 was delimited to the first half of the catalytic region. Interestingly, this region is highly conserved among other members of the Mt HMGR family. The delimited 181–amino acid region of HMGR1 required for NORK binding differs only in three amino acids from the weakly interacting HMGR2, two of which are homologous replacements (Ile-264 to Val and Ala-287 to Ser), while Leu-220 to Asn is a nonhomologous one and most likely responsible for the drastically reduced binding ability of HMGR2 to NORK. HMGR3 interacting even weaker with NORK differs from HMGR1 in seven amino acids, corresponding to three nonhomologous exchanges, Glu-180/Ser, Ser-303/Asn, Thr-315/Ile, and to four homologous ones, Arg-177/Lys, Ile-264/Val, Ala-287/Ser, and Thr-343/Ser. In noninteracting HMGR4, there are three nonhomologous replacements, Gln-283 to Leu, Ser-303 to Asn, and Met-308 to Ile, and three homologous ones, Ile-264/Val, Ala-287/Ser, and Leu-288/Ile. The same Gln-283/Leu and Met-308/Ile replacements are also present in noninteracting HMGR5, which possesses several other sequence differences as well. These Gln and Met residues are unaffected in the weakly interacting HMGR2 and HMGR3, suggesting that Gln-283 and Met-308 in HMGR1 are essential for interaction with NORK, while Lys-220 is crucial for the efficient binding.
Receptor-like kinases are the predominant class of cell surface receptors in plants. Their biological functions have been assigned in a wide range of processes, including development and interactions with the environment (Becraft, 2002; Morris and Walker, 2003). Nevertheless, the signaling cascades activated by plant receptor-like kinases remain poorly or partially defined. We expected to obtain a signal transduction protein as an interacting partner for the NORK receptor-like kinase. Finding instead the enzyme HMGR as partner was unforeseen. However, this interaction can be meaningful for different reasons as HMGR is highly regulated, for example, by phosphorylation by kinases or by specific subcellular compartmentalization (Stermer et al., 1994; Chappell, 1995; Leivar et al., 2005).
As only the active NORK kinase showed interaction with HMGR1 in the Y2H assay, kinase activity might be required for autophosphorylation of NORK or phosphorylation of HMGR1, which may lead to inactivation of the enzyme (Goldstein and Brown, 1990). While NORK kinase activity resulted in autophosphorylation, no phosphorylation of HMGR1 was detected in in vitro kinase assays (data not shown). Thus, binding of NORK to HMGR1 might keep HMGR1 in an active unphosphorylated form that could be critical for nodulation. On the other hand, autophosphorylation of NORK might provoke a conformation change required for the binding of HMGR1.Thus, NORK may function as a scaffold protein and attract the HMGR1 enzyme to a particular subcellular localization at the plasma membrane or infection thread membrane levels.
HMGR1 Activity Is Required for Nodulation in M. truncatula
The requirement of HMGR1 for nodulation is supported by pharmacological inhibition of HMGR activity during nodulation. Lovastatin is one of the most frequently used inhibitors of HMGRs. We showed that lovastatin at low concentrations, without visible effects on growth and development of M. truncatula, significantly reduced nodulation, confirming that nodule development indeed depends on an HMGR activity. The need for HMGR1 was further supported by RNAi experiments, which showed that an ∼60% reduction in the HMGR1 transcript levels in M. truncatula roots strongly decreased nodule formation. These data collectively support a key function in nodulation for HMGR1 in a NORK-dependent manner. NORK is also required for mycorrhizal symbiosis, but there is little evidence for the involvement of HMGR1 in mycorrhizal development. One indication of a possible role in mycorrhization is that Mt HMGR1 (represented by EST AW584544) is listed among genes that were transiently upregulated in the initial stages of the mycorrhizal symbiosis (Liu et al., 2003).
Where Does NORK Interact with HMGR1?
Both NORK and HMGR1 are expressed in the root and during the early stages of nodule development. HMGR1 transcripts, like NORK transcripts, are present in the epidermal cells of the nodulation-competent root zone. These overlapping expression patterns support the notion that NORK and HMGR1 function in the same cells. NORK-GFP is plasma membrane bound (Limpens et al., 2005). The topology of NORK, similar to other receptor like kinases, predicts intracellular localization of the kinase domain. Bacterial HMGRs are localized in the cytoplasm, whereas mammalian HMGRs possess eight transmembrane domains and are ER bound. Plant HMGRs have two transmembrane domains, and the different HMGR isoforms are believed to localize to discrete and distinct domains of the ER or ER-derived vesicles (McCaskill and Croteau, 1998; Leivar et al., 2005). The topological model of HMGR suggests that the short stretch between the transmembrane domains is in the lumen, while the N terminus and the C-terminal catalytic domain are exposed to the cytosol (McCaskill and Croteau, 1998; Leivar et al., 2005).
