Abstract
A rice (Oryza sativa) Rac/Rop GTPase, Os Rac1, is involved in innate immunity, but its molecular function is largely unknown. RAR1 (for required for Mla12 resistance) and HSP90 (a heat shock protein 90 kD) are important components of R gene–mediated disease resistance, and their function is conserved in several plant species. HSP90 has also recently been shown to be important in mammalian innate immunity. However, their functions at the molecular level are not well understood. In this study, we examined the functional relationships between Os Rac1, RAR1, and HSP90. Os RAR1-RNA interference (RNAi) rice plants had impaired basal resistance to a compatible race of the blast fungus Magnaporthe grisea and the virulent bacterial blight pathogen Xanthomonas oryzae. Constitutively active Os Rac1 complemented the loss of resistance, suggesting that Os Rac1 and RAR1 are functionally linked. Coimmunoprecipitation experiments with rice cell culture extracts indicate that Rac1 forms a complex with RAR1, HSP90, and HSP70 in vivo. Studies with Os RAR1-RNAi and treatment with geldanamycin, an HSP90-specific inhibitor, showed that RAR1 and HSP90 are essential for the Rac1-mediated enhancement of pathogen-associated molecular pattern–triggered immune responses in rice cell cultures. Furthermore, the function of HSP90, but not RAR1, may be essential for their association with the Rac1 complex. Os Rac1 also regulates RAR1 expression at both the mRNA and protein levels. Together, our results indicate that Rac1, RAR1, HSP90, and HSP70 form one or more protein complexes in rice cells and suggest that these proteins play important roles in innate immunity in rice.
INTRODUCTION
Plants use two innate immune systems to respond to pathogen infection, known as pathogen-associated molecular pattern (PAMP)–triggered innate immunity and effector-triggered immunity (Zipfel and Felix, 2005; Chisholm et al., 2006; Jones and Dangl, 2006). PAMP-triggered innate immunity is induced by recognition of PAMPs by transmembrane receptors and gives early responses to pathogen infection. Effector-triggered immunity involves resistance (R) proteins as receptors that specifically recognize pathogen effectors, either directly or indirectly, and usually induces hypersensitive cell death (HR). The major R proteins are intracellular receptors and contain nucleotide binding site (NBS) and leucine-rich repeat (LRR) domains. Although there has been extensive research on plant innate immunity, the molecular mechanisms of these two immune responses are not well understood.
RAR1 (for required for Mla12 resistance) is an important component of R gene–mediated disease resistance (Shirasu et al., 1999; Tornero et al., 2002; Muskett et al., 2002), which contains two zinc binding finger motifs termed CHORD-I and CHORD-II (Cys- and His-rich domains) (Shirasu et al., 1999). In plants, RAR1 interacts directly with SGT1 (for suppressor of the G2 allele of skp1) and HSP90 (Azevedo et al., 2002; Liu et al., 2002, Takahashi et al. 2003). The CHORD-I domain of RAR1 interacts directly with the N terminus of HSP90, and SGT1 binds the CHORD-II domain of RAR1. SGT1 is required for disease resistance mediated by diverse R proteins (Austin et al., 2002; Azevedo et al., 2002; Liu et al., 2002, Peart et al., 2002) and is also required for the function of an SCF (for Skp1/Cullin/F-box protein) (Kitagawa et al., 1999; Matsuzawa and Reed, 2001). HSP90 is an abundant, highly conserved, ATP-dependent molecular chaperone that is essential for eukaryotic cell viability (Picard, 2002). More recently, RAR1, SGT1, and HSP90 were shown to play an important role in regulating the stability of R proteins that contain the NBS-LRR domain (Bieri et al., 2004; Holt et al., 2005; Azevedo et al., 2006).
HSP90 and SGT1 play a key role in the activation of mammalian immune responses induced by the Nod-like receptor (NLR) protein family, a group that contains an NBS-LRR domain (Hahn, 2005; da Silva Correia et al., 2007; Mayor et al., 2007). SGT1 and HSP90 interact with NLR proteins to form an inflammasome complex and are required for activation of the complex. Therefore, regulatory networks for immune responses in plants and mammals share at least some common components, and understanding the molecular mechanisms involved in protein complexes containing R proteins or NLR proteins, SGT1, and HSP90 is becoming crucial in the study of innate immunity in higher eukaryotes.
