Abstract
Reactive oxygen species (ROS) produced by NADPH oxidase play critical roles in various cellular activities, including plant innate immunity response. In contrast with the large multiprotein NADPH oxidase complex of phagocytes, in plants, only the homologs of the catalytic subunit gp91phox and the cytosolic regulator small GTPase Rac are found. Plant homologs of the gp91phox subunit are known as Rboh (for respiratory burst oxidase homolog). Although numerous Rboh have been isolated in plants, the regulation of enzymatic activity remains unknown. All rboh genes identified to date possess a conserved N-terminal extension that contains two Ca2+ binding EF-hand motifs. Previously, we ascertained that a small GTPase Rac (Os Rac1) enhanced pathogen-associated molecular pattern–induced ROS production and resistance to pathogens in rice (Oryza sativa). In this study, using yeast two-hybrid assay, we found that interaction between Rac GTPases and the N-terminal extension is ubiquitous and that a substantial part of the N-terminal region of Rboh, including the two EF-hand motifs, is required for the interaction. The direct Rac–Rboh interaction was supported by further studies using in vitro pull-down assay, a nuclear magnetic resonance titration experiment, and in vivo fluorescence resonance energy transfer (FRET) microscopy. The FRET analysis also suggests that cytosolic Ca2+ concentration may regulate Rac–Rboh interaction in a dynamic manner. Furthermore, transient coexpression of Os Rac1 and rbohB enhanced ROS production in Nicotiana benthamiana, suggesting that direct Rac–Rboh interaction may activate NADPH oxidase activity in plants. Taken together, the results suggest that cytosolic Ca2+ concentration may modulate NADPH oxidase activity by regulating the interaction between Rac GTPase and Rboh.
INTRODUCTION
Reactive oxygen species (ROS) produced by NADPH oxidase have been shown to play many important roles in signaling and development in plants, such as plant defense response, cell death, abiotic stress, stomatal closure, and root hair development (Baxter-Burrell et al., 2002; Torres et al., 2002; Foreman et al., 2003; Kwak et al., 2003; Yoshioka et al., 2003; Jones et al., 2007). Plant NADPH oxidase genes, termed rboh (for respiratory burst oxidase homolog), encoding homologs of the mammalian NADPH oxidase catalytic subunit gp91phox, have been isolated from many plants species, including rice (Oryza sativa), Arabidopsis thaliana, tobacco (Nicotiana benthamiana), and potato (Solanum tuberosum) (Groom et al., 1996; Keller et al., 1998; Torres et al., 1998; Yoshioka et al., 2001; Yoshioka et al., 2003; Sagi and Fluhr, 2006). In phagocytes, the NADPH oxidase forms a multiprotein complex consisting of gp91phox, p22phox, p47phox, p67phox, p40phox, and the small GTPase Rac2 (Babior, 2004). By contrast, from Arabidopsis and rice genome sequencing, with the exception of rboh and Rac (also known as Rop) (Gu et al., 2004), plants do not possess the homologs of other subunits of the mammalian NADPH oxidase complex (Torres and Dangl, 2005). Furthermore, unlike the mammalian gp91phox, plant Rboh proteins possess an extended N terminus, which contains two Ca2+ binding EF-hand motifs. Recently, it has been shown that the proteins Nox5, Duox1, and Duox2, which are nonphagocytic NADPH oxidase (Nox) proteins, all possess an extended N terminus, which contains EF-hand motifs (Geiszt and Leto, 2004; Lambeth, 2004; Torres and Dangl, 2005). However, little is known about the regulation of these newly discovered animal Nox proteins and the function of their N-terminal EF-hand motifs.
In Arabidopsis, 10 rboh genes are known, and among these, rbohD are rbohF are shown to be involved in ROS production during pathogen infection (Torres et al., 2002, 2006). These two rboh genes are reported to function in abscisic acid–induced ROS generation required for the stomatal closure in guard cells (Kwak et al., 2003). During root hair development an rboh gene, rbohC, also termed RDH2, is responsible for ROS production required for root hair elongation (Foreman et al., 2003). In Nicotiana benthamiana, virus-induced gene silencing of rbohA and rbohB reduced ROS production and resistance to infection by Phytophthora infestans (Yoshioka et al., 2003). Furthermore, ROS may interact with nitric oxide in triggering the hypersensitive reaction, a form of programmed cell death that effectively restricts pathogen growth (Delledonne et al., 2001; Zaninotto et al., 2006). Suppression of tomato rboh gene expression by the antisense approach reduced ROS production in leaf and caused abnormal morphologies in leaves and flowers (Sagi et al., 2004). Despite the accumulating evidence for the involvement of Rboh in ROS production in various aspects of signaling and development in plants (Apel and Hirt, 2004, Gapper and Dolan, 2006), the regulation of plant Rboh remains unclear (Apel and Hirt, 2004; Torres and Dangl, 2005). In the absence of other homologs of the mammalian Nox subunits, the small GTPase Rac/Rop becomes a prime candidate for being a regulator of plant NADPH oxidase.
In rice, a constitutively active (CA) form of Rac1 has been shown to increase ROS production, and a dominant-negative (DN) form of Rac1 causes reduction of ROS levels (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002). DN-OsRac1 was recently shown to decrease ROS production and tobacco mosaic virus resistance in Nicotiana tabacum plants carrying the N resistance gene (Moeder et al., 2005). Regulation of ROS levels by transient expression of mutated cotton (Gossypium hirsutum) Rac genes in soybean (Glycine max) and Arabidopsis cultured cells has been demonstrated (Potikha et al., 1999). In an in vitro study using Arabidopsis cell extracts, the GTP-bound Rop2 protein increased ROS production, while the GDP-bound form decreased it, suggesting a direct role of Rop GTPase in ROS production (Park et al., 2004). In Arabidopsis, Jones et al. (2002) showed that Rop2 GTPase also is a regulator of root hair development, suggesting that it may regulate Rboh activity in root hair development. Interestingly, a negative regulator of Rho GTPase, Rho GTPase GDP dissociation inhibitor, is involved in focusing AtrbohC-mediated ROS production in the regulation of lateral hair development (Carol et al., 2005). Furthermore, analysis of another negative regulator of Rop, Rop GTPase activating protein 4, also indicates that Rop is involved in regulation of ROS production in Arabidopsis response to oxygen deprivation (Baxter-Burrell et al., 2002).
However, direct evidence showing a causal relationship between Rac/Rop and Rboh in the regulation of ROS production is lacking at present. To complicate the matter, apparently, in phagocyte, Rac2 activates gp91phox through p67phox, which acts as an adaptor in between the two proteins (Babior, 2004). However, the homolog of p67phox is not found in plants. In numerous studies involving ROS signaling in plants, the participation of Ca2+ has also been implicated (Jabs et al., 1997; Blume et al., 2000; Pei et al., 2000; Baxter-Burrell et al., 2002; Foreman et al., 2003; Kwak et al., 2003). How Ca2+ signaling integrates into Rac/Rop and ROS signaling to modulate plant defense response and development remains unknown.
To address this question, in this study, we tested whether Rac GTPase of rice can directly bind with the N-terminal region of Rboh proteins using yeast two-hybrid, in vitro pull-down assay, a nuclear magnetic resonance (NMR) titration experiment, and in vivo fluorescence resonance energy transfer (FRET) microscopy. The biological significance of Rac–Rboh interaction was shown by an agroinfiltration assay. Results of these studies indicate that direct Rac–Rboh interaction is important for activating NADPH oxidase activity. Furthermore, our studies also suggest how Rac/Rop and Ca2+ may interact with the NADPH oxidase to modulate ROS production.
