Abstract
The Arabidopsis thaliana accession Shahdara was identified as a rare naturally occurring mutant that does not liberate seed mucilage on imbibition. The defective locus was found to be allelic to the mum2-1 and mum2-2 mutants. Map-based cloning showed that MUCILAGE-MODIFIED2 (MUM2) encodes the putative β-d-galactosidase BGAL6. Activity assays demonstrated that one of four major β-d-galactosidase activities present in developing siliques is absent in mum2 mutants. No difference was observed in seed coat epidermal cell structure between wild-type and mutant seed; however, weakening of the outer tangential cell wall by chemical treatment resulted in the release of mucilage from mum2 seed coat epidermal cells, and the mum2 mucilage only increased slightly in volume, relative to the wild type. Consistent with the absence of β-d-galactosidase activity in the mutant, the inner layer of mucilage contained more Gal. The allocation of polysaccharides between the inner and outer mucilage layers was also modified in mum2. Mass spectrometry showed that rhamnogalacturonan I in mutant mucilage had more branching between rhamnose and hexose residues relative to the wild type. We conclude that the MUM2/BGAL6 β-d-galactosidase is required for maturation of rhamnogalacturonan I in seed mucilage by the removal of galactose/galactan branches, resulting in increased swelling and extrusion of the mucilage on seed hydration.
INTRODUCTION
Some plant species, including Arabidopsis thaliana, produce myxospermous seed, where the epidermal cells of the seed coat contain a large quantity of a complex, pectinaceous polysaccharide, or mucilage that is released from the seed coat upon hydration. Seed mucilage production in Arabidopsis is part of a coordinated differentiation process that occurs in the outer cell layer of the seed coat (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). After fertilization, during embryo development, and subsequent seed maturation, the seed coat is formed from two integuments of maternal origin that surround the embryo sac. The inner integument is the site of synthesis of tannins that are responsible for the characteristic brown coloration of Arabidopsis seed. The outer integument becomes myxospermous and accumulates large quantities of pectic mucilage. Differentiation of the epidermal cells involves a highly regulated series of events, including growth, morphogenesis, cell wall thickening, mucilage biosynthesis, and secretion of mucilage into the apoplast (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). The resulting polygonal epidermal cells have thickened radial cell walls and a central volcano-shaped structure, the columella, surrounded by dehydrated mucilage.
Upon imbibition, the mucilage is extruded and completely envelops the seed. In Arabidopsis, the mucilage released is composed of two layers: a water-soluble outer layer and an adherent inner layer (Western et al., 2000; Macquet et al., 2007). Although the major component of both is the pectin rhamnogalacturonan I (RG-I), the composition and structural properties of the two layers differ (Macquet et al., 2007). The outer layer appears to be exclusively RG-I with an average molar mass of ∼600 kD, whereas the inner layer has a very high molar mass and can be separated into two domains: the outer containing galactan and arabinan as minor constituents and the inner associated with cellulose and small amounts of galactan. Immunolabeling experiments also suggest the presence of small stretches of homogalacturonan (HG), with different degrees of methyl-esterification, within the RG-I pectin domains of the inner mucilage (Macquet et al., 2007).
The production of mucilage by the seed is a significant metabolic investment, and yet potential physiological roles remain hypothetical. Mucilage has been proposed to regulate germination by acting as an oxygen barrier (Witztum et al., 1969) or to enable germination under conditions of reduced water potential (Penfield et al., 2001). Other possible functions include seed dispersal (Young and Evans, 1973; Gutterman and Shem-Tov, 1996) or the protection of the germinating seedling (Western et al., 2000). The fact that the inner and outer layers of seed mucilage show differences in their composition and structural properties suggests that the role of each could be different and adjusted to a particular aspect of mucilage function. In particular, the tight linkage of the inner layer of mucilage to the seed coat and its adhesive qualities agree well with a role in seed dispersal, whereas the water-soluble nature of the outer mucilage would allow its diffusion around the seed and could stimulate or inhibit the growth of bacteria and fungi, thus preparing the rhizosphere for seedling germination (Macquet et al., 2007).
A number of maternally inherited seed mutants affected in mucilage production have been identified and these can be classed into four categories. The first comprises two mutants, mucilage-modified3 (mum3) and mum5, that produce mucilage but with altered mucilage composition or structural properties (Western et al., 2001; Macquet et al., 2007). The mutants in the second, mum1 and mum2, appear to be defective for mucilage release because although mucilage is accumulated in the epidermal cells of the seed coat, this is not extruded on imbibition (Western et al., 2001). The third group of mutants is defective in mucilage extrusion because of reduced mucilage accumulation, and this is associated with changes in columellae structure. Mutants in this group include transparent testa glabra1 (ttg1) (Koornneef, 1981), ttg2 (Johnson et al., 2002), glabra2 (gl2) (Koornneef et al., 1982), aberrant testa shape (ats) (Léon-Kloosterziel et al., 1994), mum4 (Western et al., 2001), myb61 (Penfield et al., 2001), and tt8 and egl3 (Nesi et al., 2000; Zhang et al., 2003). The final category is the most severely affected in the apetala2 (ap2) mutant mucilage and columellae are completely absent (Jofuku et al., 1994). The genes affected in ap2, ttg1, ttg2, gl2, myb61, tt8, egl3, and ats mutants play roles in transcriptional control (Jofuku et al., 1994; Rerie et al., 1994; Walker et al., 1999; Nesi et al., 2000; Penfield et al., 2001; Johnson et al., 2002; Zhang et al., 2003; McAbee et al., 2006). In only one mutant, mum4, does the defective gene correspond to a downstream target encoding a polysaccharide biosynthesis enzyme, UDP-rhamnose synthase (Usadel et al., 2004; Western et al., 2004; Oka et al., 2007).
Although a number of enzymes involved in complex polysaccharide metabolism have been purified and their corresponding genes characterized, their precise function in planta often remains obscure due to the lack of associated phenotypes in mutants or transgenic plants. This is true of plant β-d-galactosidases (EC 3.2.1.23), a large group of enzymes that can release galactose from various substrates, such as pectin, arabinogalactans, and galactolipids (Dey and Del Campillo, 1984). Despite the identification of β-d-galactosidase activities in a variety of plant extracts, the in vivo substrate is often unknown as activity has only been demonstrated using artificial substrates. β-d-galactosidases can be separated into at least two groups based on their substrate specificity: the first specifically acts on β-(1→4) galactan, whereas the second prefers β-(1→3) and β-(1→6) galactooligosaccharides (Kotake et al., 2005). The first group of β-d-galactosidases has been implicated in the degradation of β-(1→4) galactan side chains from pectin during fruit softening and is thought to play a role in cell wall loosening (Carey et al., 1995; Smith et al., 2002), and a radish (Raphanus sativus) β-d-galactosidase from the second group, Rs BGAL1, has been shown to hydrolyze β-(1→3)- and β-(1→6)-galactosyl residues from arabinogalactan protein in vitro (Kotake et al., 2005). β-d-galactosidases are also required for the degradation of xyloglucan (Iglesias et al., 2006), but so far only in vitro activities have been demonstrated.
Here, we performed a screen of Arabidopsis accessions collected from different geographical locations with the aim of identifying new elements involved in the formation of myxospermous epidermal cells and establishing comparative studies based on the environmental conditions in which mucilage-producing and nonmucilage-producing accessions are found. A naturally occurring Arabidopsis accession was identified that does not liberate mucilage on imbibition, and the mutated locus was found to be allelic to that affected in the mum2 mutant (Western et al., 2001). The MUM2 gene was identified by positional cloning and encodes a putative glycosyl hydrolase from protein family 35, BGAL6. The MUM2/BGAL6 protein was shown to be required for the removal of galactose side branches from RG-I, thus modifying the hygroscopic properties of the inner mucilage layer.
RESULTS
Identification of a Naturally Occurring Arabidopsis Mutant Affected in Mucilage Secretion
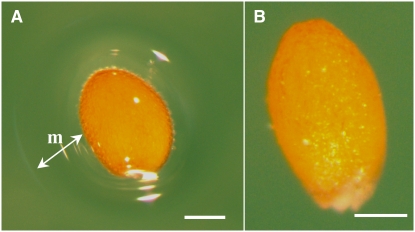
A visual screen was performed to identify accessions from the Versailles Biological Resource Centre (http://dbsgap.versailles.inra.fr/vnat/) whose seeds did not liberate mucilage when sown on agarose plates. Of the 309 seed lots tested, only the Shahdara accession did not release a refractive halo of mucilage (cf. Figures 1A and 1B). This accession had been harvested near the Shokhdara river in Tajikistan at an altitude of ∼3400 m.
Figure 1.
The Shahdara Accession Does Not Liberate Mucilage on Imbibition.
(A) A seed from the Col-0 accession with a refractive halo of mucilage around the seed. m, mucilage.
(B) A seed from the Shahdara accession.
Sterilized seeds were imbibed for 16 h on 0.5% (w/v) agarose.
Bars = 150 μm.
The absence of mucilage release in the Shahdara accession was reminiscent of that of two of the mum mutants (Western et al., 2001) whose gene identity is unknown. Crosses were performed with alleles of these two mutants, mum1-1, mum2-1, and mum2-2, and progeny examined for phenotype complementation. Only descendants from the cross between Shahdara and mum1-1 released mucilage on imbibition, indicating that the defect in the Shahdara accession was in the MUM2 locus.