Rac/Rop small GTPases are highly conserved in the plant kingdom and constitute the sole group of Rho family small GTPases in plants (Gu et al., 2004). There are seven Rac/Rop genes in rice (Oryza sativa; Miki et al., 2005) and 11 in Arabidopsis thaliana (Gu et al., 2004). Os Rac1, which is located in the plasma membrane, is involved in ROS production through an NADPH oxidase and in the initiation of cell death during defense signaling (Kawasaki et al., 1999; Ono et al., 2001). Os Rac1 functions as a positive regulator of NADPH oxidase activation in PAMP signaling (Kawasaki et al., 1999) and at the same time suppresses expression of a scavenger metallothionein (Wong et al., 2004) in transient accumulation of reactive oxygen species (ROS). Expression of constitutively active (CA)-OsRac1 enhances resistance to rice blast and bacterial blight infections (Ono et al., 2001, Suharsono et al., 2002), whereas a dominant negative (DN) version of Os Rac1 (DN-OsRac1) suppresses HR induced by incompatible races of rice blast fungus, indicating that Os Rac1 is one of the key regulators of rice innate immunity (Ono et al., 2001). Os Rac1 regulates the stability and activation of Os MAPK6, a rice mitogen-activated protein kinase, by sphingolipid elicitors (Lieberherr et al., 2005). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is activated by Rac1 in rice innate immunity (Kawasaki et al., 2006), and DN-OsRac1 inhibits N-mediated resistance to tobacco mosaic virus infection in tobacco (Nicotiana tabacum; Moeder et al., 2005). There are a number of other studies aimed at understanding the molecular mechanisms of rice innate immunity (Yang et al., 2004; Gu et al., 2005; Wang et al., 2006; Takahashi et al., 2007), but the details of how these pathways are regulated remain unknown.
In this study, we investigate the role of RAR1 and HSP90 in rice immune responses, especially in relation to the key regulator Rac GTPase. We show that both RAR1 and HSP90 play a critical role in rice innate immunity and that they apparently form a complex with Rac GTPase. Furthermore, loss of HSP90 function impairs immune responses as well as the formation of a protein complex with Rac GTPase-RAR1-HSP90. These results indicate a critical role for the functional protein complex containing Rac GTPase, RAR1, and HSP90 in rice innate immunity.
RESULTS
Os RAR1 Is Involved in Basal Resistance against Rice Blast and Bacterial Blight Infection in Rice
RAR1 is required for the function of multiple R genes, such as Mla, RPM1, RPS2, RPS5, and N genes (Shirasu et al., 1999; Tornero et al., 2002; Muskett et al., 2002; Austin et al., 2002; Azevedo et al., 2002; Liu et al., 2002; Holt et al., 2005). Therefore, we were interested in determining whether RAR1 is involved in disease resistance in rice.
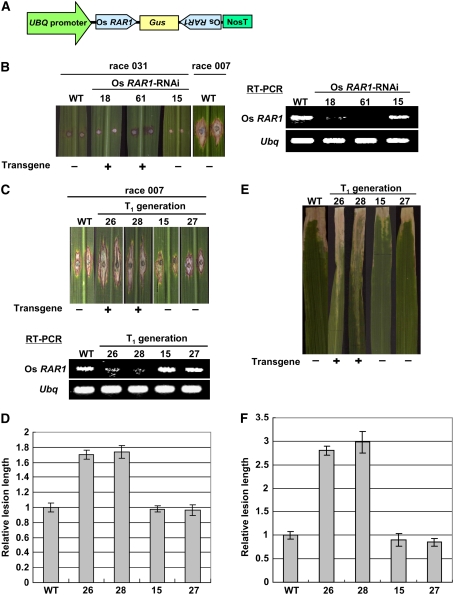
To investigate the function of RAR1 in rice disease resistance, we knocked down RAR1 expression with RNA interference (RNAi) (Figures 1A and 1B; see Supplemental Figure 1A online). Generally, Os RAR1 expression was greatly decreased in Os RAR1-RNAi lines (Figure 1; see Supplemental Figure 2 online). Japonica rice var Kinmaze, which was used for the production of transgenic plants, carries the Pi-a blast resistance gene that is incompatible with race 031 but is compatible with race 007 of Magnaporthe grisea. Two T1 transgenic lines carrying the Os RAR1-RNAi construct (Figure 1B, lines 18 and 61) showed no difference in response to infection with race 031 from the wild-type plant or a T1 segregant without a transgene (Figure 1B, line 15), making it unlikely that Pi-a–mediated blast resistance requires RAR1. To further test for possible Os RAR1 requirements in R gene–mediated blast resistance, we performed the same infection analysis using the Os RAR1-RNAi transformants in the Kanto IL 5 and Kanto IL 14 backgrounds that carry Pi-z and Pi-b blast resistance genes, respectively. These experiments clearly showed that RAR1 is not required by either of these two R genes for blast resistance (data not shown). That Os RAR1 is not required for the three R genes for rice blast resistance is consistent with previous results indicating that RAR1 is required for many but not all R genes examined (Shirasu and Schulze-Lefert, 2003).