RESULTS
Rice rboh Gene Family
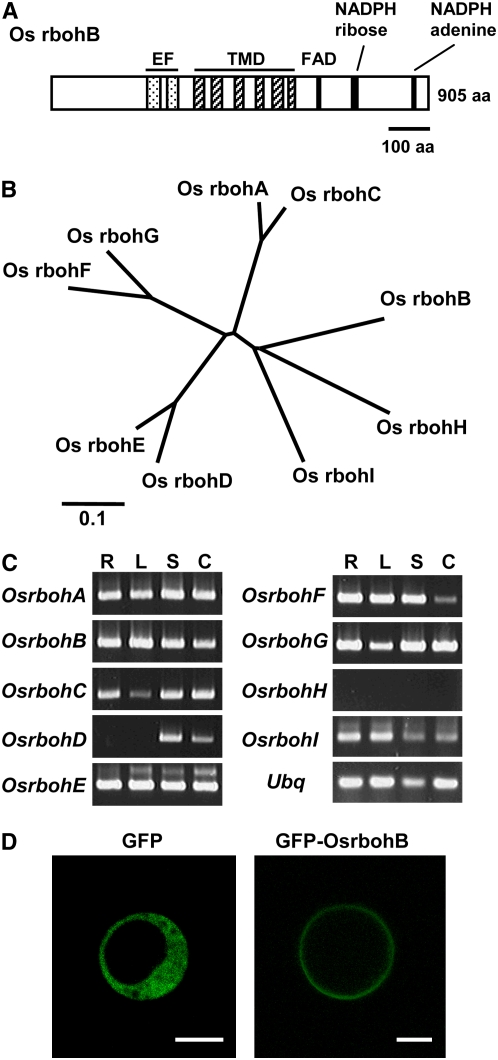
Rice rbohA was the first rboh gene isolated from a plant (Groom et al., 1996). Subsequently, rboh genes have been isolated from numerous plant species. The structure of all rice rboh genes is highly conserved, and all Rbohs have a long N-terminal extension containing two EF-hand motifs (Figure 1A) (Torres and Dangl, 2005). These EF-hand motifs were shown to bind Ca2+ in vitro (Keller et al., 1998). We searched the rice genome database for members of the rboh gene family and identified nine genes, including rbohA, which has been reported previously (Keller et al., 1998) (Figures 1A and 1B). All Os rboh genes, with the exception of Os rbohD and rbohH, are constitutively expressed in each of the three organs analyzed (Figure 1C). Transient expression of a green fluorescent protein (GFP)-OsrbohB fusion gene in rice protoplasts clearly showed that Os rbohB protein is localized to the plasma membrane of rice cells (Figure 1D).
Figure 1.
Rboh Proteins of Rice.
(A) Schematic representation of Os rbohB, showing EF-hand motifs (EF), six transmembrane domains (TMD), and FAD- and NADPH binding regions. aa, amino acids.
(B) An unrooted phylogenetic tree of rice Rbohs analyzed by ClustalW.
(C) Tissue-specific expression of rice rboh. RT-PCR was performed with 25 cycles for Ubiquitin (Ubq) and 30 cycles for all Os rboh, except for Os rbohH, where 35 cycles were performed. R, root; L, leaf; S, shoot; C, calli.
(D) Localization of GFP-OsrbohB to the plasma membrane of rice protoplast. Bars = 10 μm.
Interaction of Os Rac and Rboh in Yeast Two-Hybrid Assays
Although numerous studies suggested that Rac/Rop GTPases regulate NADPH oxidase activity in various plant systems, direct evidence of the regulatory function is still lacking. The observations that the Arabidopsis and rice genomes do not contain any gene homologous to the cytosolic components of the phagocyte NADPH oxidase and that plant Rboh proteins contain a long N-terminal extension in the cytoplasm prompted us to examine whether these N-terminal extensions may directly bind with Rac GTPase. Previously, we demonstrated that a rice Rac GTPase, Os Rac1, regulates ROS levels in transformed rice cells and plants (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002).
To understand how Rac/Rop GTPases regulate NADPH oxidase activity in plants, we performed yeast two-hybrid assays to test the possibility that Os Rac proteins interact with the N-terminal region of plant Rboh proteins (Figure 2). For these experiments, we chose four rice rboh genes, rbohA, rbohB, rbohC, and rbohD, and two potato rboh genes, St rbohA and rbohB (Yoshioka et al., 2001). We cloned the sequences corresponding to the N-terminal region of these Rboh proteins, which encode the cytosolic domains that contain EF-hand motifs. To look for possible interaction between the N-terminal region of the Rboh proteins with Rac proteins, we cloned each of the seven members of the Os Rac gene family (Kawasaki et al., 1999; Miki et al., 2005) and examined the encoded proteins for interaction with the N-terminal region of the six Rboh proteins.
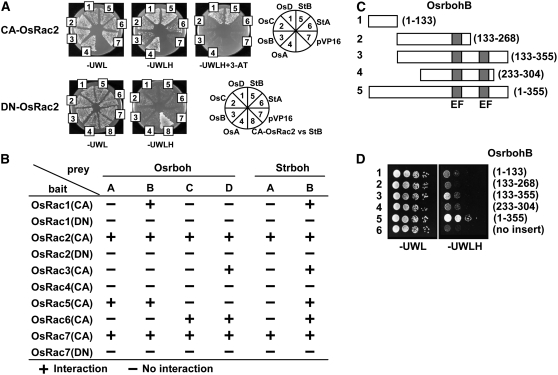
Figure 2.
Yeast Two-Hybrid Analysis of Os Rac and the N-Terminal Region of Rbohs.
(A) Representative plates showing interaction between Os Rac and rboh by yeast two-hybrid assay. CA-OsRac2 interacts with various Rboh but not DN-OsRac2. OsA, OsB, OsC, OsD, StB, StD, and pVP16 denote Os rbohA, Os rbohB, Os rbohC, Os rbohD, St rbohB, St rbohD, and empty prey vector, respectively. Growth on selective plates without His (−H) or without His (−H) plus 3-aminotriazole (3-AT) indicates a positive interaction.
(B) Interaction of rice Racs with N-terminal region of rice and potato Rbohs.
(C) Schematic representation of Os rbohB showing the size and position of each fragment of the N-terminal region of Os rbohB used for the assays.
(D) Interaction of various fragments of the N-terminal region of Os rbohB with Os Rac2.
CA forms of all seven Os Rac genes and DN forms of Os Rac1, Rac2, and Rac7 were tested. Representative results of yeast two-hybrid assays between Os Rac2 and four Os rboh and two St rboh genes are shown in Figure 2A and Supplemental Figure 1 online. Typically, CA-OsRac2 and CA-OsRac7 were able to bind with most of the rice Rboh proteins and the two potato Rboh proteins (Figures 2A and 2B). Results of the yeast two-hybrid assays indicate that CA-OsRac proteins, except for Os Rac4, which did not interact with any of the Rboh examined, interacted with two to six of the Rboh proteins examined (Figure 2B). Importantly, the DN form of Os Rac1, Rac2, and Rac7 did not interact with any of the six plant Rboh proteins tested (Figures 2A and 2B). Interestingly, St rbohB interacted with all Os Rac proteins tested except for Os Rac4, and OsRac2 interacted with all Rboh proteins tested. Each of the six plant Rboh proteins interacted with at least two Os Rac proteins (Figure 2B). Furthermore, the interaction is specific for the CA-OsRac proteins, suggesting that Rboh proteins are novel effectors of Rac proteins in plants.
To further analyze regions of the N-terminal extension that interact with Os Rac proteins, five fragments covering various regions of the N-terminal extension of Os rbohB, Os rbohD, and St rbohB were examined for their ability to interact with Os Rac2 (Figures 2C and 2D; see Supplemental Figure 1 online). For St rbohB, amino acids 104 to 320, which contain two EF-hand motifs and some additional amino acids in both the N- and C-terminal directions, could interact with Os Rac2 (see Supplemental Figure 1 online). However, for Os rbohB and Os rbohD, smaller fragments containing the two EF-hand motifs, showed interaction with OsRac2, albeit weakly (Figures 2C and 2D; see Supplemental Figure 1 online). Together, these experiments indicate that a large portion of the N-terminal extension, including the two EF-hand motifs, is required for the interaction with Os Rac GTPase.