The Mt HMGR1-YFP fusion protein did not localize to known organelles in Arabidopsis protoplasts; rather, it was found on vesicle-like structures. At HMGR1 was found on similar structures by imaging of a GFP fusion protein and immunolocalization of endogenous At HMGR1 (Leivar et al., 2005). The nature of the HMGR vesicles is undefined, but these subcellular compartments most likely originate in the ER (Leivar et al., 2005). Thus, the localization of Mt HMGR1 could be bona fide and not an artifact of the 35S promoter overexpression of the fusion protein.
If NORK is plasma membrane associated and HMGR1 is mainly on vesicles, how do they interact? Although they are anchored in different membranes, the interacting domains of both proteins are cytosolic and could be brought together by intracellular movement of the HMGR vesicles. Mutual recognition of the binding partners might also trigger controlled fusion of the plasma membrane with the vesicles, creating specific microdomains or compartments. Such membrane fusions are fundamental in eukaryotic cells and crucial for the transfer of proteins and lipids between different compartments and for exo- and endocytosis (Battey et al., 1999).
What Isoprenoid Metabolites Are Regulated by NORK-HMGR Signaling?
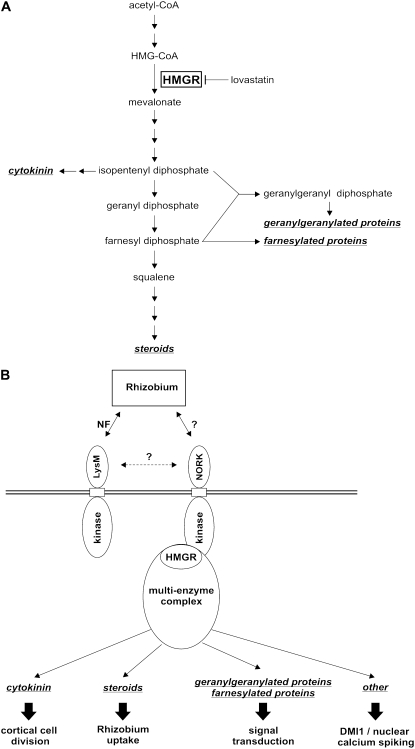
Thousands of isoprenoid compounds (derived from the common five-carbon building unit isopentenyl diphosphate and its isomer dimethylallyl diphosphate) have been identified with essential functions in many aspects of life. Cholesterol is probably the best known and most studied derivative of the isoprenoid pathway. Plants produce an exceptionally high variety of isoprenoids and isoprenoid derivatives, many of them playing key roles in plant growth and development, photosynthesis, or resistance to pests. In plants, as in bacteria, two pathways for isoprenoid biosynthesis exist (Rodríguez-Concepción and Boronat, 2002). The mevalonic acid (MVA) pathway is cytosolic, and HMGR is considered to be the rate-limiting step. The methylerythritol 4-phosphate pathway is active in plastids. The methylerythritol 4-phosphate pathway assures synthesis of gibberellins, abscisic acid, carotenoids, and chlorophyll. Important metabolites derived from the MVA pathway, and thus under the control of HMGR, are cytokinins, phytosteroids, including brassinolins and the farnesyl, or geranylgeranyl moieties of the isoprenyl lipid protein modifications (Figure 7A).
Figure 7.
MVA Pathway in Plant Cells and Putative Roles for HMGR in Nodulation.
(A) The steps of the cytoplasmic isoprenoid synthesis with indication of the relevant intermediates and important end products (in italics and underlined). The enzymatic reaction catalyzed by HMGR (boxed) is shown as well as inhibition of HMGR by lovastatin.
(B) Model for potential implications of HMGR1 via NORK in nodule development. See text for details.
In contrast with animals, plants have multiple HMGR isoforms. Their differential expression (Choi et al., 1992; Enjuto et al., 1995), subcellular localization (McCaskill and Croteau, 1998; Leivar et al., 2005), and mutant phenotypes (Suzuki et al., 2004) indicate that the distinct isoforms have metabolic specialization and, thus, that they may be involved in different branches of the mevalonate pathway and the production of different end products. This is likely achieved by metabolic channeling, meaning that cooperating enzymes are organized into macromolecular complexes in which biosynthetic intermediates are transferred between catalytic sites without diffusion into the bulk phase of the cell (Chappell, 1995). NORK interacts with HMGR1 and only weakly, or not at all, with other HMGRs. This indicates that the Nod factor signaling pathway activates or recruits a particular isoprenoid pathway. What kind of isoprenoid metabolites can be regulated by NORK-HMGR1 and involved in nodulation? Based on the metabolites of the MVA pathway and the physiology of nodule formation, we propose four alternative hypotheses.