Figure 1.
RAR1 Is Involved in Basal Resistance against Rice Blast and Bacterial Blight Infection in Rice.
(A) Diagram of the Os RAR1-RNAi construct.
(B) Transgenic plants expressing Os RAR1-RNAi were inoculated with an incompatible race 031 of the rice blast fungus M. grisea. Lesions are shown on leaf blades 12 d after inoculation. WT indicates untransformed wild type plant, and 18, 61, and 15 are T1 segregants derived from an Os RAR1-RNAi plant. Response of leaf blades of the wild type Kinmaze with infection by the compatible race 007 was used as a control for this experiment. Total RNA was extracted from leaves of the wild type Kinmaze and Os RAR1-RNAi plants. Os RAR1 mRNA was amplified by RT-PCR using specific primers. The 17S rRNA primers were included in the PCR reactions as an internal control.
(C) Response of transgenic plants expressing Os RAR1-RNAi to infection with the compatible race 007 of M. grisea. WT indicates untransformed control plant, and 26, 28, 15, and 27 are T1 segregants derived from an Os RAR1-RNAi plant. Photographs were taken 12 d after infection. Presence or absence of the Os RAR1-RNAi transgene are indicated by + and −, respectively. RNA analysis was performed as indicated in (B).
(D) Quantitative analysis of disease lesions induced by infection with a compatible race of rice blast fungus shown in (C). Leaves were inoculated with compatible race 007 of M. grisea. Bars indicate se obtained from 6 to 20 measurements.
(E) Response of transgenic plants expressing Os RAR1-RNAi to infection with X. oryzae pv oryzae race1 (T7174). WT indicates untransformed control plants, and 26, 28, 15, and 27 are T1 segregants derived from an Os RAR1-RNAi plant. Photographs were taken 12 d after infection. Presence or absence of the Os RAR1-RNAi transgene are indicated by + and −, respectively.
(F) Quantitative analysis of disease lesions induced by infection with a compatible race of bacterial blight shown in (E). Leaves were inoculated with the X. oryzae race 1. Bars indicate se obtained from four to nine measurements.
To determine whether RAR1 is involved in basal resistance to virulent blast isolates, Os RAR1-RNAi plants were infected with compatible race 007. Leaves of untransformed control plants developed typical disease symptoms (Figures 1C and 1D, race 007/WT). Of the four segregants in the T1 generation, the two that carried the Os RAR1-RNAi transgene had larger lesions (Figures 1C and 1D, lines 26 and 28), whereas the two segregants that carried no transgenes had lesion lengths similar to the wild-type control (Figures 1C and 1D, lines 15 and 27). Results of infections assays with T0 transgenic plants are shown in Supplemental Figures 1A to 1C online. Furthermore, infection of T1 generation plants with the rice bacterial blight pathogen Xanthomonas oryzae pv oryzae race 1 (T7174), which is also compatible with var Kinmaze, gave results similar to blast infection (Figures 1E and 1F). Results of infection assays with T0 transgenic plants are shown in Supplemental Figures 1D and 1E online. These results demonstrate that RAR1 is involved in basal resistance to both rice blast fungus and bacterial blight. This is consistent with the previous reports that RAR1 functions in the basal resistance of Arabidopsis to Pseudomonas syringae pv tomato (Holt et al., 2005) and of mlo barley (Hordeum vulgare) to M. grisea (Jarosch et al., 2005).