Interaction of Os Rac1 and Os rbohB in Vitro
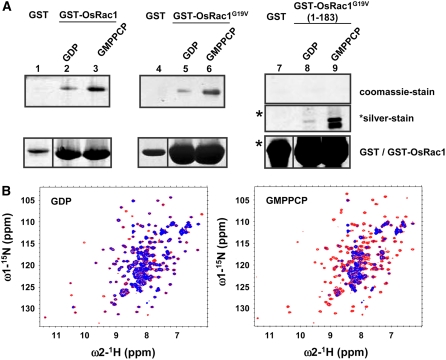
To confirm the interaction of Os Rac1 with the N-terminal region of Os rbohB proteins, recombinant proteins were produced in Escherichia coli and purified proteins, and glutathione S-transferase (GST) pull-down assay and an NMR titration experiment were performed. The N-terminal region of Os rbohB (residues 1 to 355) consists of two major protease-resistant fragments (see Supplemental Figure 2 online). One of these protease-resistant fragments, which is located at the C terminus and possesses two EF-hand motifs (residues 138 to 313), was used for the binding experiments. Although the equivalent Os rbohB fragment (133 to 355) (Figures 2C and 2D) used in the yeast two-hybrid assay showed weak interaction, this Os rbohB fragment (133 to 355) purified from E. coli showed the best stability and reproducibility compared with other Os rbohB fragments, 1 to 355, 135 to 335, 135 to 330, 138 to 316, 138 to 313, and 138 to 311 (data not shown); therefore, it was used in subsequent in vitro assays. As shown in Figure 3A, binding of Os Rac1 and the CA form of Os Rac1 (Os Rac1G19V) was observed with Os rbohB (138 to 313). Binding of Os rbohB (138 to 313) to the CA form of Os Rac1 (lacking 31 C-terminal residues), Os Rac1G19V (1 to 183), was also detected by silver staining but not by Coomassie blue staining. Because silver staining is much more sensitive than Coomassie blue staining, the results suggest that the binding of full-length Os Rac1 is much stronger than the C-terminal deletion mutant. Interestingly, Os Rac1 showed stronger binding to Os rbohB (138 to 313) in the presence of GMPPCP, a nonhydrolyzable analog of GTP that mimics GTP binding. This binding preference of Os Rac1 is consistent with the NMR data, as shown in Figure 3B.
Figure 3.
Interaction of Os Rac1 with Os rbohB (138 to 313) in Vitro.
(A) GST pull-down assay. Os rbohB (138 to 313) was incubated with the GDP-bound form of GST-OsRac1 (lane 2), the GMPPCP-bound form of GST-OsRac1 (lane 3), the GDP-bound form of GST-OsRac1G19V (lane 5), the GMPPCP-bound form of GST-OsRac1G19V (lane 6), the GDP-bound form of GST-OsRac1G19V (1 to 183) (lane 8), the GMPPCP-bound form of GST-OsRac1 (lane 9), or GST alone (lanes 1, 4, and 7). Bound proteins were analyzed by SDS-PAGE and stained with Coomassie blue (lanes 1 to 9; top panel) or silver (lanes 7 to 9; middle panel). In the bottom panel, the protein input lanes 1 to 6 and lanes 7 to 9 (as indicated by asterisks) were stained with Coomassie and silver, respectively.
(B) Overlay view of Os Rac1 1H-15N HSQC spectra in the presence of Os rbohB (138 to 313) at a molar ratio of 1:2 (15N-labeled Os Rac1/rbohB (138 to 313) (blue) or in the absence of Os rbohB (138 to 313) (red) for GDP-bound (left) and GMPPCP-bound forms (right) of Os Rac1.
In NMR spectroscopy, the 1H-15N HSQC (heteronuclear single quantum coherence) experiment is very useful for studying protein–protein interaction by comparing the two-dimensional spectrum recorded with or without the binding partner. On the 1H-15N HSQC spectrum shown in Figure 3B, each dot represents the NMR peak from the NH group of each amino acid, which possesses the corresponding 1H and 15N chemical shift values. Intermolecular interaction is monitored by the peak broadening and/or position shift of each peak of the HSQC spectrum. In the case of the GDP-bound form, the 1H-15N HSQC spectrum of the 15N- labeled Os Rac1 did not change in the presence of Os rbohB. By contrast, significant signal broadenings induced by direct Os Rac1–rbohB (138 to 313) interaction were observed for the GMPPCP-bound form. The Kd values were not determined explicitly due to signal broadenings but were estimated to be in the range >10−3 M and 10−4 ∼ 10−5 M for the GDP- and GMPPCP-bound forms, respectively, because the GDP-bound form generated very small spectral changes at the molecular ratio of 1:1 and protein concentration of 0.14 mM, and the GMPPCP-bound form generated large spectral changes, which saturated at the molecular ratio of 1:2. The binding interface was not determined because Os Rac1 was not sufficiently stable for peak assignment at 303K. These data indicate that the N-terminal region of Os rbohB (residues 138 to 313), which contains two EF-hand motifs, binds the GMPPCP-bound form of Os Rac1 preferentially and that the hypervariable C-terminal region of Os Rac1 also contributes to Os Rac1–rbohB interaction.
FRET Analysis of Os Rac1–rbohB Interaction in Rice Protoplasts
To investigate the interaction of Os Rac and Rboh proteins in vivo, we employed FRET technology. FRET analysis has been increasingly used for studies in protein–protein interactions in vivo (Miyawaki et al., 1997; Jares-Erijman and Jovin, 2003; Vogel et al., 2006), including plant–pathogen interaction (Bhat et al., 2005). In this study, we modified an intramolecular FRET system called Raichu (Ras and interacting protein chimeric unit), originally developed as biosensors for studying the activation of various small GTPases in mammalian cells (Mochizuki et al., 2001; Itoh et al., 2002), for use in rice cells. In this system, the donor and acceptor fluorophores and the two interacting proteins reside in the same molecule; therefore, their molar ratio of the individual unit is the same irrespective of expression level. Thus, this reduces error caused by difference in the levels of donor and acceptor fluorophores. The constructs used were essentially identical to those used by Itoh et al. (2002), except that the carboxyl end and the polybasic region of human Rac in the original Raichu vector (Itoh et al., 2002) were replaced by the 30–amino acid polybasic C-terminal region of Os Rac1. In addition, the human PAK and the Rac sequences used in Raichu were replaced with the N-terminal region of Os rbohB (1 to 355) and Os Rac1 (1 to 181) sequences, respectively. The polybasic C-terminal region of Os Rac1 efficiently targeted the chimeric protein to the plasma membrane in the rice protoplast (Figure 4B). The core construct was fused with a maize (Zea mays) ubiquitin promoter, which has been shown to be highly active in rice cells (Wong et al., 2004).
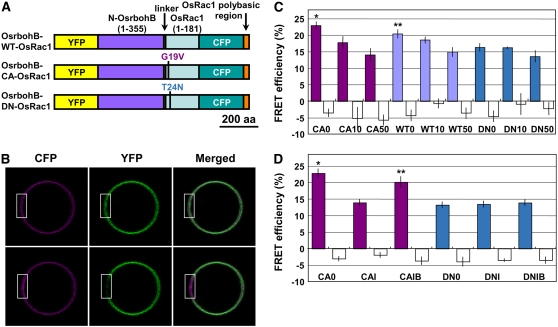
Figure 4.
In Vivo FRET Analysis in Transiently Transformed Rice Protoplasts.
(A) Schematic representation of Os Rac1-Rboh intramolecular FRET constructs used for transient assays.
(B) CFP and YFP fluorescence of rice protoplast expressing Os Rac1-Rboh FRET construct before (top panels) and after (bottom panels) YFP photobleaching. The region marked in white was used for YFP photobleaching.
(C) and (D) Colored bars indicate calculated mean FRET efficiencies of FRET constructs containing CA-OsRac1 (CA), wild type-OsRac1 (WT), or DN-OsRac1 (DN). CaCl2 (0, 10, or 50 mM) or calcium ionophore A23187 (1 μM) with (IB) or without (I) the cell-permeant calcium ion chelator AM-BAPTA (10 mM) was added into the incubation medium R2P, which contained 1 mM CaCl2, and was added after protoplast transformation. A23187 with (IB) or without (I) AM-BAPTA was added to the incubation medium 60 to 90 min before microscopy. Open bars indicate mean background FRET efficiencies, which represent the percentages of change in CFP fluorescence caused by the imaging process, without acceptor photobleaching (Bhat et al., 2004). In (C), single asterisk and double asterisks indicate significant difference from DN0 by t test at P < 0.005 and P < 0.05, respectively. Values of CA10, WT10, DN10, and DN50 were not significantly different from that of DN0 (P > 0.05). In (D), single asterisk and double asterisks indicate significant difference from DN0 by t test at P < 0.0001 and P < 0.005, respectively. Error bars indicate se (n = 21 to 28).