Cytokinin is produced by isopentenyl transferases from adenosine phosphate and dimethylallyl pyrophosphate, a product from the mevalonate pathway (Figure 7A). Therefore, cytokinin synthesis can be suppressed by lovastatin in cultured Nicotiana tabacum BY-2 cells (Crowell and Salaz, 1992). Cytokinin is known to play a major role in nodule initiation. Cytokinin likely is produced in Nod factor–stimulated root hairs and may serve as a secondary, mobile signal to the root cortical cells. Perception of cytokinin stimulates cell division in the cortex and formation of the nodule primordium (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007). Thus, it is possible that following Nod factor stimulation, the recruitment of HMGR1 by NORK in the root epidermis leads to the production of cytokinins for paracrine signaling and the stimulation of cortical cell divisions (Figure 7B). If HMGR1 is the isoform that promotes cytokinin synthesis, then downregulation of HMGR1 would be expected to block cell division and to inhibit nodule formation, which was indeed the case in the HMGR1 RNAi plants. On the other hand, analysis of M. truncatula DMI3 gain-of-function mutants suggests that DMI3 activity is sufficient to initiate nodule development in a wild-type background. To our knowledge, it has not been shown that DMI3 gain-of-function mutants are able to induce nodulation in the absence of DMI2 or HMGR1. Although the requirement of both DMI3 and NORK-HMGR1 activities for cytokinin production seems unexpected, this hypothesis cannot be excluded.
Besides being part of the early signaling pathway operative in root hairs, NORK is also involved in later stages of nodule formation and, more precisely, in controlling the endocytotic uptake of rhizobia from infection threads into host cells (Capoen et al., 2005; Limpens et al., 2005). Endocytosis in plant cells is still poorly understood (Battey et al., 1999). The relationships between the endocytotic uptake of rhizobia and other endocytotic processes are unknown. In mammalian cells, where endocytosis is far better described, this process requires steroids and cholesterol in specialized membrane microdomains named lipid rafts (Heese-Peck et al., 2002; Thomsen et al., 2002; Pichler and Riezman, 2004). Such lipid rafts play an essential role in endocytosis but also in signaling processes and cytoskeleton organization (Funatsu et al., 2000; Brdickova et al., 2001). The presence of lipid rafts in the plasma membrane of M. truncatula has been demonstrated recently, and these structures were found to be associated with a complete redox system at the plasma membrane level (Lefebvre et al., 2007). Mammalian pathogens, including Brucella species, which are closely related to rhizobia and whose uptake and intracellular life have common features with rhizobia in legume nodules (Batut et al., 2004), often hijack the cholesterol-dependent endocytotic pathway for their uptake into host cells (Gatfield and Pieters, 2000; Shin et al., 2000; Mañes et al., 2003; Batut et al., 2004). Thus, a second possible function of NORK-HMGR1 could be to direct a metabolic complex for steroid synthesis in the membrane of infection threads and thereby promoting locally the production and accumulation of steroid compounds that then mediates endocytotic uptake of the rhizobia from the infection threads (Figure 7B). However, the significant decrease in nodule number of RNAi roots suggests a role for NORK-HMGR1 at an earlier stage than rhizobial release from infection threads. Possibly the NORK-HMGR complex is active at the stage of root hair infection, when rhizobia, entrapped in a root hair curl, induce an invagination of the plasma membrane to initiate infection thread growth (Figure 7B).
In animals, inactivation of HMGR has profound effects on development and cell differentiation because of its effect on protein isoprenylation. Loss of HMGR activity, by mutation or pharmacologically, leads to the absence of farnesyl or geranylgeranyl posttranslational modifications and consequently mislocalization of important signaling proteins, such as small G-proteins involved in development. These lipid modifications are needed for membrane anchoring of these proteins and thus for their proper subcellular localization (Van Doren et al., 1998; Santos and Lehmann, 2004; Thorpe et al., 2004; Yi et al., 2006; D'Amico et al., 2007). Protein farnesylation and geranylgeranylation is also conserved in plants and is known to affect the subcellular localization and functioning of signaling proteins, such as ROP/RAC small G-proteins, transcription factors, cell cycle regulators, and others (Yalovsky et al., 1999, 2000; Crowell, 2000; Lavy et al., 2002; Galichet and Gruissem, 2003, 2006). NORK could act as a scaffold attracting HMGR and other enzymes of the protein isoprenylation pathway to engage signaling proteins locally in a Nod factor signaling complex (Figure 7B).