Os Rac1 and RAR1 Are Functionally Linked in Basal Resistance to Rice Blast
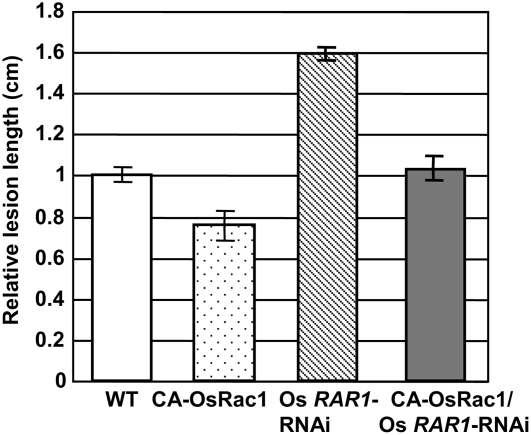
Our previous studies have shown that Rac1 is an important component of disease resistance in rice (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002). Therefore, to examine whether Rac1 and RAR1 are functionally linked in rice disease resistance, we generated transgenic rice carrying both CA-OsRac1 and Os RAR1-RNAi by introducing the Os RAR1-RNAi construct into CA-OsRac1 transgenic calli and regenerating plants for infection experiments (Figure 2). As previously reported, CA-OsRac1 rice plants were more resistant to a compatible race of rice blast (Ono et al., 2001). Transgenic plants carrying both the CA-OsRac1 and Os RAR1-RNAi constructs were more resistant to rice blast than Os RAR1-RNAi plants and had the same level of resistance as untransformed control plants (Figure 2), indicating that RAR1 is required for Rac1-mediated basal resistance to rice blast. Similar results were obtained for basal resistance to bacterial blight (data not shown).
Figure 2.
Quantitative Analysis of Disease Lesions Induced by Infection with a Compatible Race of M. grisea.
Leaves of the nontransgenic wild type, CA-OsRac1, Os RAR1-RNAi, and double transgenic CA-OsRac1/Os RAR1-RNAi plants were used in three independent experiments (mean ± se; n = 12 to 61). Double transgenic plants were made by introducing the Os RAR1-RNAi construct into the CA-OsRac1 transgenic line. The double transgenic plants were more resistant to race 007 of M. grisea than Os RAR1-RNAi plants and more susceptible than CA-OsRac1 plants.
Attempts to generate Os SGT1-RNAi rice cell cultures and whole plants by the same transformation protocol used for generating Os RAR1-RNAi cell culture and plants were unsuccessful. This is likely due to the fact that Os SGT1 is an essential single-copy gene in rice. Therefore, the function of SGT1 in rice innate immunity has not as yet been determined by genetic methods.
Os Rac1 Forms a Complex with RAR1, HSP90, and HSP70 in Vivo
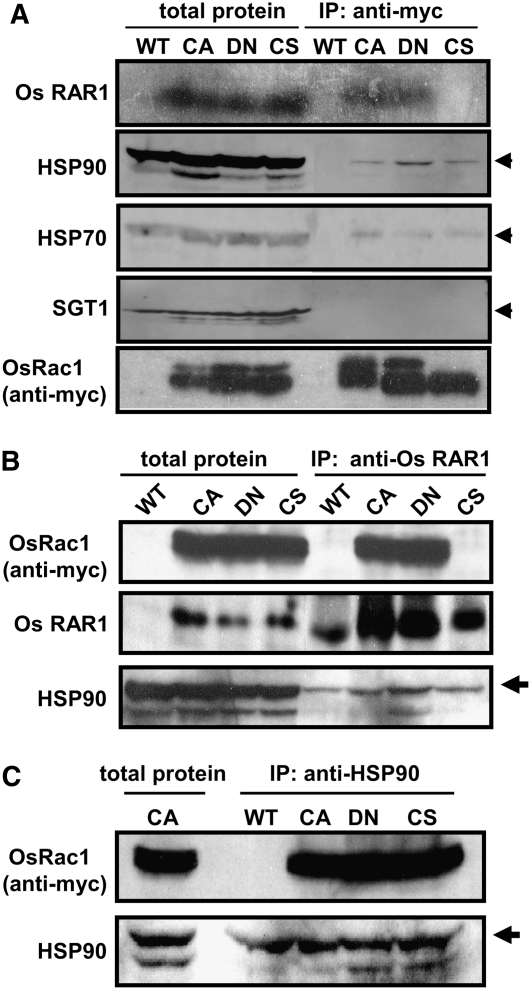
Because Os Rac1 and Os RAR1 are functionally linked in basal resistance, we examined whether RAR1 and Rac1 interact in vivo. For an analysis of RAR1 interaction with Rac1 in vivo, suspension cell cultures expressing myc-tagged CA-OsRac1, DN-OsRac1, and CS-OsRac1 driven by the maize (Zea mays) Ubiquitin promoter (Lieberherr et al., 2005) were used for coimmunoprecipitation (co-IP) experiments. Results of co-IP experiments using anti-myc and