To test the interaction of Os Rac1with the N-terminal region of Os rbohB, wild-type, CA-, and DN-OsRac1 were individually cloned into the core construct (Figure 4A). We measured the FRET efficiencies after photobleaching of yellow fluorescent protein (YFP) at the plasma membrane. Typically, after YFP photobleaching, an increase in cyan fluorescent protein (CFP) emission was detected in the plasma membrane of the rice cells transformed with the Osrboh-CA-OsRac1construct (Figure 4B). CA-OsRac1 showed the highest FRET efficiency, and DN-OsRac1 showed the lowest efficiency (Figure 4C). The FRET efficiency of DN-OsRac1 may represent the basal level of FRET caused by the proximity of the CFP and YFP proteins in this chimeric molecule. These results are comparable to those previously obtained for the interactions between small GTPases and their respective downstream effectors using a similar FRET system in animal cells (Mochizuki et al., 2001; Itoh et al., 2002). Therefore, our results suggest that Os Rac1 and Os rbohB interact with each other directly in a GTP-dependent manner in rice cells.
Previous studies have shown that cytosolic Ca2+ accumulation is required for ROS production in plant cells (Jabs et al., 1997; Sagi and Fluhr, 2001; Kurusu et al., 2005). To test the effect of Ca2+ on the interaction of Os Rac1 and Os rboh in vivo, FRET analyses were performed with or without the addition of Ca2+ to the protoplast incubation medium. Surprisingly, FRET efficiencies were decreased in a concentration-dependent manner for assays involving the interaction between the CA and wild-type forms of Os Rac1 and the N-terminal region of Os rbohB but not in the assay involving DN-OsRac1 (Figure 4C). To confirm this result, similar assays were performed with the addition of the calcium ionophore A23187 with or without the cell-permeable calcium ion chelator AM-BAPTA (Figure 4D). Results show that the addition of calcium ionophore A23187 significantly represses the FRET efficiency for the interaction of CA-OsRac1 and the N terminus of Os rbohB but not for DN-OsRac1. Furthermore, this repression was attenuated by the addition of AM-BAPTA. Therefore, the results support the notion that cytosolic Ca2+ elevation inhibits the Os Rac1–rbohB interaction.
Derepression of Ca2+-Mediated Inhibition of Os Rac1–rbohB Interaction by EF-Hand Motif Mutation
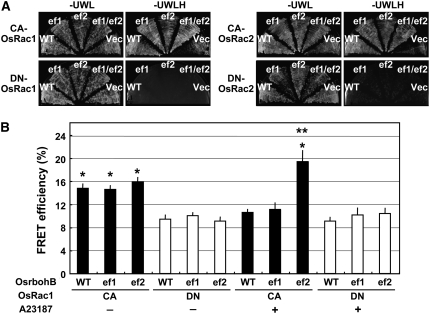
To further study if Ca2+ binding by the EF-hand motifs of the N-terminal region of rbohB is involved in Rac1–rbohB interaction, we mutated the conserved EF-hand motifs of Os rbohB, substituting the Asp residues at positions 242 and 286, generating the mutants OsrbohB D242A (ef1) and D286A (ef2) (Gutierrez-Ford et al., 2003). Mutations in these conserved Asp residues, which participate in Ca2+ coordination, are known to disrupt Ca2+ binding (see Supplemental Figure 3 online). The N-terminal region of Os rbohB (1 to 355) carrying the ef1, ef2, and ef1/ef2 mutations were cloned in the prey vector pVP16, and yeast two-hybrid assays were performed with the CA or DN form of Os Rac1 or Rac2 as baits (Figure 5A). Results of the assays showed that ef1 and ef2 mutations did not affect interaction of Os Rac1-rbohB and Os Rac2-rbohB in yeast, thus suggesting that Ca2+ binding by the two EF-hand motifs may not be required for the interaction of Os Rac1 or Os Rac2 with Os rbohB, even though the region spanning rbohB (133 to 355) may be required for Rac1–rbohB interaction (Figures 2 and 3).
Figure 5.
Effect of EF-Hand Mutations on Os Rac1–rbohB Interaction.
(A) Yeast two-hybrid analysis of Os Rac and the N-terminal region of Rbohs and its EF-hand mutants. Representative plates of yeast two-hybrid assay showing interaction between CA and DN forms of Os Rac1 (left) and Os Rac2 (right) with the N-terminal region of Rbohs and its EF-hand mutants. Growth on selective plates without His (−H) indicates a positive interaction.
(B) In vivo FRET analysis of Os Rac1-rbohB EF-hand mutant FRET constructs. Bars indicate calculated mean FRET efficiencies obtained from rice protoplasts expressing FRET constructs containing CA-OsRac1 (CA) or DN-OsRac1 (DN) with (+) or without (−) calcium ionophore A23187 (1 μM). Imaging was performed with low (0.5 to 1.5%) laser output, resulting in a mean background FRET efficiency of −2.3% ± 0.6% (data not shown). The single asterisk and double asterisks indicate significant difference from Os rbohB (WT)-DN-OsRac1 (minus A23187) (lane 4) (P < 0.001) and Os rbohB (ef2)-CA-OsRac1 (minus A23187) (lane 3) (P < 0.05), respectively. Error bars indicate se (n = 25 to 44).
WT, ef1, ef2, ef1/ef2, and Vec denote Os rbohB (wild type), Os rbohB D242A, Os rbohB (D286A), Os rbohB D242A D286A, and the empty prey vector pVP16, respectively.
Next, we introduced the ef1 and ef2 mutations into the Os Rac1-rbohB FRET constructs and performed FRET analysis in rice protoplasts in the presence or absence of the calcium ionophore A23187 (Figure 5B). Results of the FRET analysis showed that in the absence of A23187, for the CA-OsRac1 construct with the ef1 or ef2 mutation (Figure 5B, lane 2 and 3), the FRET efficiencies produced were similar to that of Os rbohB (Figure 5B, lane 1). Thus, the results are consistent with that of the yeast two-hybrid assay (Figure 5A). Interestingly, in the presence of A23187, the CA-OsRac1 construct with the ef1 mutation (Figure 5B, lane 8) responded similarly as OsrbohB (lane 7), but the construct with the ef2 mutation (lane 9) showed significant increase in FRET efficiency compared with Os rbohB (lane 7) and ef2 without A23187 treatment (lane 3). Preliminary results from the ef1 ef2 double mutant indicated that it also responded to A23187 similarly as in the ef2 mutant (data not shown). Therefore, the rise in FRET efficiency of the ef2 mutant indicates that the cytosolic Ca2+ elevation induced by A23187 failed to suppress the Os Rac1–rbohB interaction in the ef2 mutant and that the ef2 mutation derepressed Ca2+-mediated inhibition of the Os Rac1–rbohB interaction. For the DN-OsRac1 constructs with or without A23187 treatment, Os rbohB (wild type), ef1, and ef2 produced similar FRET efficiencies. Taken together, the results suggest that the second EF-hand motif (EF2) in Os rbohB may be required to suppress Os Rac1 binding at high cytosolic Ca2+ concentration.
Coexpresssion of Os Rac1 and rbohB Enhanced ROS Production in N. benthamiana
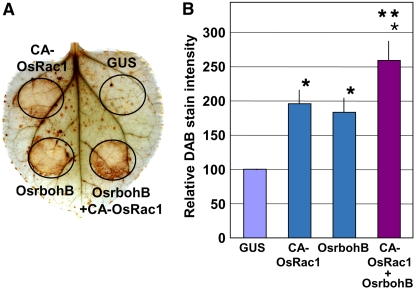
Although both Rac/Rop (Kawasaki et al., 1999; Ono et al., 2001; Park et al., 2004; Jones et al., 2007) and Rboh (Foreman et al., 2003; Yoshioka et al., 2003; Sagi et al., 2004; Torres et al., 2006) have been shown to be involved in ROS production in plants, a direct functional link between Rac/Rop and Rboh is lacking. To address the biological significance of Rac–Rboh interaction, we used the Agrobacterium tumefaciens–mediated transient expression assay (agroinfiltration) to test if transient coexpression of Os Rac1 and rbohB would lead to enhanced ROS production. Results of the agroinfiltration showed that transient expression of Os Rac1 or Os rbohB alone was sufficient to induce ROS production, and this ROS production was significantly enhanced in CA-OsRac1 and Os rbohB coexpression (Figure 6) but not in DN-OsRac1 and OsrbohB coexpression (see Supplemental Figure 5 online). This suggests that CA-OsRac1 and Os rbohB may work synergistically in ROS production. However, we cannot rule out the possibility that this enhanced ROS production was due the additive effect of CA-OsRac1 and Os rbohB and not the regulatory effect of Rac on Rboh.