Map-Based Cloning of At MUM2
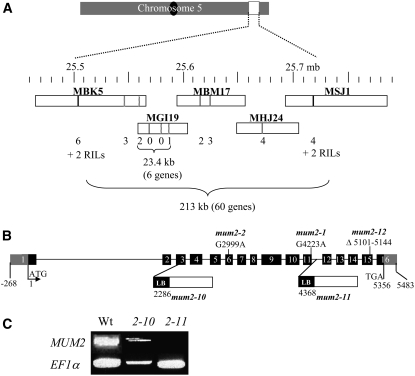
A recombinant inbred line (RIL) population was generated from a cross between Shahdara and the Bay-0 accession, and F6 plants were genotyped for a set of 38 physically anchored microsatellite markers (Loudet et al., 2002). By analyzing the mucilage phenotype in F7 seed lots from 411 RILs and comparison to their available genotyping data, the MUM2 locus was localized to the end of the lower arm of chromosome 5 between the markers MSAT5.12 (21,425 kb) and MSAT5.19 (25,924 kb). The frequency of inheritance of the Shahdara mucilage phenotype compared with that of Bay-0 indicated that it was inherited as a single recessive locus. Fine mapping was undertaken and the mapping interval reduced to five BAC clones encompassing a 213-kb interval (Figure 2A). Because only four RILs recombined in this chromosome region, additional recombinants were identified from a F2 population of Bay-0 × Shahdara and their F3 progeny examined for mucilage release. The new recombinants enabled the chromosome region containing the MUM2 locus to be reduced to a 23.4-kb region containing six annotated genes (Figure 2A). Database analyses identified mutant lines with T-DNAs inserted in four of these genes. Homozygous lines were obtained and analyzed for their ability to release mucilage on imbibition. Two lines did not release mucilage, and both of these had an insertion in the gene At5g63800. Sequencing of the At5g63800 gene in the Shahdara accession revealed the presence of a 44-bp deletion in exon 15, from Leu-662 onwards. This deletion would cause a frame-shift mutation changing the next 23 amino acids followed by the introduction of a stop codon. Crosses between Shahdara and the two T-DNA lines did not result in genetic complementation and confirmed that MUM2 corresponds to At5g63800. In addition, sequencing of At5g63800 in the mum2-1 and mum2-2 mutants identified point mutations that would convert Gly-513 into Arg and Gly-238 to Glu, respectively. Ten mum2 alleles have been identified (Dean et al., 2007); the mum2-10 allele corresponds to one of the T-DNA lines used in this study, so the second was termed mum2-11 and the mutation in the Shahdara accession termed mum2-12 (Figure 2B). Analysis of the expression of MUM2 in the T-DNA insertion mutants detected transcripts in both mutants with RT-PCR using primers situated in exons 1 and 2 (data not shown). By contrast, using primers downstream in exons 12 and 14, transcripts were only detected in mum2-10 extracts, and mum2-11 appeared to be null (Figure 2C).
Figure 2.
Map-Based Cloning of At MUM2 and Characterization of the mum2 Mutants.
(A) Physical map of the lower arm of chromosome 5 showing the 213-kb region (black lines) in which the MUM2 gene was localized. Fine mapping using new simple sequence length polymorphism (SSLP) markers (gray lines) or single nucleotide polymorphism markers (hatched lines) are indicated in the BAC clone boxes. The number of recombinants identified for each marker, in 411 RILs or 2880 F2 plants, is indicated underneath each.
(B) Schematic representation of the structure of MUM2 (At5g63800) and the sites and nature of the mum2 mutations.
(C) Effect of mum2 mutations on MUM2 gene expression. RT-PCR analysis was performed using primers in exons 12 and 14. A control experiment was performed with primers for the EF1α-4a gene transcript. 2-10, mum2-10; 2-11, mum2-11.
The Protein Encoded by At MUM2 Belongs to Glycosyl Hydrolase Protein Family 35
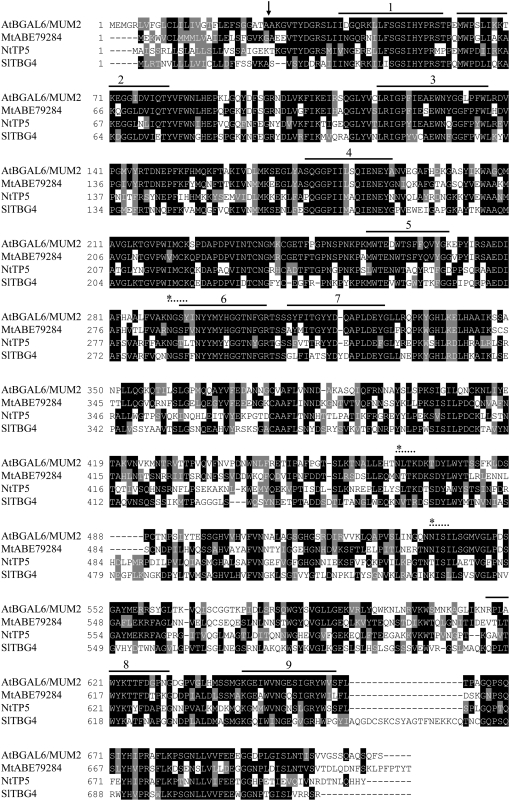
MUM2 contains 16 exons (Figure 2B), and the predicted open reading frame encodes a protein of 718 amino acids, with a predicted molecular mass of 79.7 kD. Sequence analysis showed that the protein contained the nine hallmark blocks of similarity found in protein family 35 glycosyl hydrolases (Figure 3), including the Prosite signature PS01182 in block 4. In Arabidopsis, this protein family has 18 members, and the protein encoded by At5g63800 has been termed BGAL6. The MUM2/BGAL6 protein showed high similarity to known or putative β-galactosidases from other plant species (Figure 3), such as Medicago truncatula, tomato (Solanum lycopersicum), and tobacco (Nicotiana tabacum) (62, 48, and 48% identity or 70, 60, and 57% similarity, respectively). The most similar Arabidopsis family 35 glycosyl hydrolase to BGAL6, At1g77410 (50% identity and 58% similarity) (Iglesias et al., 2006), has a C-terminal extension of 110 amino acids compared with MUM2/BGAL6, which corresponds to a Gal binding lectin-like domain (Prosite signature SSF49785). These domains subsequently can be removed by posttranslational processing (Kotake et al., 2005); the absence of such a domain in the MUM2/BGAL6 protein suggests that the two proteins could have either different functions or kinetic properties. Indeed, a mutant with a T-DNA insertion in the At1g77410 gene was not defective for mucilage release from seeds on imbibition (data not shown). The first 28 amino acids of the MUM2/BGAL6 protein probably correspond to a hydrophobic leader sequence, and analysis with PSORT predicts the mature protein to be exported to the apoplastic space (Iglesias et al., 2006); the resulting mature protein would be 76.8 kD with a pI of 9.04. Finally, three potential N-glycosylation sites are present in the MUM2/BGAL6 protein, the first situated just before block 6 being conserved between those plant sequences similar to MUM2/BGAL6.
Figure 3.
Amino Acid Alignment of At BGAL6/MUM2 with Known or Putative β-Galactosidases from Other Plants.
Alignment of the At BGAL6/MUM2 predicted amino acid sequence with sequences from M. truncatula (Mt), tobacco (Nt), and tomato (Sl). At BGAL6/MUM2, predicted Arabidopsis protein BAB10473; Mt ABE79284, M. truncatula predicted protein ABE79284; Nt TP5, predicted tobacco protein CAC13966; Sl TBG4, tomato β-galactosidase/exo-β-(1→4)-galactanase AAC25984. Sequence alignment was performed using the MultAlin program (Corpet, 1988) using the Blosum-62-12-2 symbol comparison table. The figure was drawn using Boxshade version 3.21 (K. Hofmann and M.D. Baron; http://www.ch.embnet.org/software/BOX_form.html) with a similarity threshold of 60% and the default similarities (FYW, IVLM, RK, DE, GA, TS, and NQ). Completely conserved residues across two or more sequences are shaded black, and similar residues conserved across two or more sequences are shaded gray. The arrow indicates the At BGAL6/MUM2 putative signal sequence cleavage site, the asterisks and dotted lines indicate the site of putative N-glycosylation sites, and the solid lines indicate the position of nine blocks of similarity found in family 35 glycosyl hydrolases.
At MUM2 Is Expressed in the Developing Testa
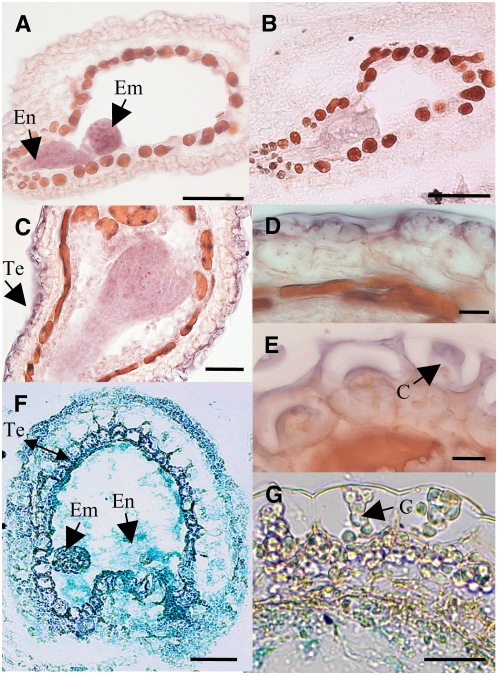
Transcripts corresponding to At MUM2 have been detected by quantitative RT-PCR analysis in a variety of plant organs, including young and mature leaves, roots, upper and basal stem regions, flowers, and siliques, with the highest levels of expression found in siliques (Iglesias et al., 2006). To obtain more detailed information concerning the spatial and temporal expression of MUM2 within seed tissue, in situ hybridization was performed at different stages of seed development. At early developmental stages, labeling was detected in all seed tissues but was particularly strong in the seed endosperm around the suspensor and in the globular embryo (Figures 4A and 4B). The hybridization signal decreased in the embryo and endosperm at the heart stage while becoming strong in the testa (Figure 4C). The testa remained strongly labeled at the torpedo stage, and expression was clearly highest in the epidermal cell layer of the outer integument (Figure 4D). In mature embryos, no expression was detectable, while the epidermal cell layer still showed some labeling (Figure 4E), but at later developmental stages, no hybridization signal was detectable in any tissue (data not shown). Expression patterns observed in transgenic plants from five independent transformants expressing the β-glucuronidase (GUS) reporter gene under the control of the MUM2 promoter region (Figures 4F and 4G) were similar to those obtained by in situ hybridization, although differences in staining intensity between the embryo, endosperm, and testa were not apparent.
Figure 4.
Spatial and Temporal Expression Patterns of the At MUM2 Gene in Developing Seeds.
(A) to (E) In situ hybridization using an At MUM2–specific probe: hybridization with antisense probe ([A] and [C] to [E]) and control hybridization with sense probe (B).
(F) and (G) Histochemical analysis of GUS reporter gene expression from the At MUM2 promoter. Expression in seeds at the globular embryo stage ([A], [B], and [F]), seeds with heart-stage embryo (C), epidermal cells of seeds with torpedo stage embryo (D), and epidermal cells of seeds with a mature embryo at mid-seed development ([E] and [G]). En, endosperm; Em, embryo; Te, testa. Bars = 50 μm in (A) to (C) and (F), 10 μm in (D) and (E), and 25 μm in (G).