Finally, the production of isoprenoid compounds could assure the signaling between the plasma membrane–localized NORK and the nuclear-localized DMI1 protein (Riely et al., 2007) (Figure 7B). Based on homologies with bacterial ion channels (such as MthK), DMI1 is predicted to be a nuclear ion channel, and its activity may be regulated by calcium, NAD, or other unknown ligands (Ané et al., 2004; Riely et al., 2007). The presence of a complete redox system in M. truncatula lipid rafts (Lefebvre et al., 2007) suggests that redox signals and NAD production could be the missing link between NORK and DMI1 and more generally between the site of Nod factor perception and nuclear calcium spikes. It is also possible that DMI1 ligands may derive from the mevalonate pathway, which would provide a direct connection between DMI1 and HMGR activities.
To elucidate the mode of action of HMGR1, more information is needed about the different branches of the mevalonate pathway and the end products produced by the different HMGR isoforms. In addition, the step in nodule formation and Nod factor signaling that is affected by HMGR1 must be defined more precisely.
METHODS
Nodulation Assays
Medicago truncatula ecotype Jemalong (line A17) and ecotype R108 were used for nodulation assays. Seeds were immersed for 4 min in pure H2SO4 for scarification and sterilized with 8% (w/v) calcium hypochlorite solution for 20 min. Seeds were then placed for 3 d at 4°C in the dark and germinated overnight at room temperature in the dark. Freshly germinated seedlings were transferred to square plates containing BNM nodulation growth medium (Ehrhardt et al., 1992). Eight seedlings were placed per plate and the lower part of the plate was wrapped in aluminum foil to keep the roots in the dark. The plates were placed vertically in a growth room at 24°C in 16-h-light/8-h-dark cycle conditions. After 2 d of growth, the plants were transferred to fresh BNM plates containing lovastatin at the final concentrations as indicated in Figure 5. The 12.5 mM lovastatin (Sigma-Aldrich) stock solution was prepared freshly in ethanolic NaOH (15% [v/v] ethanol and 0.25% [w/v] NaOH) and incubated at 60°C for 1 h for hydrolyzing the lactone ring. After 1 d of growth on lovastatin-containing plates, the roots of the plants were inoculated with Sinorhizobium meliloti Sm1021 that was grown overnight in TA liquid medium (1% tryptone, 0.1% yeast extract, 0.5% NaCl, 1 mM MgSO4, and 1 mM CaCl2) at 30°C with 150 rpm up to OD600 ≈ 1.0 and then washed twice with distilled water and resuspended in water at OD600 ≈ 0.05. The number of lateral roots and the number of nodules were counted at different times as indicated in Figure 5. For measurement of the length of the main root, the growth path of the root was indicated on the Petri dish with a pencil and scanned and measured with Adobe Photoshop 7.0 tools.
Y2H Screening and Pairwise Assays
For the Y2H screenings and pairwise interactions, bait constructs were cloned in the vector pBD-GAL4 (Stratagene). The bait sequences for the Mt NORK genes and homologous kinases were obtained by PCR amplification of root or nodule cDNA using oligos with EcoRI and XhoI linkers for cloning in pBD-GAL4. Truncated versions of NORK-IC (NORK-ICΔ1 and NORK-ICΔ2) were generated with restriction enzymes. The Y2H cDNA library in pAD-GAL4 was made of young nodules and roots of M. truncatula line R108 (Györgyey et al., 2000). The truncated forms of Mt HMGR1ΔN, HMGR1ΔNΔC1, and HMGR1ΔNΔC2 were created by PCR coupled with oligos for restriction enzyme cloning. Screening was performed according the manufacturer's procedure (Stratagene). Colonies were collected and plated on SD-WLHA medium. Similar transformation and selection procedures were used for direct interaction studies.
Transfection of Arabidopsis Protoplasts and Protein Extraction
For transient expression of proteins, HMGR1ΔN, NORK-IC, and GFP were cloned under the control of the 35S promoter in the pRT104 vector (Topfer et al., 1987) and tagged with the HA or c-Myc epitopes. A cell suspension culture derived from Arabidopsis thaliana ecotype Columbia seedlings was grown in Murashige and Skoog (MS) complete liquid media containing sucrose (30 g/L), kinetin (14 μg/L), and 2,4-D at 23°C with 135 rpm shaking under continuous light and weekly subcultured (15 mL of culture in 85 mL of fresh medium). Forty milliliters of a 5-d-old cell suspension were treated with cell wall digestion solution (0.1 g/mL of cellulase Serva R10, 0.02 g/mL of macerozyme Yakult, 4.13 g/L of MS, 0.34 M glucose, and 0.34 M mannitol, pH 5.5) overnight at room temperature in the dark. Cells were centrifuged and the pellet was washed with culturing medium (4.13 g/L of MS, 0.34 M glucose, and 0.34 M mannitol, pH 5.5). Protoplasts were then separated from the debris using a density gradient (4.13 g/L MS and 30 g/L sucrose, pH 5.5) coupled to centrifugation. For each transformation, 15 μg of plasmid DNA was added to 106 protoplasts, and transformation was performed by 20 min of incubation with polyethylene glycol solution (25% [w/v] PEG 6000, 0.45 M mannitol, and 0.1 M calcium nitrate, pH 9) in the dark. Finally, protoplasts were rinsed with 0.275 M Ca(NO3)2 and incubated overnight in the dark in the culturing medium. Protoplasts were then pelleted and resuspended in extraction buffer (25 mM Tris-HCl, pH 7.7, 10 mM MgCl2, 75 mM NaCl, 15 mM β-glycerophosphate, 0.1% Tween 20, 10% glycerol, 1 mM DTT, 1 mM NaF, 0.5 mM NaVO3, and 0.5 mM PMSF), vortexed, and frozen in liquid nitrogen. Soluble proteins were separated from the debris by centrifugation at 4°C.