Figure 6.
Transient Coexpression of Os Rac1 and Os rbohB Enhances ROS Production in N. benthamiana.
(A) The 3,3′-diaminobenzidine (DAB)–stained N. benthamiana leaves transiently transformed with β-glucuronidase (GUS) (P35S-GUS), Os Rac1 (P35S-CA-OsRac1), and Os rbohB (P35S-OsrbohB).
(B) DAB staining intensity indicating in situ ROS levels of agroinfiltrated N. benthamiana leaves. Bars indicate calculated mean relative DAB stain intensity based on the stain intensity of the control GUS. The single asterisk and double asterisks indicate significant difference from the control GUS (lane 1) (P < 0.05) and CA-OsRac1 (lane 2) or OsrbohB (lane 3) (P < 0.05), respectively. Error bars indicate se (n = 19).
DISCUSSION
All plant Rbohs identified to date possess an N-terminal extension, which is not found in the mammalian phagocyte gp91phox, and plant genomes lack the homologs of the regulatory subunits of the phagocytic Nox complex, except for Rac GTPase. These observations suggest that the N-terminal extension of Rboh may be important for the regulation of its activity. This prompted us to investigate if Rac GTPase directly regulates Rboh activity in plants.
Results of our yeast two-hybrid assay suggest that direct interaction between Rac and the N-terminal region of Rboh is ubiquitous in plants. The yeast two-hybrid assay showed that most rice Rac proteins could directly interact with multiple Rbohs, including potato Rboh. By further dissecting the N-terminal region, the yeast two-hybrid assay revealed that a large portion of the N-terminal region, extending beyond the two EF-hand motifs, is required for the interaction with Os Rac GTPase. In fact, in addition to the two EF-hand motifs, analysis for conserved domains by PROSITE (Hulo et al., 2006) predicted at the cutoff level of 4.862 the weak possibility of a partial EF-hand motif at residues 212 to 228, which lies immediately upstream of the first EF-hand motif (229 to 264) (see Supplemental Figure 3 online). The PROSITE default cutoff is 5.0. Furthermore, the residues 212 to 228 are also found within the major protease-resistant fragments (see Supplemental Figure 2 online) that are required for interaction with Os Racs (Figure 2; see Supplemental Figure 1 online). This indicates that a large portion of the N-terminal extension may contain motifs that are required for interaction with Rac GTPases.
In plants, the N-terminal region of Rboh, which contains two EF-hand motifs, may function as a bridge between Rac and Rboh, in effect acting as a substitute to the mammalian p67phox homolog. A number of Rac-interacting domains have been reported (Cotteret and Chernoff, 2002). Our work demonstrates direct binding of a Rac GTPase to an EF-hand motif–containing protein. Furthermore, results of the yeast two-hybrid assay, in vitro pull-down assays, NMR titration, and FRET analysis all showed that Rboh preferentially binds to the GTP-bound or CA form of Rac, suggesting that the interactions are selective for the type of Rac and Rboh. Apparently, the binding of Rboh is very sensitive to the bound cofactor of Rac. Collectively, the results also suggest that plant Rac GTPase may function as a molecular switch for regulating Rboh activity, as in the case of human Rac and gp91phox. Os Rac proteins are highly homologous except for the insert region and the hypervariable C-terminal polybasic region (Miki et al., 2005). Interestingly, GST-OsRac1G19V (1 to 183) with a truncated C-terminal polybasic region showed lower binding affinity to Os rbohB (138 to 313) than that of GST-OsRac1G19V, while retaining the GMPPCP binding preference. Therefore, the hypervariable polybasic region may contribute to Rboh binding.
The selectivity of Rac–Rboh interactions also provides a clue to resolve the seemingly opposing role of different Rac GTPases in plant defense signaling. Although Os Rac1 is a positive regulator of defense in rice (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002; Wong et al., 2004), Hv RacB in barley (Schultheiss et al., 2002) and Nt Rac5 in tobacco (Morel et al., 2004) are negative regulators of defense response. In this study, the yeast two-hybrid assay showed that all Os Rac proteins, except for Os Rac4, interact with more than one Rboh proteins. However, we do not rule out the possibility that Os Rac4 may interact with other Rboh proteins that are yet to be tested.
FRET microscopy has emerged as a useful approach to study small GTPase-mediated signal transduction in mammalian cells (Mochizuki et al., 2001; Itoh et al., 2002). Here, we used it to study the interactions of Rboh and Rac GTPase in rice cells. The original Raichu-Rac construct contained a human Rac1 C terminus to localize the chimeric protein at the plasma membrane in animal cells (Itoh et al., 2002). However, in rice cells, the human Rac1 C terminus mislocalized the chimeric protein to the nucleus, instead of the plasma membrane (data not shown). This mislocalization may be due to the similarity of the C terminus of human Rac1 to the nuclear localization signal, as in many animal small GTPases (Williams, 2003). Therefore, in our constructs for FRET analysis, we replaced the human Rac1 C-terminal sequences with that of the C terminus Os Rac1 (residues 185 to 214), and this modification correctly targeted the chimeric to the plasma membrane, the site where Os Rac1 (Ono et al., 2001) and Rboh function (Figure 1D). Results of the FRET analysis supported the results of yeast two-hybrid and in vitro protein–protein interaction assays that Os Rac1 interacts with the N-terminal region of Os rbohB in vivo.
Previous studies have established the requirement of cytosolic Ca2+ accumulation in the initiation of oxidative burst (Blume et al., 2000; Grant et al., 2000; Sagi and Fluhr, 2001; Kurusu et al., 2005). This conclusion is mainly deduced from the inhibition of elicitor-stimulated oxidative burst, particularly the second accumulation phase in the biphasic ROS production profile, by calcium chelators or channel blockers in cell cultures. Although addition of calcium ionophore A23187 enhanced elicitor-stimulated ROS production (Bindschedler et al., 2001; Davies et al., 2006), addition of the calcium ionophore alone did not significantly stimulate ROS production (Jabs et al., 1997; Bindschedler et al., 2001; Davies et al., 2006). Using an in-gel assay, one study showed that Ca2+ alone is sufficient to stimulate membrane extracts to produce ROS in an NADPH-dependent manner, but the identity of the Rboh remains unknown (Sagi and Fluhr, 2001). However, as far as we know, purified Rboh protein was not used in the study, and this finding has not been confirmed by other studies. Alternatively, as plants possess many Rbohs in their genomes, we cannot eliminate the possibility that some Rbohs may be regulated differently from Os rbohB, and Ca2+ may be required for their activation. In mammalian Nox5, binding of Ca2+ to its N-terminal region, which contains four EF-hand motifs, induces a conformational change that is thought to activate the enzyme (Banfi et al., 2004). Although the role of calcium in regulating the oxidative burst remains unclear, it is possible that a conformation change in the N-terminal region of Rboh is a prerequisite to the activation of its enzymatic activity. The possible existence of other modulators of Rboh also needs to be considered.
Phosphorylation is thought to play important roles in the activation of the oxidative burst (Lamb and Dixon, 1997; Wojtaszek, 1997; Scheel, 1998; Nurnberger and Scheel, 2001). Calcium-dependent protein kinases (CDPKs) are known to be involved in plant response to biotic and abiotic stresses (Grant and Loake, 2000; Cheng et al., 2002; Ludwig et al., 2004) in which ROS also play important roles (Baxter-Burrell et al., 2002; Apel and Hirt, 2004). Recently, Nuhse et al. (2007) and Kobayashi et al. (2007) showed that the phosphorylation of Rboh is important for activation of its enzymatic activity. Kobayashi et al. (2007) also showed that phosphorylation was mediated by CDPK. Interestingly, both studies suggested that phosphorylation of Rboh is not sufficient for full activation of Rboh. On the other hand, emerging genetic evidence is pointing to the involvement of small GTPases in the regulation of ROS production in plants (Kawasaki et al., 1999; Ono et al., 2001; Baxter-Burrell et al., 2002; Nakanomyo et al., 2002; Carol et al., 2005). In this study, our agroinfiltration assay showed that coexpression of Os Rac1 and Os rbohB enhanced ROS production. Furthermore, our in vitro NADPH oxidase assay using plasma membrane fractions isolated from tobacco Bright Yellow 2 cultured cells also indicated that addition of GTP-bound Os Rac1 stimulated superoxide production (data not shown). Therefore, CDPKs and Rac GTPases may modulate Rboh enzymatic activity. However, the functional relation between the phosphorylation of Rboh and the interaction of Rboh and Rac GTPase remain unknown.