At MUM2 Is Expressed in Specific Cell Types in Non-Seed Tissues
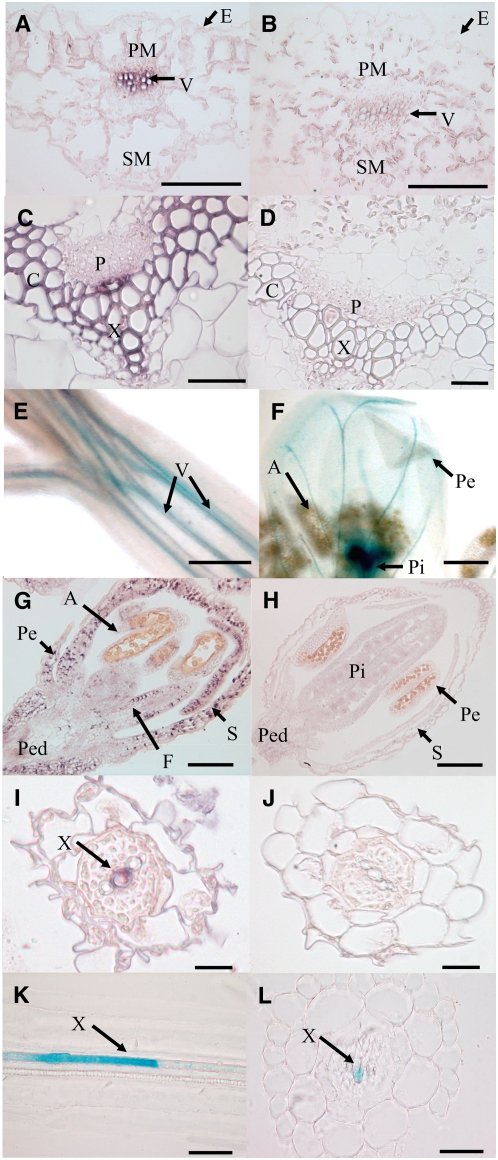
The presence of MUM2 transcripts in leaves, roots, stems, and flowers (Iglesias et al., 2006) suggested that it is expressed ubiquitously. Analysis of expression by in situ hybridization and GUS reporter gene expression confirmed that MUM2 was expressed in all plant organs but often appeared restricted to certain cell types within tissues. In leaves, the protoxylem of the vascular bundles was labeled (Figures 5A and 5B); however, in mature leaves as this transformed into dead metaxylem, the expression was reduced to isolated cells within the vascular tissue (data not shown). In upper and basal stem regions, MUM2 mRNA was detected in protoxylem (Figures 5C to 5E; data not shown). Expression was also detected in collenchyma once developed in older parts of the stem (Figures 5C and 5D). In flowers, expression was observed in most tissues (Figures 5F to 5H) and was very strong in the vascular tissue of the style just after fertilization (Figure 5F) and young anthers (data not shown). Finally, weak expression was also visualized in root xylem (Figures 5I and 5J) but often in isolated cells with reticulate or pitted wall thickenings (Figures 5K and 5L) characteristic of metaxylem (Gorenflot, 1986). As such cells are dead, the labeled cells probably corresponded to cells in the process of differentiating into metaxylem.
Figure 5.
At MUM2 Expression in Non-Seed Tissues.
(A) to (D) and (G) to (J) In situ hybridization using an At MUM2–specific probe.
(A), (C), (G), and (I) Hybridization with antisense probe.
(B), (D), (H), and (J) Control hybridization with sense probe.
(E), (F), (K), and (L) Histochemical analysis of GUS reporter gene expression from the At MUM2 promoter.
(A) and (B) Transverse sections of leaves.
(C) and (D) Transverse sections of basal stems.
(E) and (F) Whole mount of upper stem and flowers.
(G) and (H) Transverse sections of flowers.
(I), (J), and (L) Transverse sections of roots.
(K) Longitudinal section of a root.
A, anther; C, collenchyma; E, epidermis; F, filament; P, phloem; Pe, petal; Ped, pedicel; Pi, pistil; PM, palisade mesophyll; S, sepal; SM, spongy mesophyll; V, vascular tissue; X, xylem. Bars = 250 μm in (E), 200 μm in (F), 100 μm in (A), (B), (G), and (H), 50 μm in (I) to (L), and 20 μm in (C) and (D).
Structure and Development of mum2 Mutant Seed Coats
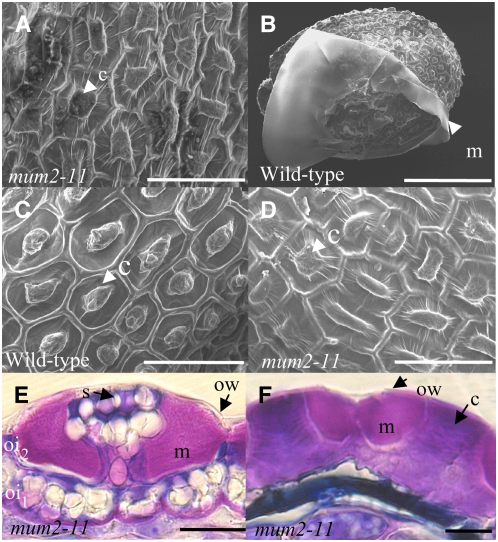
To gain more information about the cause of the absence of mucilage release on imbibition, the structure of mum2 mutant seed coats was studied in more detail. Scanning electron microscopy was performed on mature seeds either before or after imbibition. The epidermal cells of both dry wild-type and mutant seeds exhibited the typical morphology of hexagonal cells with a central columella and were equivalent in size (Figure 6A; data not shown). In imbibed wild-type seeds, the released mucilage was observed as a white film attached to the seed coat (Figure 6B), and the hollow around the columella appeared deeper as the dehydrated mucilage was no longer present (Figure 6C). In agreement with the absence of mucilage release, the seed coat of imbibed mum2 mutant seeds showed no difference from that of dry seeds (Figure 6D). Thus, in contrast with previously identified mutants affected in mucilage release, the epidermal cells of mature mum2 mutants showed no obvious structural defect.
Figure 6.
Mucilage Accumulation and Epidermal Seed Coat Cell Structure in the mum2 Mutant.
(A) to (D) Scanning electron micrographs of the surface of wild-type and mum2-11 mutant seeds.
(E) and (F) Sections of developing seeds stained with toluidine blue.
(A) mum2-11 mature dry seed.
(B) and (C) Imbibed mature wild-type seed.
(D) Imbibed mature mum2-11 seed.
(E) Seed coat from mum2-11 seed at torpedo embryo stage.
(F) Seed coat from mum2-11 seed at late maturation stage.
c, columella; m, mucilage; s, starch granules; ow, outer tangential cell wall; oi2, epidermal layer of the outer integument; oi1, underlying layer of the outer integument. Bars = 50 μm in (A), (C), and (D), 200 μm in (B), and 10 μm (E) and (F).
To determine whether the mum2 mutants were affected in mucilage accumulation, epidermal cells were examined for developmental defects by light microscopy on seed sections. Mucilage is accumulated in the epidermal layer of the seed coat during seed development (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000), and the process of differentiation of epidermal cells starts at the globular stage with cells becoming highly vacuolated and accumulating starch granules. Mucilage begins to accumulate in the apoplastic space from the heart stage onwards, and this is accompanied by the gradual formation of a central column of cytoplasm. When embryos have reached the upturned-U stage, a cellulosic cell wall begins to appear around the cytoplasm, including the central column, and continues until no starch granules remain. The resulting cell contains a columella made up entirely of cell wall material and a thickened inner tangential cell wall. No difference was observed in the timing or process of differentiation between wild-type and mum2 mutant seeds (data not shown). Furthermore, no structural difference was apparent, and mucilage was clearly accumulated in the apoplastic space (Figures 6E and 6F); the mucilage staining intensity with toluidine blue was equivalent in the wild type and mutants. Comparisons were not possible from 15 to 16 d after flowering onwards as mucilage was released from wild-type seeds during the fixing procedure, whereas that of the mum2 mutants remained in the epidermal cells.
β-d-Galactosidase Activity Is Absent from mum2 Mutants
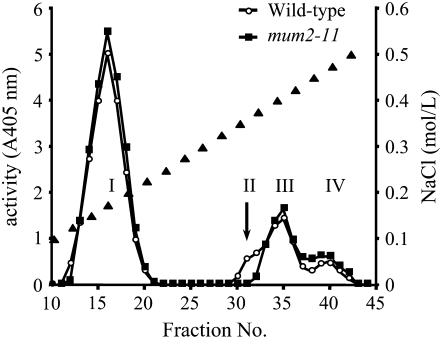
To verify the putative function attributed to MUM2/BGAL6, β-d-galactosidase activity was assayed in wild-type and mum2-10 and mum2-11 protein extracts from developing siliques. Extracts were partially purified prior to enzyme assays using cation-exchange chromatography, and the resulting fractions were assayed for β-d-galactosidase activity using the artificial substrate p-nitrophenol-β-d-galactopyranoside. Four major peaks of β-d-galactosidase activity were observed in wild-type extracts, but the activity of the minor peak, number II, was absent from mum2 extracts (Figure 7; data not shown), indicating that the lesion in this mutant affected a β-d-galactosidase activity in siliques.
Figure 7.
Analysis of β-d-Galactosidase Activity in the mum2-11 Mutant Using Cation-Exchange Chromatography.
Elution profile of protein extracts from wild-type (open circles) and mum2-11 (closed squares) developing siliques separated by carboxymethyl-sepharose chromatography. The NaCl discontinuous gradient is indicated by black triangles. β-d-galactosidase activity was assayed at 37°C for 30 min. Four major peaks of activity were found, indicated by roman numerals, and the arrow indicates the peak of activity absent in mum2-11 extracts.
To determine whether glycosyl hydrolase activities are associated with seed mucilage, in situ activity assays were performed on mature seeds using the artificial substrates 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) or 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-GlcA). No ClBr-indigo was released when X-GlcA was used as a substrate, indicating the absence of glucuronidase activity within the mucilage (data not shown). By contrast, blue labeling was observed with the X-Gal substrate and clearly showed that an endogenous β-galactosidase activity was present in seed mucilage (see Supplemental Figure 1A online). As mucilage is not released from the mum2 mutants, the eventual loss of endogenous activity could not be examined.