Coimmunoprecipitation and Protein Gel Blot Analysis
For each coimmunoprecipitation reaction, 300 μL of total protein extract at 5 μg/μL were mixed to 500 ng of rat anti-HA antibody (clone 3F10; Roche) and 80 μL of magnetic micro beads bound to G-protein (MACS Protein G Microbeads; Amersham). The mixture was agitated overnight at 4°C. Coimmunoprecipitations were performed on MACS columns (Amersham) as suggested by the manufacturer. The different protein fractions were separated on 10% SDS-PAGE in electrophoresis buffer (25 mM Tris-HCl, pH 8.3, 250 mM glycine, and 0.1% SDS) and then transferred to PVDF membranes (Amersham) in transfer buffer (48 mM Tris-HCl, pH 8.3, 39 mM glycine, and 20% ethanol). Membranes were then saturated with blocking solution (50 mM Tris-HCl, pH 7.4, 25 mM NaCl, 25 mM KCl, 0.05% Tween 20, and 5% defatted milk powder). The mouse anti-HA antibody (12CA5; Boehringer Mannheim) or mouse anti-c-Myc antibody (9E10; Boehringer-Mannheim) were applied at 0.08 μg/mL into the blocking solution overnight with slow shaking at 4°C. Secondary sheep antibody coupled to horseradish peroxidase (IgG anti-mouse A-5906; Sigma-Aldrich) was diluted 1:30,000 into the blocking solution and applied to the membrane. Chemoluminescence was finally detected with the commercial ECL detection kit (Amersham).
Subcellular Localization of Mt HMGR1 in Arabidopsis Protoplasts
The full-length Mt HMGR1 open reading frame, fused to the YFP open reading frame, was cloned in pRT104 under the control of 35S promoter. The fusion protein was transiently expressed in Arabidopsis protoplasts as described above. Three days after transfection, protoplasts were observed by confocal microscopy.
RT-PCR and in Situ Hybridization
In three independent experiments, total RNA was isolated from M. truncatula (Jemalong) roots, nodules, leaves, stems, and flowers by the RNeasy plant mini kit (Qiagen) for cDNA synthesis. To remove traces of genomic DNA, equal amounts of total RNA were treated with DNase (FPLC pure; Amersham) that was subsequently heat inactivated. cDNAs were synthesized by retrotranscription in the presence of Powerscript reverse transcriptase (Clontech), RNase inhibitor (RNasin; Promega), and oligo(dT) primers. Diluted fractions were used in PCR (EUROBIO TAQ polymerase). HMGR and NORK cDNAs were amplified in 35 cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min). Mtc27 constitutive marker was amplified in 25 cycles. PCR products were analyzed by the Fisher Scientific Bioblock gel documentation system.
In situ hybridizations were performed according to de Almeida Engler et al. (2001). DIG-labeled specific antisense or sense riboprobes were generated for Mt HMGR1 (226 bp from the 5′ untranslated region) and for Mt NORK (381-bp fragment). Hybridization signals were revealed with DIG antibodies and alkaline phosphatase–conjugated secondary antibodies. Signals were observed with a light microscope and CCD camera.
RNAi
The 5′ untranslated region with a short part of the coding region was amplified from Mt HMGR1, resulting in a 226-bp PCR fragment (same fragment as for the in situ hybridization). As a control, the corresponding fragment was amplified from Mt HMGR4, generating a PCR product of 158 bp. The PCR products were first cloned in pENTR/D TOPO (Invitrogen). LR recombination was performed with the modified pK7GWIWG2(II) binary vector containing the constitutively expressed fluorescent DsRED1 marker (Limpens et al., 2004). Agrobacterium rhizogenes MSU440 was transformed with these constructs and the empty vector. A. rhizogenes–mediated hairy root transformation was done according to Boisson-Dernier et al. (2001). DsRED1 fluorescence in the roots was observed with confocal microscopy.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: EU302813 (Mt HMGR1), EU302814 (Mt HMGR2), EU302815 (Mt HMGR3), EU302816 (Mt HMGR4), EU302817 (Mt HMGR5), AJ418369 (Mt NORK), AF492655 (Lj SYMRK), and AY751547 (Sr SYMRK).