Our yeast two-hybrid assay and in vivo FRET analyses (Figures 4 and 5) suggest that cytosolic Ca2+ elevation suppresses the Rac–Rboh interaction and that Ca2+ binding by the two EF-hand motifs is not required for the Os Rac1–rbohB interaction. Furthermore, the mutation in the second EF-hand motif (EF2) of Os rbohB derepressed Ca2+-mediated inhibition of OsRac1 binding at elevated cytosolic Ca2+ concentration. These results indicate that Ca2+ possesses a negative role in Rboh regulation.
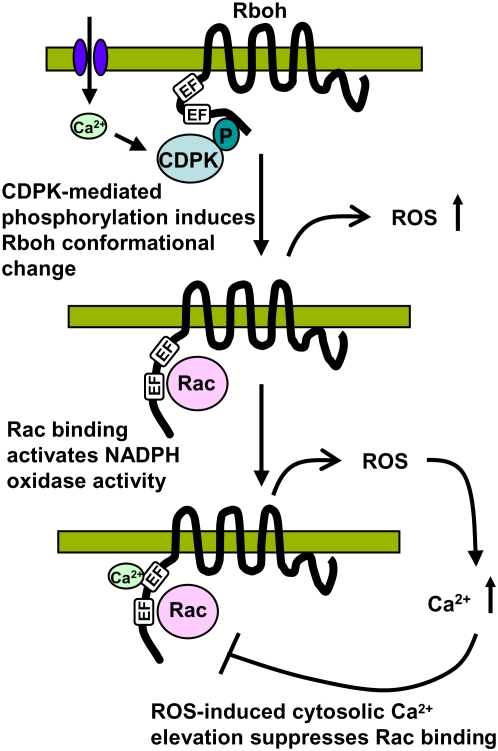
To account for the positive role of Ca2+ in the initiation of Rboh-mediated ROS production and the suppression of Rac–Rboh interaction by cytosolic Ca2+ elevation, we propose a model where the cytosolic Ca2+ accumulation plays a dual role in the oxidative burst (Figure 7). In the initial stage of the oxidative burst, cytosolic Ca2+ influx may be required by CDPK to phosphorylate the N-terminal region of Rboh, thus inducing a conformational change. This conformational change may be brought about by Ca2+-dependent phosphorylation by CDPK. The conformational change may release Rboh from an autoinhibitory state.
Figure 7.
Model of Plant NADPH Oxidase Regulation.
Initial cytosolic Ca2+ influx activates CDPK, which phosphorylates the N-terminal region of Rboh, leading to a conformation change that facilitates Rac GTPase binding of Rboh, leading to the activation of ROS production. Subsequently, the ROS produced may induce cytosolic Ca2+ elevation, which inhibits Rac binding, thus terminating the oxidative burst.
In the second stage, the conformational change in the N terminus may expose it for the interaction with Rac GTPase. The binding of Rac GTPase to the vicinity of the EF-hand motifs in the N-terminal region of Rboh may bring it into close proximity to the catalytic region of Rboh, thereby allowing the N-terminal region to substitute the function of p67phox protein in interaction between human Rac2 and gp91phox. As indicated by the agroinfiltration assay, Rac GTPase may possess the ability to stimulate the NADPH oxidase activity of Rboh by direct interaction. Subsequently, the increase in ROS production may induce a second phase of cytosolic Ca2+ accumulation by stimulating the opening of plasma membrane calcium channels (Pei et al., 2000). Alternatively, the persistent cytosolic Ca2+ accumulation may be caused by a Ca2+-activated Ca2+ release mechanism, resulting in the release of Ca2+ from internal stores (Ward and Schroeder, 1994; Sagi and Fluhr, 2006).
Finally, the cytosolic Ca2+ may reach a threshold where it begins to inhibit the Rac–Rboh interaction, such that the Rac-activated ROS production by Rboh is inhibited; thus, the oxidative burst is terminated. Therefore, the subsequent cytosolic Ca2+ elevation may serve as a negative feedback mechanism to dynamically modulate the length of time and possibly the intensity of oxidative burst. This feedback mechanism may operate to restrict cell death at infection sites as recently shown in Arabidopsis Rboh (Torres and Dangl, 2005). Our findings provide a possible mechanism not only for the regulation of NADPH oxidase for deliberate generation of ROS in plants during defense, abiotic stress, and development but also a possible mode of the regulation for nonphagocytic EF-hand motif–containing NADPH oxidases found in human.
METHODS
Comparison of Predicted Amino Acid Sequences of Rice Rboh
To identify rboh genes, the amino acid sequence of rice (Oryza sativa) rbohA was used as a query for BLASTN searches of the rice genome databases (http://riceblast.dna.affrc.go.jp/) and the rice full-length cDNA database (KOME; http://cdna01.dna.affrc.go.jp/cDNA/). Conserved domains of predicted amino acid sequences were analyzed by PROSITE with the psscan.pl program (http://www.expasy.org/ftp/databases/prosite/tools/ps_scan) (Hulo et al., 2006). Predicted amino acid sequences of Rbohs were aligned using ClustalW and neighbor-joining methods.
Yeast Two-Hybrid Assays
The bait vectors carrying CA and DN mutants of Os Rac1 have been described previously (Kawasaki et al., 2006). The CA and DN mutants of six members (Rac2 to Rac7) of the Os Rac family were produced by substitution of the amino acids corresponding to CA and DN mutants of Os Rac1. Plant Rac GTPases undergo posttranslational lipid modification at their C-terminal Cys residue for anchoring the protein to the plasma membrane (Sorek et al., 2007). Therefore, C-terminal Cys-to-Ser mutagenesis was performed on the CA and DN mutants by PCR amplification, and then the amplified fragments were subcloned into the bait vector, pBTM116. The resultant mutants were designated as OsRac2-G16V, OsRac2-T21N, OsRac3-G17V, OsRac4-G17V, OsRac5-G15V, OsRac6-G15V, and OsRac7-G15V. The N-terminal regions of Rbohs were inserted into the prey vector pVP16. The combinations of the bait and prey vectors were introduced into cells of Saccharomyces cerevisiae L40. The interaction was analyzed based on requirement of His for yeast growth as described previously (Kawasaki et al., 2006).
Sample Preparation of Os rbohB (138 to 313)
Os rbohB (1 to 355) was expressed in Escherichia coli, purified, and subjected to limited proteolysis with trypsin or α-chymotrypsin. The proteinase-resistant fragments were identified by N-terminal sequencing and mass spectroscopy. DNA encoding Os rbohB (138 to 313) was cloned into the pET32c vector (Novagen), expressed as a Thioredoxin (Trx)-6×His tag fusion protein in E. coli, and purified by affinity, anion exchange, and size exclusion chromatography. The Trx-His tag was removed with recombinant Enterokinase (Novagen).
GST Pull-Down Assay
GST-OsRac1, GST-OsRac1G19V, and GST-OsRac1G19V (1 to 183) were expressed in E. coli, purified, and incubated with 4 mM GDP (Sigma-Aldrich) or GMPPCP (a GTP analog; Sigma-Aldrich) in 20 mM Bis-Tris, pH 6.8, 5 mM EDTA, and 0.4 mM DTT. Nucleotide exchange was stopped on ice by adding MgCl2 to 20 mM. This reaction was repeated twice. After absorbing the GST-tagged proteins onto 10 to 20 μL of glutathione sepharose resin (Amersham Biosciences), 30 to 40 μL of 0.6 mM purified OsrbohB (138 to 313) was applied and incubated at room temperature for 1 h. After the beads were washed, bound proteins were analyzed by SDS-PAGE by staining with Coomassie Brilliant Blue or silver stain.
NMR Titration Experiment
DNA encoding Os Rac1 was cloned into the pET32a vector (Novagen) and expressed as a Trx-His tag fusion protein in E. coli growing in minimal medium containing 0.5 g/L 15N-ammonium chloride. Os Rac1 was purified by affinity and size exclusion chromatography, and the tag was removed with enterokinase. The nucleotide exchange reaction was performed as described above. All NMR experiments were recorded on a Bruker Avance500 NMR spectrometer at 303K using 0.14 mM 15N OsRac1 in 50 mM Bis-Tris, pH 6.8, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, and 10% D2O. The binding of Os Rac1 to Os rbohB was monitored by 1H-15N HSQC experiments, whereby nonlabeled Os rbohB (138 to 313) was titrated into concentrations ranging from 0.014 to 0.24 mM. Peak broadenings were saturated at a molar ratio of 1:2 (OsRac1:OsrbohB) for the GMPPCP-bound form but not for the GDP form.