anti-RAR1 antibodies indicate that CA-OsRac1 and DN-OsRac1 associate with endogenous Os RAR1, but there is no association in extracts from CS-OsRac1, which has impaired membrane localization, suggesting that membrane localization of Rac1 is required for its association with RAR1 (Figure 3A, top panel). However, in these and other co-IP experiments, we always detected two protein bands for Rac1 by anti-myc antibody (Figure 3A, bottom panel; Lieberherr et al., 2005). However, the reasons for this observation are not known.
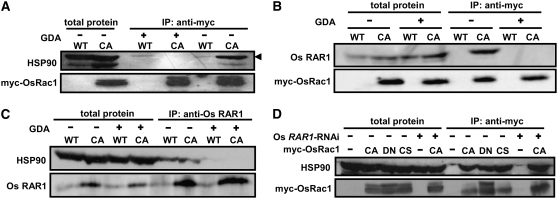
Figure 3.
Os Rac1 Forms a Complex with RAR1, HSP90, and HSP70 in Vivo.
(A) Co-IP of OsRac1 and RAR1, HSP90, and HSP70. Total protein extracts from Os Rac1 transgenic mutants were incubated with anti-myc antibody and protein A Sepharose beads. Precipitates were washed, collected by centrifugation, and separated by SDS-PAGE. Total extracted and immunoprecipitated samples from wild type cell culture were used as a control. Immunoblot analyses were performed with anti-RAR1 antibody. Os RAR1 was detected in CA and DN-OsRac1 immune complexes but not in C212S-OsRac1 (top panel). All three Os Rac1 mutants contained HSP90 (second panel). HSP70 was also detected in all three Os Rac1 mutants. Os SGT1 was not detected in Os Rac1 protein complexes (bottom panel).
(B) Immunoprecipitation with anti-RAR1 antibody and immunobloting with Os Rac1 indicates the association of RAR1 and Rac1 in complexes from CA- and DN-OsRac1 cells but not from CS-OsRac1 cells. Signals were also detected from crude extracts of transgenic but not of the nontransgenic wild-type cell culture (top panel). As a positive control, RAR1 was detected in crude extracts as well as in precipitated complexes from all cell cultures (middle panel).
(C) Confirmation of HSP90 association with Rac1 in vivo. Immunoprecipitation was performed with anti-HSP90 antibody and immunoblotted with anti-Os Rac1 (top panel) and anti-HSP90 (bottom panel) antibodies.
To confirm the association of Os Rac1 and Os RAR1, we immunoprecipitated RAR1 in protein extracts from three transgenic Rac1 cell cultures with anti-RAR1 antibody and then examined the precipitate with anti-myc antibody (Figure 3B). Os Rac1 was found in the Os RAR1 complex in CA- and DN-OsRac1 extracts but not in the CS-OsRac1 mutant, whereas RAR1 protein was detected in co-IPs from three Rac1 mutants and the untransformed control (Figure 3B, top two panels). These results indicate that RAR1 and Rac1 are part of the same protein complex in rice cell cultures.