Acknowledgments
The Sr SymRK cDNA was a kind gift of Ward Capoen (University of Ghent, Belgium) and the root hair cDNA was kindly provided by Francisco Merchan (Institut des Sciences du Végétal). We thank Julie Cullimore for critical reading of the manuscript. This work was supported by the European Union FP6 Grain Legume Integrated Program (E.K. and Z.K.), a Hatch grant (J.-M.A.), the “Ministère de l'Education Nationale de la Recherche et de la Technologie” (G.L.), and by the Hungarian Grants NKFP 4/031/2004, OTKA T046819, and GVOP 3.1.1-2004-05-0101/3.0 (G.E. and G.B.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Eva Kondorosi (eva.kondorosi@isv.cnrs-gif.fr).
References
- Alberts, A.W., et al. (1980). Mevinolin: A highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc. Natl. Acad. Sci. USA 77 3957–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ané, J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. [DOI] [PubMed] [Google Scholar]
- Arrighi, J.F., et al. (2006). The Medicago truncatula LysM motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battey, N.H., James, N.C., Greenland, A.J., and Brownlee, C. (1999). Exocytosis and endocytosis. Plant Cell 11 643–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut, J., Andersson, S.G., and O'Callaghan, D. (2004). The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2 933–945. [DOI] [PubMed] [Google Scholar]
- Bauer, P., Ratet, P., Crespi, M., Schultze, M., and Kondorosi, A. (1996). Nod factors and cytokinins induce similar cortical cell divisions, amyloplast deposition and MsEnod12A expression patterns in alfalfa roots. Plant J. 10 91–105. [Google Scholar]
- Becraft, P.W. (2002). Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 18 163–192. [DOI] [PubMed] [Google Scholar]
- Bersoult, A., Camut, S., Perhald, A., Kereszt, A., Kiss, G.B., and Cullimore, J.V. (2005). Expression of the Medicago truncatula DMI2 gene suggests roles of the symbiotic nodulation receptor kinase in nodules and during early nodule development. Mol. Plant Microbe Interact. 18 869–876. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier, A., Chabaud, M., Garcia, F., Bécard, G., Rosenberg, C., and Barker, D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14 695–700. [DOI] [PubMed] [Google Scholar]
- Brdickova, N., Brdicka, T., Andera, L., Spicka, J., Angelisova, P., Milgram, S.L., and Horejsi, V. (2001). Interaction between two adapter proteins, PAG and EBP50: A possible link between membrane rafts and actin cytoskeleton. FEBS Lett. 507 133–136. [DOI] [PubMed] [Google Scholar]
- Capoen, W., Goormachtig, S., De Rycke, R., Schroeyers, K., and Holsters, M. (2005). SrSymRK, a plant receptor essential for symbiosome formation. Proc. Natl. Acad. Sci. USA 102 10369–10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira, R., Galera, C., de Billy, F., Penmetsa, R.V., Journet, E.P., Maillet, F., Rosenberg, C., Cook, D., Gough, C., and Dénarié, J. (2000). Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12 1647–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell, J. (1995). The biochemistry and molecular biology of isoprenoid metabolism. Plant Physiol. 107 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D., Ward, B.L., and Bostock, R.M. (1992). Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell 4 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, J.B., and Long, S.R. (1994). Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell 6 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell, D.N. (2000). Functional implications of protein isoprenylation in plants. Prog. Lipid Res. 39 393–408. [DOI] [PubMed] [Google Scholar]
- Crowell, D.N., and Salaz, M.S. (1992). Inhibition of growth of cultured tobacco cells at low concentrations of lovastatin is reversed by cytokinin. Plant Physiol. 100 2090–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico, L., Scott, I.C., Jungblut, B., and Stainier, D.Y. (2007). A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Curr. Biol. 17 252–259. [DOI] [PubMed] [Google Scholar]
- de Almeida Engler, J., De Groodt, R., Van Montagu, M., and Engler, G. (2001). In situ hybridization to mRNA of Arabidopsis tissue sections. Methods 23 325–334. [DOI] [PubMed] [Google Scholar]
- D'Haeze, W., and Holsters, M. (2002). Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12 79R–105R. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Atkinson, E.M., and Long, S.R. (1992). Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256 998–1000. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kaló, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966. [DOI] [PubMed] [Google Scholar]