The Defect in mum2 Mutants Affects Mucilage Swelling
The absence of mucilage release from epidermal seed coat cells in mum2 seeds could be due to increased mechanical resistance or permeability of the outer tangential cell wall retaining the mucilage or modification of the hydrophilic potential of the mucilage such that its expansion is reduced. No obvious structural change was observed in the outer tangential cell wall thickness or staining properties in the mum2 mutants (Figure 6; data not shown). To determine if the defect was nonetheless due to an alteration in the outer cell wall, chemical treatments were employed with the aim of weakening or removing it and allowing mucilage release. Treatment with hot hydrochloric acid (0.05 M, 85°C) followed by sodium hydroxide (0.3 M) resulted in mucilage release from epidermal cells of the mum2-11 mutant. It is important to note that seeds treated in this way required washing prior to subsequent staining or labeling, so the mucilage observed only corresponded to the inner adherent mucilage layer. Staining with ruthenium red showed that although the mum2-11 mucilage was no longer retained by the cell wall, its appearance was different from that of wild-type mucilage released from seeds treated sequentially with acid and alkali (see Supplemental Figure 1B online); the mucilage only swelled a little, blistering slightly out of the epidermal layer (Figure 8; see Supplemental Figures 1B to 3 online). This indicated that either less mucilage was in fact present in the epidermal cells or that its hygroscopic properties were altered.
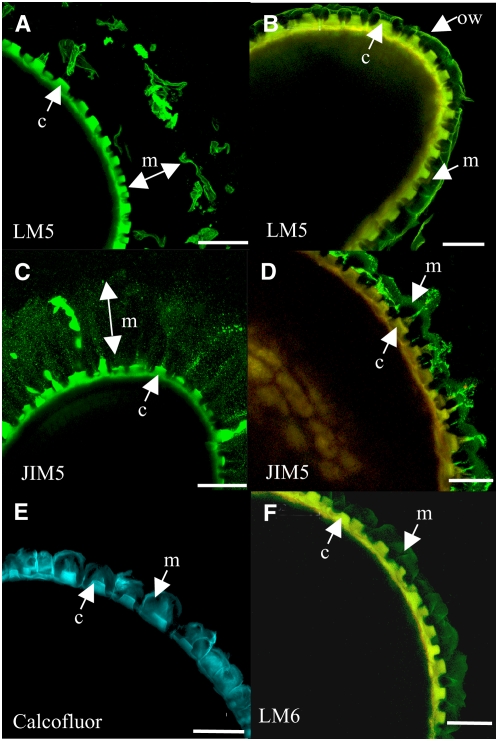
Figure 8.
Labeling of Seed Mucilage Released from Acid- and Alkali-Treated Seeds with Antipectin Antibodies Observed as Immunofluorescence.
Whole seeds were treated sequentially with acid and then alkali to release mucilage from epidermal cells and then labeled with LM5 ([A] and [B]), JIM5 ([C] and [D]), Calcofluor (E), and LM6 (F). Seeds were either from the wild type ([A] and [C]) or mum2-11 ([B] and [D] to [F]). Confocal microscopy was used to take optical sections passing through seeds. c, columella; m, mucilage; ow, outer tangential cell wall. Bars = 70 μm in (A) and (C), 50 μm in (B), (D), and (F), and 60 μm in (E).
Analysis of the Inner Layer of mum2 Mucilage by Immunolabeling with Antipectin Antibodies
The absence of swelling of mum2 mucilage when the outer tangential cell wall was removed would suggest that the composition of the inner mucilage is different in the mutant and that removal of galactose increases the hydrophilic potential of the mucilage. In situ analysis with the antibody LM5, which recognizes (1→4)-β-galactan (Jones et al., 1997), has shown that the inner layer of wild-type mucilage contains a small amount of galactan (Macquet et al., 2007). Immunolabeling was performed to determine whether the amount of galactan present in mucilage released by chemical treatment of mum2 seeds was different from that of the wild type. The acid and alkali treatment eliminated the LM5 labeling observed in the inner layer of wild-type mucilage, except for that associated with large debris; this appeared to correspond to pieces of outer tangential cell wall (Figure 8A). Despite the removal of labeling by acid treatment, mum2-11 mucilage still showed a strong signal with LM5 (Figure 8B), being strongest at the outer edge of the released mucilage, perhaps from labeling of outer tangential cell wall material that adhered to the mucilage. Labeling of the underlying mucilage was also clearly visible.
Cellulose, arabinan, and methyl-esterified HG have also been shown to be minor components of the inner mucilage layer (Macquet et al., 2007). The mucilage released from mum2 seeds was thus compared with wild-type seeds treated with acid and alkali for these components. The limited amount of labeling observed for wild-type seed mucilage with LM6, which recognizes (1→5)-α-arabinan (Willats et al., 1998), was eradicated by the acid/alkali treatment, in a similar manner to LM5 labeling, with only cell wall debris visible (data not shown). This debris was again visible in wild-type mucilage labeled with Calcofluor or the antibody recognizing moderately methyl-esterified HG, JIM5 (Willats et al., 2000) (Figure 8C; data not shown), but otherwise the staining of the inner mucilage was equivalent to that for wild-type mucilage released by imbibition in water (Macquet et al., 2007). The mucilage of mum2-11 mutants showed labeling with JIM5, Calcofluor, and LM6 (Figures 8D to 8F). Although LM6 labeling was relatively uniform through the mucilage, JIM5 labeling seemed to be restricted to a region underneath the layer of outer cell wall debris. Therefore, two major differences were observed between wild-type and mum2-11 acid and alkali-treated seeds, both LM5 and LM6 labeled the inner mucilage, suggesting increased galactan and arabinan residues in the mucilage pectin or modified accessibility.
Sugar Analysis of the Soluble Outer Layer and Adherent Inner Layer of mum2 Mucilage
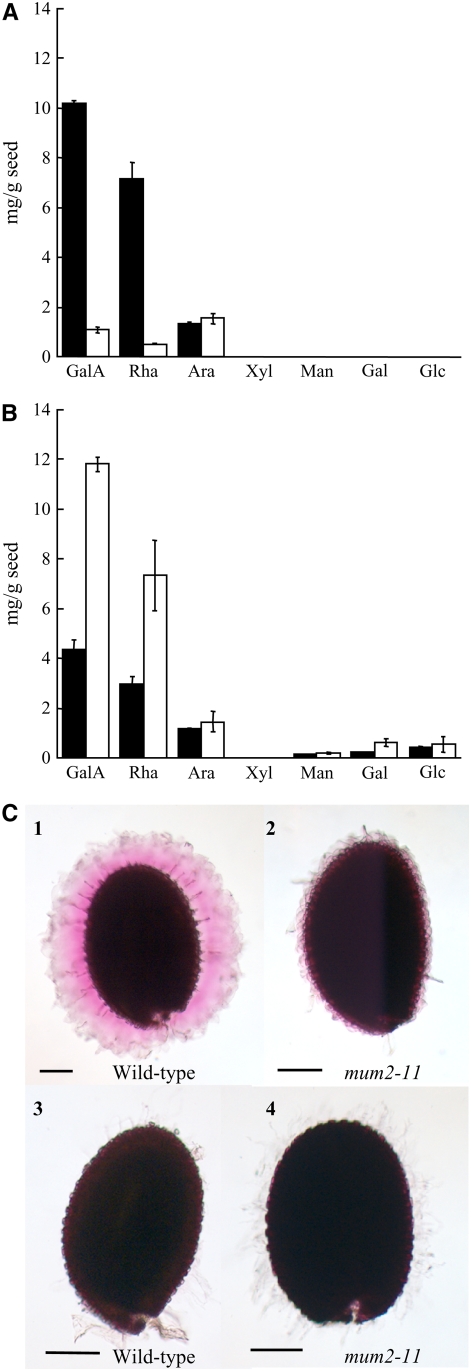
Extracts obtained from wild-type and mutant seeds on sequential treatment with acid and alkali were dialyzed and analyzed for their sugar composition. It has been shown previously that treatment of wild-type seeds with acid or alkali does not extract the inner mucilage layer (Macquet et al., 2007). Furthermore, the extraction procedure used here, which did not employ lateral shaking, resulted in a wild-type extract that was similar to that of water-extracted mucilage (Macquet et al., 2007) with respect to the amounts of Rha and GalA; thus, these soluble extracts can be considered to represent the outer mucilage layer. As expected, in the wild type, the major components were Rha and GalA, with Ara as a minor sugar (Figure 9A). In the mum2-11 mutant, Rha and GalA levels were very low compared with those of the wild type, although Ara levels were similar (Figure 9A). As the Ara abundance was stable in the wild type and mum2, whereas Rha levels were reduced, this Ara is likely to be present as free arabinan chains. A small amount of Gal was also detectable in the mum2 soluble mucilage in contrast with that of the wild type. Strikingly, the total amount of sugars extracted from mum2-11 was 80% lower, indicating that the mutant contained less soluble mucilage than the wild type. The mutation in mum2, therefore, affected both the amount and composition of soluble mucilage.
Figure 9.
Sugar Analysis of Extracts and Ruthenium Red Staining of Acid- and Alkali-Treated Wild-Type and mum2-11 Seeds before and after Enzymatic Hydrolysis of the Inner Layer of Seed Mucilage.
(A) and (B) Amounts of indicated sugars in soluble extracts (A) or hydrolysates from RGase digestion (B). Black bars, the wild type; white bars, mum2-11. Values from digest hydrolysates were calculated after deduction of the amount of the different individual sugars released by buffer alone. Only those sugars present at significant levels are presented. Error bars represent sd values (n = 2).
(C) Mucilage remaining around seeds after digestion with pectolytic enzymes was visualized by ruthenium red staining of the wild type (left) and mum2-11 (right). (1) and (2) undigested control; (3) and (4) digested with RGase, endo-PGase II, and cellulase in ad-mixture. All experiments were performed in duplicate. Bars = 150 μm.
Like the inner layer of wild-type mucilage, that released from mum2 seeds on acid and alkali treatment remained attached to the seed. Analysis of the composition of the inner mucilage layer was achieved for the wild type using enzymatic digestion (Macquet et al., 2007) and demonstrated that the major component was RG-I. The same technique was employed on acid- and alkali-treated seeds to determine mucilage sugar composition. Seeds were digested with rhamnogalacturonan hydrolase (RGase), endo-polygalacturonase II (endo-PGase II), or cellulase either individually or together in an ad-mixture. Ruthenium red staining of seeds after digestion was similar to that observed previously for wild-type water-extracted seeds (Macquet et al., 2007) with the inner mucilage layer of both the wild type and mutant digested extensively by RGase and the ad-mixture and little effect of endo-PGase II or cellulase (Figure 9C; data not shown). The sugar composition of mum2-11 extracts after digestion with RGase or ad-mixture was significantly enriched in Rha and GalA compared with the wild type (Figure 9B; data not shown), indicating that despite the width of the inner mucilage layer appearing reduced on ruthenium red staining, the amount of polysaccharides present was in fact higher. The amount of Gal and Ara present in the inner layer was also higher (Figure 9B). Nonetheless, when the sugars extracted from both mucilage layers were added together, levels were only ∼10% lower in the mum2-11 mutant compared with the wild type (Figures 9A and 9B). It is noteworthy that the molar ratio of Gal/100 Rha was much higher for mum2-11 than for the wild type, 7.2 compared with 2.0, respectively.