Confocal Laser Microscopy and Acceptor Photobleaching FRET Analysis
Rice protoplasts were prepared and transformed by electroporation as described previously (Wong et al., 2004). After electroporation, the protoplasts were incubated at 30°C for 12 to 18 h before confocal laser microscopy. Intracellular localization of GFP fluorescence was analyzed using a LSM510 microscope (Carl Zeiss) with a 488-nm argon ion laser as excitation source. Acceptor photobleaching FRET analysis was performed using a LSM510 META microscope (Carl Zeiss) as described previously (Karpova et al., 2003; Bhat et al., 2004). Acceptor photobleaching was performed using a 514-nm argon ion laser with 25 to 50 cycles of repeated scans at 75 or 50% output for Figures 4 and 5, respectively. Genetically modified GFP variants, SECFP, and Venus (designated as CFP and YFP, respectively) were used as donor and acceptor fluorophores, respectively (Mochizuki et al., 2001). The CFP- and YFP-tagged proteins, expressed in rice protoplasts, were coexcited by the 458-nm line laser, and the emitted fluorescence was collected in the lambda spectrum mode of the LSM510 software version 2 (Carl Zeiss) for Figure 4 and LSM510 software version 3 for Figure 5. CFP and YFP emission spectra were linearly unmixed using relevant reference spectra. Background fluorescence was subtracted, and FRET efficiency was calculated according to the published procedures (Bhat et al., 2004).
Mutagenesis of the EF-Hand Motifs of Os rbohB
Site-directed mutagenesis of the EF-hand motifs of Os rbohB was performed using the Quikchange kit (Stratagene) according to the manufacturer's instructions. D242A (ef1) and D286A (ef1) mutations were generated using the following primer pairs: F- D242A (5′-CATTCTTTGACATGGTTGCCAAGAACGCTGATGG-3′) and R-D242A (5′-CCATCAGCGTTCTTGGCAACCATGTCAAAGAATG-3′); F-D242A (5′-ATGGAAGAGCTTGCCCCTACAAACTTGGGATAC-3′) and R- D242A (5′-TCCCAAGTTTGTAGGGGCAAGCTCTTCCATAATGAG-3′). Mutations were confirmed by DNA sequencing.
Agroinfiltration of Nicotiana benthamiana and Detection of ROS
Agroinfiltration of N. benthaniana was performed as described previously (Kobayashi et al., 2007) with modifications. Agrobacterium tumefaciens strain GV3010 harboring the helper plasmid pSoup (Hellens et al., 2000) and binary plasmids carrying the the cDNA of Os Rac1, Os rbohB, flag-tagged St rbohB, or the control GUS were used to infiltrate 5-week-old N. benthamiana leaves (Kobayashi et al., 2007). Agrobacterium culture was resuspended in a buffer containing 10 mM MgCl2, 10 mM MES, pH 5.6, and 150 μM acetosyringone at OD600 of 0.5, incubated at 23°C for 4 to 5 h, and diluted to OD600 of 0.1 in the same buffer before infiltrating into the fourth and fifth leaves of N. benthaniana using a needleless 1-mL syringe.
To visualize ROS in situ, DAB staining was performed as described by Thordal-Christensen et al. (1997), and the staining was evaluated as described by Torres et al. (2002) with modifications. The agroinfiltrated plants were kept in a growth chamber at 75% humidity for 2 d after agroinfiltration. The agroinfiltrated leaves were detached and incubated in 1 mg/mL DAB solution for 8 h. Then, the leaves were fixed with a solution of 3:1:1 ethanol/acetic acid/glycerol. DAB-stained leaves were scanned, and pixel intensity of agroinfiltrated regions was quantified by the ImageJ (National Institutes of Health) software. The mean pixel intensity from three spots outside the infiltrated regions of each leaf was used for background subtraction, and main veins were avoided in all regions selection. The relative DAB stain intensity was calculated based on the pixel intensity of the control GUS-agroinfiltrated region of each leaf to facilitate comparison of the DAB staining between different leaves.
Accession Numbers
Sequence data for various annotated Rbohs can be found in the GenBank/EMBL data libraries under the following accession numbers: Os rbohA (NM_191558), Os rbohB (AK065117), Os rbohC (AK120905), Os rbohD (AK072353), Os rbohE (AK100241), Os rbohF (XM_482730), Os rbohG (AK120739), Os rbohH (ABA99453), Os rbohI (ABA94089), St rbohA (AB050660), and St rbohB (AB050661).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Interaction of Various Fragments of the N-Terminal Region of Os rbohD or St rbohB with Os Rac2.
Supplemental Figure 2. Proteolysis of the N-Terminal Region of Os rbohB (1 to 355).
Supplemental Figure 3. 1H-15N HSQC Spectra of Uniformly 15N-Labeled Os Rac1 with Different Concentrations of Os rbohB (138 to 313).
Supplemental Figure 4. Partial Amino Acid Alignment of Os rbohB and Calcineurin B.
Supplemental Figure 5. Transient Coexpression of DN-OsRac1 and Os rbohB Does Not Enhance ROS Production in N. benthamiana.
Supplemental Table 1. Amino Acid Alignments of Rice Rbohs Used to Generate Figure 1B.
Supplementary Material
Acknowledgments
We thank Michiyuki Matsuda for the Raichu plasmid, Megumi Iwano for technical assistance with FRET microscopy, and Damien Lieberherr for assistance with PROSITE analysis. This research was supported by Grants-in-Aid from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project IP4001) and the Japan Society for Promotion of Science (13GS0023) to K.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ko Shimamoto (simamoto@bs.naist.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. [DOI] [PubMed] [Google Scholar]
- Babior, B.M. (2004). NADPH oxidase. Curr. Opin. Immunol. 16 42–47. [DOI] [PubMed] [Google Scholar]
- Banfi, B., Tirone, F., Durussel, I., Knisz, J., Moskwa, P., Molnar, G.Z., Krause, K.H., and Cox, J.A. (2004). Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5). J. Biol. Chem. 279 18583–18591. [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell, A., Yang, Z., Springer, P.S., and Bailey-Serres, J. (2002). RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296 2026–2028. [DOI] [PubMed] [Google Scholar]
- Bhat, R.A., Borst, J.W., Riehl, M., and Thompson, R.D. (2004). Interaction of maize Opaque-2 and the transcriptional co-activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol. Biol. 55 239–252. [DOI] [PubMed] [Google Scholar]
- Bhat, R.A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., and Panstruga, R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 102 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler, L.V., Minibayeva, F., Gardner, S.L., Gerrish, C., Davies, D.R., and Bolwell, G.P. (2001). Early signalling events in the apoplastic oxidative burst in suspension cultured French bean cells involve cAMP and Ca2+. New Phytol. 151 185–194. [DOI] [PubMed] [Google Scholar]
- Blume, B., Nurnberger, T., Nass, N., and Scheel, D. (2000). Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell 12 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol, R.J., Takeda, S., Linstead, P., Durrant, M.C., Kakesova, H., Derbyshire, P., Drea, S., Zarsky, V., and Dolan, L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438 1013–1016. [DOI] [PubMed] [Google Scholar]
- Cheng, S.H., Willmann, M.R., Chen, H.C., and Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129 469–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotteret, S., and Chernoff, J. (2002). The evolutionary history of effectors downstream of Cdc42 and Rac. Genome Biol. 3 2.1–2.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, D.R., Bindschedler, L.V., Strickland, T.S., and Bolwell, G.P. (2006). Production of reactive oxygen species in Arabidopsis thaliana cell suspension cultures in response to an elicitor from Fusarium oxysporum: Implications for basal resistance. J. Exp. Bot. 57 1817–1827. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 98 13454–13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446. [DOI] [PubMed] [Google Scholar]
- Gapper, C., and Dolan, L. (2006). Control of plant development by reactive oxygen species. Plant Physiol. 141 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiszt, M., and Leto, T.L. (2004). The Nox family of NAD(P)H oxidases: host defense and beyond. J. Biol. Chem. 279 51715–51718. [DOI] [PubMed] [Google Scholar]
- Grant, J.J., and Loake, G.J. (2000). Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 124 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., and Mansfield, J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23 441–450. [DOI] [PubMed] [Google Scholar]
- Groom, Q.J., Torres, M.A., Fordham-Skelton, A.P., Hammond-Kosack, K.E., Robinson, N.J., and Jones, J.D. (1996). rbohA, a rice homologue of the mammalian gp91phox respiratory burst oxidase gene. Plant J. 10 515–522. [DOI] [PubMed] [Google Scholar]