Because RAR1 interacts with HSP 90 and we confirmed interaction of rice RAR1 and HSP90 in yeast two-hybrid assays (data not shown), we tested whether HSP90 is part of the Rac1 complex in vivo. The co-IP experiments showed that HSP90 coprecipitated with Rac1 in extracts from the three myc-OsRac1 transgenic cell cultures but not from untransformed cultured cells (Figure 3A, second panel). The association between Rac1 and HSP90 was confirmed by co-IP with anti-HSP90 antibody followed by immunoblots with anti-myc antibody (Figure 3C). These results demonstrate that Rac1 and HSP90 associate with each other in vivo.
HSP70 is known to participate with HSP90 in almost all cochaperone complexes studied in eukaryotes (Pratt and Toft, 2003). Therefore, we examined whether HSP70 also coprecipitated with Rac1 by anti-myc antibody (Figure 3A, third panel). HSP70 precipitated with anti-myc, suggesting that it is part of the Os Rac1 complex. These data suggest that Rac1 forms a multiprotein complex containing RAR1, HSP90, and HSP70, which is mainly localized at the plasma membrane, although other possibilities remain to be studied.
SGT1 interacts with RAR1 and HSP90 (Shirasu and Schulze-Lefert, 2003), and the SGT1-HSP90 association was recently shown to be crucial in NLR-mediated immune responses in mammals (da Silva Correia et al., 2007; Mayor et al., 2007). However, repeated experiments clearly indicate that SGT1 does not coimmunoprecipitate with Os Rac1 (Figure 3A), Os RAR1 (Figure 3B), or HSP90 (data not shown) under the conditions used. Because two-hybrid assays with rice RAR1, SGT1, and HSP90 showed that SGT1 is able to interact with RAR1 and HSP90 (data not shown), it is possible that SGT1 interaction with the Rac1 complex may be unstable and transient in rice cell cultures. Alternatively, SGT1 may complex with RAR1 and HSP90 in some other subcellular localizations.
RAR1 and HSP90 Are Essential for Rac1-Mediated Enhancement of Sphingolipid-Triggered Immune Responses in Rice Cell Cultures
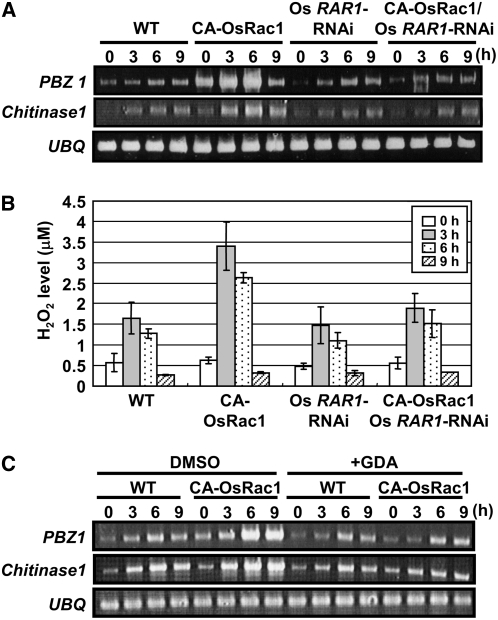
Because RAR1 and HSP90 complex with Os Rac1, we investigated whether PAMP-triggered immune responses, such as PR gene expression and ROS production, require RAR1. For this purpose, we analyzed PR gene expression in cultured cells in response to sphingolipid elicitors (SEs). SEs were isolated from the membranes of rice blast fungus and were shown to induce the accumulation of phytoalexins, cell death, increased resistance to infection by virulent pathogens, and PR gene expression (Koga et al., 1998; Umemura et al., 2000; Suharsono et al., 2002). Expression of the PR genes Probenazole1 (PBZ1) and Chitinase1 was induced in the wild type and was strongly enhanced in CA-OsRac1 cells (Figure 4A). Os RAR1-RNAi did not affect induction of PBZ1 or Chitinase1 expression. Interestingly, however, in the double transgenic CA-OsRac1/Os RAR1-RNAi cell line, PR gene expression was lower than in the CA-OsRac1 cell culture and close to wild-type levels (Figure 4A), indicating that the enhancement of PR gene expression by CA-OsRac1 was counteracted by Os RAR1-RNAi. Thus, RAR1 is essential for Rac1-mediated enhancement of PR gene expression in rice cell cultures, suggesting a functional link between Rac1 and RAR1 in PR gene induction.