To investigate the modifications that could be the target for a β-d-galactosidase activity absent in the mum2 mutant, mass spectrometry (MS) analyses were performed on the inner mucilage layer digestion products. The major constituent of the inner mucilage layer, RG-I, is composed mainly of a repeating disaccharide [→2)-α-l-Rhap-(1→4)-α-d-GalpA-(1→]n (RG backbone) and can have side branches on the Rha residues consisting of arabinan and (arabino)-galactan moieties or single sugar units. In accordance, the major signals on the full MS spectra in both the wild type and mum2-11 had mass-to-charge (m/z) ratios that are consistent with oligomers with a basic structure of alternating GalA and Rha residues (Table 1, Figure 10). The mum2-11 full MS spectrum exhibited additional peaks that might correspond to RG oligomers carrying one or two hexose residues (Schols et al., 1995; Renard et al., 1998) (Figure 10, Table 2). Detailed linkage and branching patterns were established for these different oligomers using electrospray ion trap mass spectrometry (Quéméner et al., 2003a, 2003b; Ralet et al., 2005). For unbranched RG-I oligosaccharides, fragmentation analyses conducted on the different parent ions yielded daughter ions that agreed with alternating 4-linked GalA units and 2-linked Rha units, the reducing ends being GalA units (Cancilla et al., 1998). For peaks that could correspond to oxidized and dehydrated forms of the RG oligomers carrying one hexose residue, fragmentation analyses revealed that reducing ends were GalA units (see Supplemental Figure 2A online) and that the hexose unit that was directly linked onto a Rha unit on position O-3 or O-4 was in effect oxidized and dehydrated in these compounds (see Supplemental Figures 2B and 2C online). Fragmentation analyses performed on peaks that could correspond to oxidized and dehydrated forms of the RG oligomers carrying two hexose residues revealed that only one hexose unit was directly linked onto each Rha unit (data not shown).
Table 1.
Unbranched RG-I Oligosaccharides Detected by Mass Spectrometry in Both Wild-Type and mum2-11 Hydrolysates Obtained from Digestion of the Inner Mucilage Layer with RGase
| Oligosaccharides
|
||||
|---|---|---|---|---|
| Ion Type | Rha1-GalA1 | Rha1-GalA2 | Rha2-GalA2 | Rha2-GalA3 |
| [M-H]− | 339 | 515 | 661 | 837 |
| [M-H]−Na form+ | – | 537 | 683 | 859 |
| [M-H]− 2Na form+ | – | – | – | 881 |
| [M-2H]2− | – | 257 | – | 418 |
| [M-2H]2− Na form+ | – | – | – | 429 |
| [M-3H]3− | – | – | – | 278 |
Values are m/z ratios. Single [M-H]−, double [M-2H]2−, and triple [M-3H]3− deprotonated pseudomolecular ions and some of their sodium forms were detected.
Figure 10.
The mum2-11 Mutant Mucilage Enzymatic Digest Contains Additional Ions Compared with the Wild Type Corresponding to Hexose Branching on the RG-I Backbone.
Full MS spectra of wild-type (A) and mum2-11 mutant (B) hydrolysates obtained from RGase digestion of acid- and alkali-treated seeds. Ion peaks indicated by an asterisk are only present in mum2-11 digest products; their m/z ratios are indicated in bold.
Table 2.
Branched RG-I Oligosaccharides Only Detected in mum2-11 on MS of Hydrolysates Obtained from Digestion of the Inner Mucilage Layer with RGase
| Oligosaccharides
|
||||
|---|---|---|---|---|
| Ion Type | Rha1-GalA2- Hexose1 | Rha2-GalA2- Hexose1 | Rha2-GalA2- Hexose2 | Rha3-GalA3- Hexose1 |
| [M-H]− | 677 | 823 | 985 | – |
| [M-H]− oxidized dehydrated | 657 | 803 | – | – |
| [M-H]− oxidized +32 (O2) | 707 | 853 | – | – |
| [M-H]− Na form+ oxidized +32 (O2) | 729 | – | – | – |
| [M-2H]2− | – | 411 | 492 | 572 |
| [M-2H]2− oxidized dehydrated | 328 | 401 | 482 | 562 |
| [M-2H]2− oxidized +32 (O2) | 353 | 426 | 497 | 587 |
| [M-2H]2− Na form+ oxidized +32 (O2) | – | – | – | 598 |
| [M-3H]3− oxidized dehydrated | – | – | – | 374 |
Values are m/z ratios. Single [M-H]−, double [M-2H]2−, and triple [M-3H]3− deprotonated pseudomolecular ions and some of their sodium forms were detected.
The inner mucilage layer digestion products were also analyzed for modifications by high-performance anion exchange chromatography (HPAEC). Elution volumes of the different peaks were compared with those of authentic RG-I homemade standards with different degrees of polymerization (Rha2-GalA2, Rha3-GalA3, Rha4-GalA4, and Rha5-GalA5) and to those of sugar beet (Beta vulgaris) modified hairy regions (MHRs) (molar ratio Rha/Gal/GalA of 1:1.3:2.4) of RGase digestion products. Sugar beet MHR digestion products were also analyzed by MS and yielded peaks corresponding to Rha1-GalA2, Rha2-GalA3, Rha1-GalA2-Gal1, Rha2-GalA2-Gal1, Rha2-GalA2-Gal2, Rha3-GalA3-Gal1, and Rha3-GalA3-Gal2 (data not shown). The wild-type inner mucilage layer digestion products yielded three peaks by HPAEC (see Supplemental Figure 2 online), the first two of which had retention times consistent with the Rha1-GalA2 and Rha2-GalA3 oligomers detected by MS (Figure 10, Table 1). The mum2-11 inner mucilage digest contained several additional peaks, three having retention times equivalent to peaks in the sugar beet MHR digest (see Supplemental Figure 3 online), indicating that they corresponded to Rha-GalA oligomer variants with Gal branches.
DISCUSSION
A screen of a collection of Arabidopsis accessions was performed for defects in seed mucilage release with the aim of exploiting the natural variation that exists between Arabidopsis populations. One such Arabidopsis accession, Shahdara, was identified (Figure 1) and shown to contain a natural mutation at the MUM2 locus. Map-based cloning and the identification of five independent allelic mutants defective in mucilage release with lesions in the At5g63800 gene demonstrated that this corresponded to MUM2 (Figure 2). Furthermore, the same gene identity was deduced independently for At MUM2 using a different mapping population (Dean et al., 2007). In agreement with its role in mucilage release, MUM2 was expressed in the epidermal cells of the seed coat during seed development (Figure 4). The differentiation of the epidermal cells begins immediately after fertilization and continues to the middle of the mature embryo stage (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). GUS activity staining and in situ hybridization signals showed that MUM2 was expressed in the seed coat over this period and was no longer present during late seed development when epidermal cell differentiation is complete (Figure 4; data not shown). Furthermore, the in situ hybridization signal became more intense from the heart stage onwards (Figure 4C), just preceding the appearance of mucilage accumulation in the epidermal cells of the seed coat. In contrast with wild-type seeds, developing mum2 seeds from mature cotyledon embryo stage onwards did not generally liberate mucilage from the outer cell wall when placed in aqueous conditions and suggested that cell wall or mucilage modification had occurred at this stage (Western et al., 2001). Therefore, the timing of the increase in MUM2 hybridization signal is in accordance with the predicted ensuing requirement for protein activity.
The protein encoded by At MUM2 is annotated as BGAL6 in the carbohydrate-active enzymes database (http://afmb.cnrs-mrs.fr/CAZY/) (Coutinho and Henrissat, 1999) as it is predicted to be a putative β-d-galactosidase corresponding to protein family 35 glycosyl hydrolases. In accordance, the predicted protein contains the nine similarity blocks and the Prosite signature associated with glycosyl hydrolases of protein family 35 (Figure 3). Furthermore, it showed significant similarity to other plant β-d-galactosidases, notably TBG4 from tomato, that could hydrolyze X-Gal in vitro and corresponds to β-d-galactosidase purified from tomato fruit (Smith et al., 1998). Neither of the point mutations in mum2-1 or mum2-2 was situated in a conserved domain, although both affected amino acids that were invariant between the plant β-d-galactosidases, confirming their functional importance. Interestingly, the truncated protein resulting from the Shahdara deletion would contain all of the protein family 35 similarity blocks. As some seeds produced by Shahdara liberated drops of mucilage, this would indicate that the protein produced could sometimes be functional; although its activity appeared reduced, it remains to be determined whether this is due to altered stability or modified activity. Enzyme activity tests using proteins extracted from developing siliques confirmed that one of the major peaks of β-d-galactosidase activity present in the wild type was absent from mum2 mutants (Figure 7; data not shown). The presence of an endogenous β-d-galactosidase activity in the inner layer of seed mucilage was confirmed using an in situ activity test. Activity was clearly localized within the inner mucilage, being strongest closest to the seed coat (see Supplemental Figure 1A online). It was not possible to determine whether the in situ β-d-galactosidase activity was absent from mum2 seeds as mucilage was not released, and, when performed on seeds treated sequentially with acid and alkali, activity was no longer detected in wild-type seed mucilage. The presence of the β-d-galactosidase within the mucilage concords with the predicted apoplastic location of the MUM2/BGAL6 based on structural features: a hydrophobic leader sequence, a leader sequence cleavage site, and three possible N-glycosylation sites. In addition, a MUM2:green fluorescent protein fusion was observed outside the plasma membrane (Dean et al., 2007).