- Gu, Y., Wang, Z., and Yang, Z. (2004). ROP/RAC GTPase: An old new master regulator for plant signaling. Curr. Opin. Plant Biol. 7 527–536. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Ford, C., Levay, K., Gomes, A.V., Perera, E.M., Som, T., Kim, Y.M., Benovic, J.L., Berkovitz, G.D., and Slepak, V.Z. (2003). Characterization of tescalcin, a novel EF-hand protein with a single Ca2+-binding site: Metal-binding properties, localization in tissues and cells, and effect on calcineurin. Biochemistry 42 14553–14565. [DOI] [PubMed] [Google Scholar]
- Karpova, T.S., Baumann, C.T., He, L., Wu, X., Grammer, A., Lipsky, P., Hager, G.L., and McNally, J.G. (2003). Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209 56–70. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., De Castro, E., Langendijk-Genevaux, P.S., Pagni, M., and Sigrist, C.J. (2006). The PROSITE database. Nucleic Acids Res. 34 D227–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, R.E., Kurokawa, K., Ohba, Y., Yoshizaki, H., Mochizuki, N., and Matsuda, M. (2002). Activation of Rac and Cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22 6582–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs, T., Tschope, M., Colling, C., Hahlbrock, K., and Scheel, D. (1997). Elicitor-stimulated ion fluxes and O2- from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl. Acad. Sci. USA 94 4800–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares-Erijman, E.A., and Jovin, T.M. (2003). FRET imaging. Nat. Biotechnol. 21 1387–1395. [DOI] [PubMed] [Google Scholar]
- Jones, M.A., Raymond, M.J., Yang, Z., and Smirnoff, N. (2007). NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J. Exp. Bot. 58 1261–1270. [DOI] [PubMed] [Google Scholar]
- Jones, M.A., Shen, J.J., Fu, Y., Li, H., Yang, Z., and Grierson, C.S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T., Koita, H., Nakatsubo, T., Hasegawa, K., Wakabayashi, K., Takahashi, H., Umemura, K., Umezawa, T., and Shimamoto, K. (2006). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 103 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, T., Damude, H.G., Werner, D., Doerner, P., Dixon, R.A., and Lamb, C. (1998). A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 10 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., Doke, N., and Yoshioka, H. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19 1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu, T., Yagala, T., Miyao, A., Hirochika, H., and Kuchitsu, K. (2005). Identification of a putative voltage-gated Ca2+ channel as a key regulator of elicitor-induced hypersensitive cell death and mitogen-activated protein kinase activation in rice. Plant J. 42 798–809. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.M., Leonhardt, N., Torres, M.A., Dangl, J.L., Bloom, R.E., Bodde, S., Jones, J.D., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 251–275. [DOI] [PubMed] [Google Scholar]
- Lambeth, J.D. (2004). NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4 181–189. [DOI] [PubMed] [Google Scholar]
- Ludwig, A.A., Romeis, T., and Jones, J.D. (2004). CDPK-mediated signalling pathways: Specificity and cross-talk. J. Exp. Bot. 55 181–188. [DOI] [PubMed] [Google Scholar]
- Miki, D., Itoh, R., and Shimamoto, K. (2005). RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 138 1903–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki, A., Llopis, J., Heim, R., McCaffery, J.M., Adams, J.A., Ikura, M., and Tsien, R.Y. (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388 882–887. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N., Yamashita, S., Kurokawa, K., Ohba, Y., Nagai, T., Miyawaki, A., and Matsuda, M. (2001). Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 411 1065–1068. [DOI] [PubMed] [Google Scholar]
- Moeder, W., Yoshioka, K., and Klessig, D.F. (2005). Involvement of the small GTPase Rac in the defense responses of tobacco to pathogens. Mol. Plant Microbe Interact. 18 116–124. [DOI] [PubMed] [Google Scholar]
- Morel, J., Fromentin, J., Blein, J.P., Simon-Plas, F., and Elmayan, T. (2004). Rac regulation of NtrbohD, the oxidase responsible for the oxidative burst in elicited tobacco cell. Plant J. 37 282–293. [DOI] [PubMed] [Google Scholar]
- Nakanomyo, I., Kost, B., Chua, N.H., and Fukuda, H. (2002). Preferential and asymmetrical accumulation of a Rac small GTPase mRNA in differentiating xylem cells of Zinnia elegans. Plant Cell Physiol. 43 1484–1492. [DOI] [PubMed] [Google Scholar]
- Nuhse, T.S., Bottrill, A.R., Jones, A.M., and Peck, S.C. (2007). Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger, T., and Scheel, D. (2001). Signal transmission in the plant immune response. Trends Plant Sci. 6 372–379. [DOI] [PubMed] [Google Scholar]
- Ono, E., Wong, H.L., Kawasaki, T., Hasegawa, M., Kodama, O., and Shimamoto, K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J., Gu, Y., Lee, Y., and Yang, Z. (2004). Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol. 134 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G.J., Grill, E., and Schroeder, J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406 731–734. [DOI] [PubMed] [Google Scholar]
- Potikha, T.S., Collins, C.C., Johnson, D.I., Delmer, D.P., and Levine, A. (1999). The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 119 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, M., Davydov, O., Orazova, S., Yesbergenova, Z., Ophir, R., Stratmann, J.W., and Fluhr, R. (2004). Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 16 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, M., and Fluhr, R. (2001). Superoxide production by plant homologues of the gp91(phox) NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 126 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, M., and Fluhr, R. (2006). Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel, D. (1998). Resistance response physiology and signal transduction. Curr. Opin. Plant Biol. 1 305–310. [DOI] [PubMed] [Google Scholar]
- Schultheiss, H., Dechert, C., Kogel, K.H., and Huckelhoven, R. (2002). A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol. 128 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, N., Poraty, L., Sternberg, H., Bar, E., Lewinsohn, E., and Yalovsky, S. (2007). Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol. Cell. Biol. 27 2144–2154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Suharsono, U., Fujisawa, Y., Kawasaki, T., Iwasaki, Y., Satoh, H., and Shimamoto, K. (2002). The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 99 13307–13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley powdery mildew interaction. Plant J. 11 1187–1194. [Google Scholar]
- Torres, M.A., and Dangl, J.L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8 397–403. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., Jones, J.D., and Dangl, J.L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K.E., and Jones, J.D. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14 365–370. [DOI] [PubMed] [Google Scholar]
- Vogel, S.S., Thaler, C., and Koushik, S.V. (2006). Fanciful FRET. Sci. STKE 2006: re2. [DOI] [PubMed]
- Ward, J.M., and Schroeder, J.I. (1994). Calcium-activated K+ channels and calcium-induced calcium release by slow vacuolar ion channels in guard cell vacuoles implicated in the control of stomatal closure. Plant Cell 6 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C.L. (2003). The polybasic region of Ras and Rho family small GTPases: A regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell. Signal. 15 1071–1080. [DOI] [PubMed] [Google Scholar]
- Wojtaszek, P. (1997). Oxidative burst: An early plant response to pathogen infection. Biochem. J. 322 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, H.L., Sakamoto, T., Kawasaki, T., Umemura, K., and Shimamoto, K. (2004). Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 135 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, H., Numata, N., Nakajima, K., Katou, S., Kawakita, K., Rowland, O., Jones, J.D., and Doke, N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, H., Sugie, K., Park, H.J., Maeda, H., Tsuda, N., Kawakita, K., and Doke, N. (2001). Induction of plant gp91 phox homolog by fungal cell wall, arachidonic acid, and salicylic acid in potato. Mol. Plant Microbe Interact. 14 725–736. [DOI] [PubMed] [Google Scholar]
- Zaninotto, F., La Camera, S., Polverari, A., and Delledonne, M. (2006). Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol. 141 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.