- Enjuto, M., Lumbreras, V., Marin, C., and Boronat, A. (1995). Expression of the Arabidopsis HMG2 gene, encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase, is restricted to meristematic and floral tissues. Plant Cell 7 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseling, J.J., Lhuissier, F.G., and Emons, A.M. (2004). A nonsymbiotic root hair tip growth phenotype in NORK-mutated legumes: implications for nodulation factor-induced signaling and formation of a multifaceted root hair pocket for bacteria. Plant Cell 16 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu, N., Kumanogoh, H., Sokawa, Y., and Maekawa, S. (2000). Identification of gelsolin as an actin regulatory component in a Triton insoluble law density fraction (raft) of newborn bovine brain. Neurosci. Res. 36 311–317. [DOI] [PubMed] [Google Scholar]
- Fang, Y., and Hirsch, A.M. (1998). Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol. 116 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galichet, A., and Gruissem, W. (2003). Protein farnesylation in plants–Conserved mechanisms but different targets. Curr. Opin. Plant Biol. 6 530–535. [DOI] [PubMed] [Google Scholar]
- Galichet, A., and Gruissem, W. (2006). Developmentally controlled farnesylation modulates AtNAP1;1 function in cell proliferation and cell expansion during Arabidopsis leaf development. Plant Physiol. 142 1412–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield, J., and Pieters, J. (2000). Essential role for cholesterol in entry of mycobacteria into macrophages. Science 288 1647–1650. [DOI] [PubMed] [Google Scholar]
- Goldstein, J.L., and Brown, M.S. (1990). Regulation of the mevalonate pathway. Nature 343 425–430. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo, S., Crespi, M., and Frugier, F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györgyey, J., Vaubert, D., Jimenez-Zurdo, J.I., Charon, C., Troussard, L., Kondorosi, A., and Kondorosi, E. (2000). Analysis of Medicago truncatula nodule expressed sequence tags. Mol. Plant Microbe Interact. 13 62–71. [DOI] [PubMed] [Google Scholar]
- Heese-Peck, A., Pichler, H., Zanolari, B., Watanabe, R., Daum, G., and Riezman, H. (2002). Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell 13 2664–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku, H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531. [DOI] [PubMed] [Google Scholar]
- Kaló, P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. [DOI] [PubMed] [Google Scholar]
- Lavy, M., Bracha-Drori, K., Sternberg, H., and Yalovsky, S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14 2431–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, B., Furt, F., Hartmann, M.-A., Michaelson, L.V., Carde, J.-P., Sargueil-Boiron, F., Rossignol, M., Napier, J.A., Cullimore, J., Bessoule, J.-J., and Mongrand, S. (2007). Characterization of lipid rafts from Medicago truncatula root plasma membranes: A proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 144 402–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P., Gonzalez, V.M., Castel, S., Trelease, R.N., Lopez-Iglesias, C., Arro, M., Boronat, A., Campos, N., Ferrer, A., and Fernandez-Busquets, X. (2005). Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiol. 137 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy, J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364. [DOI] [PubMed] [Google Scholar]
- Limpens, E., Mirabella, R., Fedorova, E., Franken, C., Franssen, H., Bisseling, T., and Geurts, R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102 10375–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens, E., Ramos, J., Franken, C., Raz, V., Compaan, B., Franssen, H., Bisseling, T., and Geurts, R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55 983–992. [DOI] [PubMed] [Google Scholar]
- Liu, J., Blaylock, L.A., Endre, G., Cho, J., Town, C.D., VandenBosch, K.A., and Harrison, M.J.Blob.Blob. (2003). Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15 2106–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar, D.P., Schaff, J.E., Laskey, J.G., Kieber, J.J., Bilyeu, K.D., and Bird, D.M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 38 203–214. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. [DOI] [PubMed] [Google Scholar]
- Mañes, S., del Real, G., and Martinez-A, C. (2003). Pathogens: Raft hijackers. Nat. Rev. Immunol. 3 557–568. [DOI] [PubMed] [Google Scholar]
- Marsh, J.F., Rakocevic, A., Mitra, R.M., Brocard, L., Sun, J., Eschstruth, A., Long, S.R., Schultze, M., Ratet, P., and Oldroyd, G.E. (2007). Medicago truncatula NIN is essential for Rhizobium independent nodule organogenesis induced by autoactive CCaMK. Plant Physiol. 144 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius, U., Charon, C., Rolfe, B.G., Kondorosi, A., and Crespi, M. (2000). Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol. Plant Microbe Interact. 13 617–628. [DOI] [PubMed] [Google Scholar]