In a previous study, the phenotype of mum2-1 and mum2-2 ethyl methanesulfonate–generated mutants suggested that they were defective in mucilage extrusion either due to reinforcement of the outer tangential epidermal cell wall or due to altered mucilage composition (Western et al., 2001). Nonetheless, analysis of the epidermal cells by scanning electron microscopy and light microscopy did not identify any obvious structural defect, and analysis of the monosaccharide composition of whole seeds did not find any significant difference from the wild type (Western et al., 2001). The seed coat of the potentially null mum2 T-DNA insertion mutants was also like that of the wild type, and even after imbibition, the seed coat resembled that of dry wild-type seeds, with the outer tangential cell wall intact (Figures 6A to 6D; data not shown). Furthermore, sections of developing mum2 seeds showed that the outer tangential cell wall did not appear thickened or stain differently from that of the wild type, and mucilage was secreted normally into the apoplast around the columella (Figures 6E and 6F). However, removal of the outer tangential cell wall by chemical treatment showed that the mucilage produced in the mum2 mutant was different from that of the wild type as it only swelled slightly. Analysis of the mucilage released from the mutant as either soluble mucilage in the acid/alkali extract or the inner mucilage layer by enzyme digestion after acid/alkali treatment showed that the absence of mucilage swelling was not because less mucilage was synthesized (Figures 9A and 9B). In fact, the total amount of sugars released from inner and outer mucilage added together was only slightly lower in the mutant. Interestingly, the partitioning of the sugars between the inner and outer mucilage was altered, with the mum2 mutant containing significantly more of all the major monosaccharides in the inner mucilage, indicating that there was more polysaccharide in the inner layer of mucilage in the mutant than the wild type, with a proportional decrease in the outer mucilage layer (Figures 9A and 9B). The partial degradation of pectin by avocado (Persea americana) β-d-galactosidase was previously shown to increase pectin solubility and decrease its apparent average molecular size, in part due to decreased aggregation of pectin molecules (De Veau et al., 1993). The RG-I in the inner mucilage layer of Arabidopsis has a higher average molar mass than that of the outer water-soluble layer (Macquet et al., 2007), and the altered proportions of inner and outer mucilage in the mum2 mutant could result from an absence of solubilization of RG-I by the MUM2/BGAL6 β-d-galactosidase. Results obtained by in situ immunolabeling of the inner mucilage layer were consistent with monosaccharide analyses, with the signal from antibodies against (1→4)-β-galactan and (1→5)-α-arabinan being higher in mum2 than the wild type (Figures 8A, 8B, 8F, 9A, and 9B; data not shown).
As mum2 mucilage only swelled slightly when the outer cell wall no longer exerted a restraining force (see Supplemental Figures 1B to 3 online), this indicates that the mum2 mutation is unlikely to modify the outer cell wall properties in addition to those of the mucilage. It is possible, however, that as well as affecting the swelling of the inner mucilage, the mum2 mutation reduces the mechanical pressure on the outer cell wall due to the significant decrease in the amount of soluble outer mucilage (Figure 9A). Indeed, a function of the outer mucilage layer could be to generate the force required to break the outer tangential wall.
Although the MUM2/BGAL6 protein has been shown in vitro to release Gal from an artificial substrate (Dean et al., 2007) and mum2 protein extracts lacked a β-d-galactosidase activity (Figure 7), the question remained as to whether the in vivo substrate of the MUM2/BGAL6 protein was galactan and in what form. HPAEC analyses indicated the presence of Rha-GalA-Gal oligomers in the hydrolysates of the inner layer of mum2 mucilage that were absent from wild-type extracts (see Supplemental Figure 2 online) and MS analyses of extracts obtained for mum2 inner mucilage demonstrated that the mutant contained a number of links corresponding to branching of a hexose on Rha (Table 2, Figure 10; see Supplemental Figure 2 online). As the only hexose present in higher levels in mum2-11 inner mucilage extracts was Gal (Figure 9A), the MUM2/BGAL6 protein must act in vivo on galactose/galactan side branches of RG-I. Linkage analysis of mum2-soluble mucilage found higher levels of branch point Rha and terminal Gal residues, which also indicates that RG-I branching to Gal is higher than in the wild type (Dean et al., 2007).
The demonstration of different properties for branched and unbranched RG-I is potentially of broader interest because it may be exploited in commercial applications using pectin. The expression of a fungal β-d-galactosidase in potato tubers was previously shown to reduce the galactosyl content of RG-I and increase the accessibility of pectin in the cell wall to digestion with pectolytic enzymes (Sørensen et al., 2000). In the light of our new data concerning the action of β-d-galactosidase on RG-I hygroscopic properties, it is likely that this was due to similar modifications of RG-I properties. Furthermore, it will be interesting to determine whether similar proteins in tomato bring about fruit softening via comparable changes to cell wall RG-I properties. The expression of MUM2 in non-seed tissues (Figure 5; Iglesias et al., 2006; Dean et al., 2007) suggests that MUM2/BGAL6 could play a role in the modification of cell wall RG-I elsewhere. Notably, expression was observed in the xylem of leaves, stems, roots, and flowers (Figure 5). MUM2/BGAL6 could modify RG-I in xylem cell walls and might thereby increase the porosity of these cells. In addition, a proportion of mum2 mutant siliques was observed to be shorter than those of the wild type, suggesting altered fertility (Western et al., 2001). This is coherent with the strong expression of MUM2 in the vascular tissue of the style just after fertilization (Figure 5F), possibly to facilitate growth of the elongating pollen tube by softening the cell walls.
The identification of a naturally occurring accession affected in mucilage liberation has not only enabled the identification of a polysaccharide biosynthesis enzyme involved in the modification of mucilage properties, but opens the way for studies on the physiological role of seed mucilage. Hence, it is possible that seed mucilage was deleterious for Shahdara due to some characteristics of the environment in which the population was collected and that Shahdara seed has been subjected to a selective pressure leading to the survival of seeds that do not liberate mucilage. Alternatively, this function might simply be neutral to adaptation in Shahdara's environment, and a mutation has occurred that was not counterselected. Furthermore, MUM2 was expressed in other tissues, and any natural selection of a mum2 mutation could be related to MUM2 functions in these. The population of accessions screened was not evenly distributed geographically, with only seven of the 309 seed lots tested originating from central Asia. Therefore, it is impossible to distinguish between a selected or neutral mutation in Shahdara, and the identification of further accessions affected in mucilage liberation will be necessary for correlative studies to be undertaken.
METHODS
Plant Material and Mucilage Phenotyping
Seeds from 1 AV to 319 AV, except numbers 73, 93, 148, 270, 277, 292, 293, 300, 310, and 311, were obtained from the Versailles Natural Variation Collection of Arabidopsis accessions (http://dbsgap.versailles.inra.fr/vnat/) where Shahdara (236AV) corresponds to Nottingham Arabidopsis Stock Centre (NASC) stock number N929. Seeds from the Bay-0 × Shahdara RIL population (Loudet et al., 2002; http://dbsgap.versailles.inra.fr/vnat/) were phenotyped using the F7 generation produced from plants that had been cultivated together. The mum1-1, mum2-1, and mum2-2 mutants (Col-2 accession) were obtained from NASC (http://arabidopsis.info), and the Salk T-DNA insertion lines N511436 and N610461 (Col-0 accession), corresponding to mum2-10 and mum2-11, were identified in the SIGnAL database (Alonso et al., 2003; http://signal.salk.edu). To determine whether mucilage was released, sterilized seeds were sown on 0.5% (w/v) agarose plates and examined after 16 h for the presence of a refractive halo around the seed. Crosses were performed between homozygous mutants of mum2-12 (Shahdara accession) and mum1-1, mum2-1, mum2-2, mum2-10, and mum2-11 seeds. As the seed coat is maternally derived, progeny from F2 and F3 generations were examined for genetic complementation; only seeds from the cross with mum1-1 showed phenotypic complementation.
Positional Cloning
For preliminary mapping, available genotyping data were used; this corresponded to the 420 RILs from the Bay-0 × Shahdara population analyzed at the F6 generation (Loudet et al., 2002). Subsequently for fine mapping, additional recombinants were identified by screening 2880 individuals for recombination using high-throughput PCR analysis with flanking simple sequence length polymorphism (SSLP) markers whose size polymorphisms were easily visualized by gel electrophoresis. DNA for the high-throughput PCR analysis was extracted in a 96-tube format as follows: two young leaves were harvested from each plant into 1.2-mL tubes (Costar) containing two glass beads (4-mm diameter). After freezing the plates in liquid N2, the contents were ground by the beads in a shaker (Qiagen MM300) for 3 min at 30 vibrations/s. The ground tissue was then incubated for 1 h at 65°C in 400 μL of 2% (w/v) N-cetyl-N,N,N-trimethyl-ammonium bromide, 1.4 M NaCl, 20 mM EDTA, 100 mM Tris-HCl, pH 8.0, and 0.2% (v/v) β-mercaptoethanol prior to extraction with an equal volume of chloroform:isoamyl alcohol (24:1) and centrifugation at 3000g for 5 min. The aqueous phase was then transferred to new tubes and nucleic acids precipitated with 0.7 volumes of isopropanol, with centrifuging at 3000g for 30 min. DNA pellets were washed with 70% (v/v) ethanol, air-dried at 50°C for 30 min, and resuspended in 30 μL of 10 mM Tris-HCl and 1 mM EDTA, pH 8.0. Additional SSLP or single nucleotide polymorphism markers were then used on the identified recombinants to reduce the mapping interval. Sequencing of At MUM2 was performed using the Applied Biosystems DNA sequencing kit (Bigdye Terminator version 3.0) and ABI Prism 310 genetic analyzer.
Expression Analysis
Total RNA was prepared from various frozen plant tissues using a mammalian total RNA kit (Sigma-Aldrich) following the manufacturer's protocol and including an on-column DNase I treatment (RNase-free DNase set; Qiagen). Total RNA (2 μg) was used as a template to synthesize first-strand cDNA using an oligo(dT) 18-mer primer and the SuperScript first-strand synthesis kit (Invitrogen) according to manufacturer's instructions. For the amplification of PCR products from single-stranded complementary DNA in wild-type and mum2 mutants, the following exon primers were used: At MUM2 (exons 1 and 2), forward primer 5′-GAGTAACGTACGACGGAC-3′ and reverse primer 5′-CTGTCCAAGCTTAGGCTCA-3′; At MUM2 (exons 12 and 14), forward primer 5′-GGTTAGCTTAATCAATGGAC-3′ and reverse primer 5′-GGTTCTTAATTAGTCCAGCT-3′; EF1αA4 forward primer 5′-ATGCCCCAGGACATCGTGATTTCAT-3′and reverse primer 5′-TTGGCGGCACCCTTAGCTGGATCA-3′. PCR products were analyzed on a 3% (w/v) agarose gel.
Chromatographic Separation of β-Galactosidase Proteins from Plant Extracts and Activity Assays
Protein extracts were prepared from 2 g of Arabidopsis thaliana tissue and separated by cation-exchange chromatography as described by Minic et al. (2006). One-milliliter fractions were collected, and assays for β-d-galactosidase activity performed using 50-μL aliquots from each, except for the first 10 fractions, which contain pigments that absorb at 405 nm. Reactions were performed with 2 mM p-nitrophenol-β-d-galactopyranoside (Sigma-Aldrich) and 0.1 M Na-acetate, pH 5.0, in a total volume of 0.5 mL and incubating at 37°C for 10 min to 1 h, depending on the strength of staining intensity. Reactions were stopped by the addition of 0.5 mL of 0.4 M sodium bicarbonate, and the absorbance of hydrolyzed substrate determined spectrophotometrically at 405 nm.