Figure 4.
RAR1 and HSP90 Are Essential for Enhancement of PAMP-Triggered Immune Responses Mediated by Rac1 in Rice Cell Cultures.
(A) Suppression of SE-induced PBZ1 and Chitinase 1 by Os RAR1-RNAi in CA-OsRac1 cell cultures. SE induction of PBZ1 and Chitinase 1 was examined in the wild type, CA-OsRac1, Os RAR1-RNAi, and Os RAR1-RNAi/CA-OsRac1 double mutants after 3, 6, or 9 h of treatment with SE. Activation of SE-induced PBZ1 and Chitinase1 expression in the CA-OsRac1 cell cultures (left) was suppressed by Os RAR1-RNAi (right).
(B) Suppression of SE-induced hydrogen peroxide production by Os RAR1-RNAi in the CA-OsRac1 cell culture. Levels of H2O2 were measured 3, 6, or 9 h after SE treatment.
(C) SE-induced PBZ1 and Chitinase 1 expression in rice cell cultures in the presence or absence of GDA treatment. Cell cultures of wild type and CA-OsRac1 were treated with GDA (right) or control DMSO (left) overnight with a time course of SE treatment. Expression of PBZ1 and Chitanase1 was strongly enhanced in CA-OsRac1 cells and reduced by GDA treatment (right).
SE induces H2O2 production in rice cell cultures (Suharsono et al., 2002; Wong et al., 2004; Lieberherr et al., 2005), raising the possibility that RAR1 is involved. H2O2 production was much higher in the CA-OsRac1 cell culture (Figure 4B). However, this increase in H2O2 production was diminished in the double transgenic CA-OsRac1/Os RAR1-RNAi cell cultures (Figure 4B), indicating that, like PR gene induction, RAR1 is involved in Rac1-mediated H2O2 production in rice cell cultures.
Because HSP90 is essential for disease resistance in plants (Schulze-Lefert, 2004) and for some NLR-mediated immune responses in mammals (da Silva Correia et al., 2007; Mayor et al., 2007), its role in PAMP-triggered immune responses mediated by Os Rac1 was determined using geldanamycin (GDA), an HSP90-specific inhibitor. Wild type and CA-OsRac1 cell cultures were pretreated with GDA overnight and then treated with SE for 3, 6, and 9 h. PBZ1 and Chitinase1 were induced by SE in wild-type cells, but expression was much greater in CA-OsRac1 cells. A high level of PR gene expression was observed even at time point 0 (i.e., without SE treatment) in CA-OsRac1 transgenic cells, peaking by 6 h after SE treatment. Treatment of cell cultures with GDA resulted in a substantial decrease in PBZ1 and Chitinase1 mRNA in CA-OsRac1 cell culture (Figure 4C). These results indicate that HSP90 function is essential for Rac1-mediated enhancement of PAMP signaling. However, there was no reduction in PR gene induction in wild-type cells with GDA treatment. Together, these findings show that both RAR1 and HSP90 are essential for Rac1-mediated enhancement of PAMP-triggered immune responses in rice cell cultures.
HSP90 but Not RAR1 May Be Essential for Association with the Rac1 Complex
The nucleotide-bound state of HSP90 is known to be essential for function of its client adaptor proteins in the complex (Catlett and Kaplan, 2006). Furthermore, HSP90 function was shown to be crucial for complex formation with HSP90, SGT1, and NLR proteins such as NOD1 and NALP3 and activation of immune responses in mammals (da Silva Correia et al., 2007; Mayor et al., 2007).
To determine if HSP90 is required for the association of HSP90 and RAR1, rice cell cultures were treated with 10 μM GDA, and the interaction of Os Rac1 with HSP90 was measured with co-IP. There was no HSP90 signal with GDA treatment, indicating that functional HSP90 is essential for its association with Rac1 (Figure 5A). GDA treatment also caused dissociation of RAR1 from the Rac1 complex (Figure 5B). Furthermore, anti-RAR1 antibody was used to test whether intact HSP90 is required for HSP90-RAR1 associations. GDA treatment reduced precipitation with anti-RAR1 to nearly undetectable levels, suggesting that HSP90 is essential for association HSP90 and RAR1 (Figure 5C). Finally, we tested whether intact Os RAR1 is required for association of HSP90 in the Rac1 complex using Os RAR1-RNAi/myc-CA-OsRac1 cell culture and found that RAR1 is not required for the association of HSP90 in the Rac1 complex (Figure 5D). Together, these results indicate that HSP90 function is essential for association of HSP90 and RAR1 in the Rac1 complex in rice cell cultures.