- Mitra, R.M., Gleason, C.A., Edwards, A., Hadfield, J., Downie, J.A., Oldroyd, G.E., and Long, S.R. (2004). A Ca2+ dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill, D., and Croteau, R. (1998). Some caveats for bioengineering terpenoid metabolism in plants. Trends Biotechnol. 16 349–355. [Google Scholar]
- Morris, E.R., and Walker, J.C. (2003). Receptor-like protein kinases: The keys to response. Curr. Opin. Plant Biol. 6 339–342. [DOI] [PubMed] [Google Scholar]
- Murray, J.D., Karas, B.J., Sato, S., Tabata, S., Amyot, L., and Szczyglowski, K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315 101–104. [DOI] [PubMed] [Google Scholar]
- Pichler, H., and Riezman, H. (2004). Where sterols are required for endocytosis. Biochim. Biophys. Acta 1666 51–61. [DOI] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Gronlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. [DOI] [PubMed] [Google Scholar]
- Riely, B.K., Lougnon, G., Ané, J.M., and Cook, D.R. (2007). The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J. 49 208–216. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción, M., and Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, A.C., and Lehmann, R. (2004). Isoprenoids control germ cell migration downstream of HMGCoA reductase. Dev. Cell 6 283–293. [DOI] [PubMed] [Google Scholar]
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. [DOI] [PubMed] [Google Scholar]
- Shin, J.-S., Gao, Z., and Abraham, S.N. (2000). Involvement of cellular caveolae in bacterial entry into mast cells. Science 289 785–788. [DOI] [PubMed] [Google Scholar]
- Smit, P., Limpens, E., Geurts, R., Fedorova, E., Dolgikh, E., Gough, C., and Bisseling, T. (2007). Medicago LYK3, an entry receptor in rhizobial Nod factor signaling. Plant Physiol. Published on June 22, 2007. 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed]
- Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., and Geurts, R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791. [DOI] [PubMed] [Google Scholar]
- Stacey, G., Libault, M., Brechenmacher, L., Wan, J.R., and May, G.D. (2006). Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 9 110–121. [DOI] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962. [DOI] [PubMed] [Google Scholar]
- Stermer, B.A., Bianchini, G.M., and Korth, K.L. (1994). Regulation of HMG-CoA reductase activity in plants. J. Lipid Res. 35 1133–1140. [PubMed] [Google Scholar]
- Suzuki, M., Kamide, Y., Nagata, N., Seki, H., Ohyama, K., Kato, H., Masuda, K., Sato, S., Kato, T., Tabata, S., Yoshida, S., and Muranaka, T. (2004). Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J. 37 750–761. [DOI] [PubMed] [Google Scholar]
- Thomsen, P., Roepstorff, K., Stahlhut, M., and van Deurs, B. (2002). Caveolae are highly immobile plasma membrane microdomains, which are not involved in constitutive endocytic trafficking. Mol. Biol. Cell 13 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe, J.L., Doitsidou, M., Ho, S.Y., Raz, E., and Farber, S.A. (2004). Germ cell migration in zebrafish is dependent on HMGCoA reductase activity and prenylation. Dev. Cell 6 295–302. [DOI] [PubMed] [Google Scholar]
- Tirichine, L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156. [DOI] [PubMed] [Google Scholar]
- Tirichine, L., Sandal, N., Madsen, L.H., Radutoiu, S., Albrektsen, A.S., Sato, S., Asamizu, E., Tabata, S., and Stougaard, J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 104–107. [DOI] [PubMed] [Google Scholar]
- Topfer, R., Matzeit, V., Gronenborn, B., Schell, J., and Steinbiss, H.H. (1987). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren, M., Broihier, H.T., Moore, L.A., and Lehmann, R. (1998). HMG-CoA reductase guides migrating primordial germ cells. Nature 396 466–469. [DOI] [PubMed] [Google Scholar]
- Wais, R.J., Galera, C., Oldroyd, G., Catoira, R., Penmetsa, R.V., Cook, D., Gough, C., Dénarié, J., and Long, S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky, S., Rodríguez-Concepción, M., Bracha, K., Toledo-Ortiz, G., and Gruissem, W. (2000). Prenylation of the floral transcription factor APETALA1 modulates its function. Plant Cell 12 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky, S., Rodríguez-Concepción, M., and Gruissem, W. (1999). Lipid modifications of proteins - slipping in and out of membranes. Trends Plant Sci. 4 439–445. [DOI] [PubMed] [Google Scholar]
- Yi, P., Han, Z., Li, X., and Olson, E.N. (2006). The mevalonate pathway controls heart formation in Drosophila by isoprenylation of Gγ1. Science 313 1301–1303. [DOI] [PubMed] [Google Scholar]