In Situ Glycosyl Hydrolase Activity Assay
Seeds (10 mg) were placed in microtitre plate wells containing 50 mM sodium acetate, pH 5.0, 0.1% (v/v) Triton X-100, and 2.5 mM potassium ferricyanide/potassium ferrocyanide. Substrate, 5-bromo-4-chloro-3-indolyl β-d-galactoside (Quantum Biotechnologies), or 5-bromo-4-chloro-3-indolyl β-d-glucuronide (Duchefa) in DMSO was added to 1 mg/mL and a vacuum applied for 15 min. Samples were then incubated for 24 h at 37°C before removing the reaction mixture and replacing it with absolute ethanol to stop the reaction. Samples were observed with a SMZ 1000 binocular dissecting microscope (Nikon).
Mucilage Analysis
Mucilage was released from wild-type and mum2-11 mutant samples in 0.05 M HCl (200 mg of seed/5 mL) for 30 min at 85°C followed by addition of 5 mL of 0.3 M NaOH and incubation at room temperature for 10 min. Seeds were rinsed three times with 5 mL of water, acidified to pH 4.5 with HCl, and then rinsed with 5 mL of 50 mM sodium acetate buffer, pH 4.5 (three changes). The extraction supernatant and first two rinses were combined and retained for analysis after dialysis. Digestion with RGase (0.1 nkat), endo-PGase II (0.7 nkat), and cellulase (0.9 nkat) (Maxazyme; DSM) was performed on 200 mg of seed as described by Macquet et al. (2007). Uronic acid content was determined by the automated m-hydroxybiphenyl method (Thibault, 1979), and individual neutral sugars were analyzed as their alditol acetate derivatives (Blakeney et al., 1983) by gas–liquid chromatography after hydrolysis with 2 M trifluoroacetic acid at 121°C for 2.5 h.
HPAEC of Inner Mucilage Hydrolysates
HPAEC was performed on a Waters system with pulsed amperometric detection. Enzyme digest hydrolysates were desalted using a Sephadex G-10 column (100 × 1.6 cm) with a flow rate of 1 mL/min and eluting using deionized water. Desalted enzymatic hydrolysates were injected (20 μL) onto an analytical Carbopac PA-1 column (4 × 250 mm) equipped with a Carbopac PA-1 guard column (4 × 50 mm) at 1 mL/min, pH 13. The elution was performed with three linear gradient phases of (1) 200 to 350 mM sodium acetate containing 100 mM NaOH (0 to 20 min), (2) 350 to 600 mM sodium acetate in 100 mM NaOH (20 to 40 min), and (3) 600 to 700 mM sodium acetate in 100 mM NaOH (40 to 60 min). The column was regenerated by washing with 1 M sodium acetate containing 100 mM NaOH and then re-equilibrated with the starting buffer. Sugar beet (Beta vulgaris) modified hairy regions were prepared from sugar beet pulp as previously described (Bonnin et al., 2001).
MS of Inner Mucilage Hydrolysates
Electrospray ionization ion trap mass spectrometry experiments were performed using an LCQ Advantage ion trap mass spectrometer (ThermoFinnigan) with ionization by negative electrospray. Samples were diluted to obtain a final concentration of 20 to 50 μg/mL in methanol/water 1:1 (v/v). Infusion was performed at a flow rate of 2.5 μL/min using nitrogen as a sheath gas. The MS analyses were performed under automatic gain control conditions, using a typical needle voltage of 3.8 kV and a heated capillary temperature of 185°C. More than 50 scans were summed for MS spectra acquisition.
Scanning Electron Microscopy
Dry seeds were mounted directly, whereas imbibed seeds were air-dried in a Petri dish prior to use. Samples were coated with gold in a Cryotrans CT 1500 sputter coater and observed using a Philips 525M scanning electron microscope (FEI) with an acceleration voltage of 10 kV. Images were obtained with Link ISIS 3.35 (Oxford Instruments).
Paraffin or Resin Embedding and in Situ Hybridization
To obtain silique samples, flowers were tagged on the day of flowering and sampled at the specified number of days afterwards. For sections, tissues were fixed with 4% (w/v) paraformaldehyde, 0.1% (v/v) Triton X-100, and 0.5 M sodium phosphate buffer, pH 7.0, under vacuum for 1 h at room temperature or 4°C and then overnight at 4°C. Samples were then rinsed with water and dehydrated in ethanol. The samples were then incubated in Histo-Clear II (National Diagnostics) before embedding in paraffin (Paraplast plus; Sherwood Medical) as described by Jackson (1991) or infiltrated with resin using the Technovit 7100 kit (Histo-Technik) according to manufacturer's instructions using a series of graded resin/ethanol solutions. Tissue sections (6 to 8 μm) were cut, placed on slides (Dako), and where required paraffin removed from samples with Histo-Clear followed by rehydration of samples. Sections were either used directly for histochemical staining or were pretreated as described by Jackson (1991) with modifications (Lefebvre et al., 2006). For probe preparation, cDNA fragments were amplified from single-stranded complementary DNA that had been reverse transcribed from wild-type RNA extracted from siliques as described above. One primer sequence in each PCR contained the T7 origin of transcription; for the sense probe, amplification was performed with forward primer 5′-GTAATACGACTCACTATAGGTCTTTTAAGACAGCCT-3′ and reverse primer 5′-GCTAGTGCATTGTTGACG-3′ and for the antisense probe with forward primer 5′-GGTCTTTTAAGACAGCCT-3′ and reverse primer 5′-GTAATACGACTCACTATAGCTAGTGCATTGTTGACG-3′. Transcription was performed using Riboprobe combination system SP6-T7 RNA polymerase (Promega), followed by hybridization and immunological detection as described by Lefebvre et al. (2006), except that hybridization was performed at 43°C.
Cytochemical Staining and Immunolabeling Procedures
Mucilage released from mature seeds was stained with 200 μg mL−1 ruthenium red or 25 μg mL−1 Calcofluor (fluorescent brightener 28; Sigma-Aldrich) essentially as described by Willats et al. (2001). Ruthenium red staining was observed with a light microscope (Axioplan 2; Zeiss) and Calcofluor staining with an inverted Leica TCS-SP2-AOBS spectral confocal laser microscope (Leica Microsystems) equipped with a 405-nm laser diode for excitation. Developing seed sections were stained with 0.1% (w/v) toluidine blue. Four primary monoclonal antibodies were used for immunolabeling; JIM5, JIM7, LM5, and LM6 (Plant Probes), which bind low-ester HG, high-ester HG, (1→4)-β-d-galactan, or (1→5)-α-l-arabinan, respectively (Knox et al., 1990; Jones et al., 1997; Willats et al., 2001). Intact mature seeds were treated to release mucilage as described above for mucilage analysis, before immunolabeling and observation with an inverted Leica TCS-SP2-AOBS spectral confocal laser microscope (Leica Microsystems), as described by Macquet et al. (2007). Light generated by autofluorescence is represented by red or yellow in confocal microscopy images.
Construction and Analysis of the At MUM2 Promoter:GUS Fusion
A 2-kb promoter fragment was amplified from wild-type Col-0 genomic DNA by PCR using Phusion high-fidelity DNA polymerase (Ozyme) and the following primers containing recombination sequences, forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGCTTCTCCGCAGCGGATTTC-3′ and reverse 5′-GGGGACCACTTTGTACAAGAAGCTGGGTCCTTTTCTTCTCCTTCTCTTC-3′. The resulting PCR product was then recombined into the pDONR207 vector (Invitrogen) using BP clonase according to manufacturer's instructions and transformed into Escherichia coli strain DH10B. The At MUM2 promoter fragment was then sequenced and compared with database sequence to confirm no PCR-induced errors were present before recombination into the binary vector pBI101-R1R2-GUS (F. Divol, J.C. Palauqui, and B. Dubreucq, unpublished data) using LR clonase (Invitrogen) according to the manufacturer's instructions. The resulting plasmid was then transformed into Agrobacterium tumefaciens C58C1pMP90 by electroporation. Stable transformation of wild-type Col-0 was then performed using the floral dip method of Clough and Bent (1998) and selecting kanamycin-resistant transformants. Histochemical detection of GUS expression was performed as described by Jefferson et al. (1987) on various plant tissues using 1 or 2.5 mM potassium ferricyanide/potassium ferrocyanide. Similar results were obtained with at least five independent transformants. Whole-mount tissues were photographed using either a SMZ 1000 binocular dissecting microscope (Nikon) or a light microscope (Axioplan 2). Tissue sections (8 μm) were cut from paraffin-fixed tissue and observed by light microscopy as described above.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession number At5g63800 (MUM2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Arabidopsis Seed Mucilage Contains a β-d-Galactosidase That Affects Mucilage Swelling Properties.
Supplemental Figure 2. The Hexose Unit Is Directly Linked to That of Rhamnose in Several Oligomers Present in mum2-11 Mutant Hydrolysates Obtained from RGase Digestion of Alkali- and Acid-Treated Seeds.
Supplemental Figure 3. Analysis of RGase Digestion Products from the Arabidopsis Seed Inner Mucilage Layer or Sugar Beet Modified Hairy Regions by High-Performance Anion Exchange Chromatography.
Supplemental Data Set 1. Amino Acid Alignment Presented in Figure 3.
Supplementary Material
Acknowledgments
This work was supported by a doctoral fellowship from the French Ministry of Education and Research to A.M. and an Action Concertée Incitative grant from the French Ministry of Research. We thank Michaël Anjuère for technical assistance with plant culture, Matthieu Simon for preparing seed stocks from the Versailles Natural Variation collection, Fabrice Petitpas for technical aid, and Zoran Minic for technical advice on β-d-galactosidase purification and activity assay. We are also grateful for the expertise and guidance of Nelly Wolff with scanning electron microscopy and Lionel Gissot and Olivier Grandjean with confocal microscopy, and we are indebted to Isabelle Debeaujon for stimulating discussions and advice. The Leica TCS-SP2-AOBS spectral confocal laser microscope was cofinanced by grants from Institut National de la Recherche Agronomique and the Ile-de-France region, and MS analyses were performed within the RIO platform “Biopolymers-Interaction-Structural Biology” located at Institut National de la Recherche Agronomique, Nantes (http://www.nantes.inra.fr/plateformes_et_plateaux_techniques/plateforme_bibs).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Helen M. North (helen.north@versailles.inra.fr).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Beeckman, T., De Rycke, R., Viane, R., and Inzé, D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113 139–148. [Google Scholar]
- Blakeney, A.B., Harris, P.J., Henry, R.J., and Stone, B.A. (1983). A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr. Res. 113 291–299. [Google Scholar]
- Bonnin, E., Brunel, M., Gouy, Y., Lesage-Meessen, L., Asther, M., and Thibault, J.-F. (2001). Aspergillus niger I-1472 and Pycnoporous cinnabarinus MUCL39533, selected for the biotransformation of ferulic acid to vanillin, are also able to produce cell wall polysaccharide-degrading enzymes and feruloyl esterases. Enzyme Microb. Technol. 28 70–80. [DOI] [PubMed] [Google Scholar]
- Cancilla, M.T., Penn, S.G., and Lebrilla, C.B. (1998). Alkaline degradation of oligosaccharides coupled with matrix-assisted laser desorption/ionization Fourier transform mass spectrometry: A method for sequencing oligosaccharides. Anal. Chem. 70 663–672. [DOI] [PubMed] [Google Scholar]
- Carey, A.T., Holt, K., Picard, S., Wilde, R., Tucker, G.A., Bird, C.R., Shuch, W., and Seymour, G.B. (1995). Tomato exo-(1→4)–β-D-galactanase. Isolation, changes during ripening in normal and mutant tomato fruit and characterisation of a related cDNA clone. Plant Physiol. 108 1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho, P.M., and Henrissat, B. (1999). Carbohydrate-active enzymes: An integrated database approach. In Recent Advances in Carbohydrate Bioengineering. H.J. Gilbert, G. Davies, B. Henrissat, and B. Svensson, eds (Cambridge, UK: The Royal Society of Chemistry), pp. 3–12.
- Dean, G.H., Zheng, H., Tewari, J., Young, D.S., Hwang, Y.T., Western, T.L., Carpita, N.C., McCann, M.C., Mansfield, S.D., and Haughn, G.W. (2007). The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19 4007–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veau, E.J.I., Gross, K.C., Huber, D.J., and Watada, A.E. (1993). Degradation and solubilization of pectin by β-galactosidases purified from avocado mesocarp. Physiol. Plant. 87 279–285. [Google Scholar]
- Dey, P.M., and Del Campillo, E. (1984). Biochemistry of the multiple forms of glycosidases in plants. Adv. Enzymol. Relat. Areas Mol. Biol. 56 141–249. [DOI] [PubMed] [Google Scholar]
- Gorenflot, R. (1986). Primary structure in Angiosperms. In Plant Biology – Higher Plants, Vol. 1, S.A. Masson, ed (Paris: Masson), pp. 93–101.
- Gutterman, Y., and Shem-Tov, S. (1996). Structure and function of the mucilaginous seed coats of Plantago coronopus inhabiting the Negev Desert of Israel. Isr. J. Plant Sci. 44 125–133. [Google Scholar]
- Iglesias, N., Abelenda, J.A., Roniño, M., Sampedro, J., Revilla, G., and Zarra, I. (2006). Apoplastic glycosidases active against xyloglucan oligosaccharides of Arabidopsis thaliana. Plant Cell Physiol. 47 55–63. [DOI] [PubMed] [Google Scholar]
- Jackson, D.P. (1991). In-situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and M. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku, K.D., du Boer, B.G.W., Van Montagu, M., and Okamro, J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C.S., Kolevski, B., and Smyth, D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Seymour, G.B., and Knox, J.P. (1997). Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-D-galactan. Plant Physiol. 113 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox, J.P., Linstead, P.J., King, J., Cooper, C., and Roberts, K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues or root apices. Planta 181 512–521. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. (1981). The complex syndrome of the ttg mutants. Arabidopsis Info Serv. 18 45–51. [Google Scholar]
- Koornneef, M., Dellaert, L.W.M., and Vanderveen, J.H. (1982). EMS-induced and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L) heynh. Mutat. Res. 93 109–123. [DOI] [PubMed] [Google Scholar]
- Kotake, T., Dina, S., Konishi, T., Kaneko, S., Igarashi, K., Samejima, M., Watanabe, Y., Kimura, K., and Tsumuraya, Y. (2005). Molecular cloning of a β-galactosidase from radish that specifically hydrolyzes β-(1→3)- and β-(1→6)-galactosyl residues of arabinogalactan protein. Plant Physiol. 138 1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., Nambara, E., and Marion-Poll, A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45 309–319. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel, K.M., Keijer, C.J., and Koornneef, M. (1994). A seed shape mutant of Arabidopsis that is affected in integument development. Plant Cell 6 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet, O., Chaillou, S., Camilleri, C., Bouchez, D., and Daniel-Vedele, F. (2002). Bay-0 x Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor. Appl. Genet. 104 1173–1184. [DOI] [PubMed] [Google Scholar]
- Macquet, A., Ralet, M.C., Kronenberger, J., Marion-Poll, A., and North, H.M. (2007). In-situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 48 984–999. [DOI] [PubMed] [Google Scholar]
- McAbee, J.M., Hill, T.A., Skinner, D.J., Izhaki, A., Hauser, B.A., Meister, R.J., Reddy, G.V., Meyerowitz, E.M., Bowman, J.L., and Gasser, C.S. (2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 46 522–531. [DOI] [PubMed] [Google Scholar]
- Minic, Z., Do, C.-T., Rihouey, C., Morin, H., Lerouge, P., and Jouanin, L. (2006). Purification, functional characterization, cloning, and identification of mutants of a seed-specific arabinan hydrolase in Arabidopsis. J. Exp. Bot. 57 2339–2351. [DOI] [PubMed] [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for the expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, T., Nemoto, T., and Jigami, Y. (2007). Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J. Biol. Chem. 282 5389–5403. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Meissner, R.C., Shoue, D.A., Carpita, N.C., and Bevan, M.W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéméner, B., Cabrera Pino, J.C., Ralet, M.-C., Bonnin, E., and Thibault, J.-F. (2003. a). Assignment of acetyl groups to O-2 and/or O-3 of pectic oligogalacturonides using negative electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 38 641–648. [DOI] [PubMed] [Google Scholar]
- Quéméner, B., Désiré, C., Lahaye, M., Debrauwer, L., and Negroni, L. (2003. b). Structural characterisation by both positive- and negative- ion electrospray mass spectrometry of partially methyl-esterified oligogalacturonides purified by semi-preparative high-performance anion-exchange chromatography. Eur. J. Mass Spectrom. (Chichester, Eng.) 9 45–60. [DOI] [PubMed] [Google Scholar]
- Ralet, M.-C., Cabrera, J.C., Bonnin, E., Quéméner, B., Hellìn, P., and Thibault, J.-F. (2005). Mapping sugar beet pectin acetylation pattern. Phytochemistry 66 1832–1843. [DOI] [PubMed] [Google Scholar]
- Renard, C.M.G.C., Lahaye, M., Mutter, M., Voragen, A.G.J., and Thibault, J.-F. (1998). Isolation and structural characterisation of rhamnogalacturonan oligomers generated by controlled acid hydrolysis of sugar-beet pulp. Carbohydr. Res. 305 271–280. [DOI] [PubMed] [Google Scholar]
- Rerie, W.G., Feldmann, K.A., and Marks, M.D. (1994). The GLABRA2 gene encodes a homeodomain protein required for normal trichome development in Arabidopsis. Genes Dev. 8 1388–1399. [DOI] [PubMed] [Google Scholar]
- Schols, H.A., Vierhuis, E., Bakx, E.J., and Voragen, A.G.J. (1995). Different populations of pectic hairy regions occur in apple cell walls. Carbohydr. Res. 275 343–360. [DOI] [PubMed] [Google Scholar]
- Sørensen, S.O., Pauly, M., Bush, M., Skjøt, M., McCann, M.C., Borkhardt, B., and Ulvskov, P. (2000). Pectin engineering: modification of potato pectin by in vivo expression of an endo-1,4-β-D-galactanase. Proc. Natl. Acad. Sci. USA 97 7639–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.L., Abbott, J.A., and Gross, K.C. (2002). Down regulation of tomato β-galactosidase 4 results in decreased fruit softening. Plant Physiol. 129 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.L., Starrett, D.A., and Gross, K.C. (1998). A gene coding for tomato fruit β-galactosidase II is expressed during fruit ripening. Plant Physiol. 117 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, J.-F. (1979). Automatisation du dosage des substances pectiques par la méthode au métahydroxydiphényle. Lebensm. Wiss. Technol. 12 247–251. [Google Scholar]
- Usadel, B., Kuschinsky, A.M., Rosso, M.G., Eckermann, N., and Pauly, M. (2004). RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol. 134 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Burn, J., Tan, W.L., Skinner, D.J., Martin-McCaffrey, L., Moffatt, B.A., and Haughn, G.W. (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 127 998–1011. [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Skinner, D.J., and Haughn, G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Young, D.S., Dean, G.H., Tan, W.L., Samuels, A.L., and Haughn, G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor, J.B., Symonds, V.V., Mendenhall, J., and Lloyd, A.M. (2000). Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J. 22 483–493. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., Limberg, G., Buchholt, H.C., van Alebeek, G.-J., Benen, J., Christensen, T.M., Visser, J., Voragen, A., Mikkelsen, J.D., Knox, J.P. (2000). Analysis of pectic epitopes recognised by hybridoma and phage display monoclonal antibodies using defined oligosaccharides, polysaccharides and enzymatic degradation. Carbohydr. Res. 327 309–320. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., Marcus, S.E., and Knox, J.P. (1998). Generation of a monoclonal antibody specific to (1→5)-α-l-arabinan. Carbohydr. Res. 308 149–152. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., McCartney, L., and Knox, J.P. (2001). In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213 37–44. [DOI] [PubMed] [Google Scholar]
- Witztum, A., Gutterman, Y., and Evenari, M. (1969). Integumentary mucilage as an oxygen barrier during germination of Blepharis persica (Burm) Kuntze. Bot. Gaz. 130 238–241. [Google Scholar]
- Young, J.A., and Evans, R.A. (1973). Mucilaginous seed coats. Weed Sci. 21 52–54. [Google Scholar]
- Zhang, F., Gonzalez, A., Zhao, M., Payne, C.T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in a TTG1-dependent pathways of Arabidopsis. Development 130 4859–4869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.