Figure 5.
HSP90 Function but Not Os RAR1 May Be Essential for Their Association with the Os Rac1 Complex.
(A) and (B) Wild-type and CA-OsRac1 cell cultures were treated with 10 μM GDA or DMSO overnight. Total protein extracts were incubated with anti-myc antibody and protein A Sepharose beads. Precipitates were washed, collected by centrifugation, and separated by SDS-PAGE. Immunoblot analyses were performed with anti-HSP90 (A) or anti-RAR1 (B) antibody to examine the interactions of Rac1 with HSP90 and RAR1. Signals were detected only from immunocomplexes without GDA treatment. Immunoblots with anti-myc antibody were used as a control ([A], bottom panel).
(C) Immunoprecipitation performed with anti-RAR1 antibody after GDA treatment. Immunoblot analyses were performed with anti-HSP90 antibody. HSP90 was not detected in RAR1-precipitated complex after GDA treatment.
(D) RAR1 is not required for association of HSP90 in the Rac1 complex. Total protein extracts from the wild type, CA-, DN-, or CS-OsRac1, Os RAR1-RNAi, and the CA-OsRac1/Os RAR1-RNAi double transgenic mutant were incubated with anti-myc antibody. Precipitates were washed, collected by centrifugation, and separated by SDS-PAGE. Total extracted and immunoprecipitated samples from wild-type cultured cells were used as control. Immunoblot analyses were performed with anti-HSP90 antibody to examine Os Rac1-HSP90 association. Os RAR1-RNAi in the CA-OsRac1/Os RAR1-RNAi double transgenic mutant had no effect on Rac1-HSP90 association (last column).
Os RAR1 Expression Is Regulated by Os Rac1
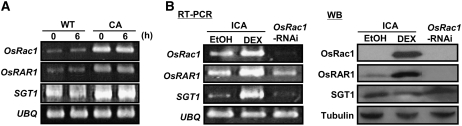
Os RAR1 protein was apparently present in higher concentrations in CA-OsRac1 cell cultures, suggesting that Os RAR1 expression is affected by Rac1. To test this hypothesis, Os RAR1 mRNA accumulation was measured by RT-PCR in wild-type and CA-OsRac1 cell cultures. Os SGT1 mRNA was also measured because SGT1 transcripts are induced by pathogen infection in rice and Arabidopsis (Cooper et al., 2003; Zimmermann et al., 2004). CA-OsRac1 overexpression cultures had much higher mRNA levels of both Os RAR1 and Os SGT1 (Figure 6A), suggesting that Os Rac1 regulates RAR1 and SGT1 expression. To further examine the regulatory relationship between Os Rac1, Os RAR1, and Os SGT1, a cell culture in which CA-OsRac1 can be induced (ICA) by treatment with dexamethasone (DEX) (Wong et al., 2004) and an Os Rac1-RNAi cell culture (Miki et al., 2005) were examined. Induction of CA-OsRac1 with DEX treatment clearly increased both transcript (Figure 6B) and protein (Figure 6B) accumulation levels of RAR1 compared with the ethanol-treated control (ICA/DEX and ICA/ethanol), and RAR1 protein was reduced in the Os Rac1-RNAi cell culture. By contrast, SGT1 mRNA accumulation correlated with Rac1 expression, but SGT1 protein levels did not change in these cell cultures (Figure 6B). These results indicate that RAR1 could be transcriptionally regulated by Rac1, but any control of SGT1 by Rac1 would be at the posttranscriptional level. The presence of some RAR1 transcript, but no protein in the Os Rac1-RNAi line, could indicate that there is another regulatory component besides Rac1 or that Rac1 also has some posttranscriptional roles in RAR1 expression. Nevertheless, these results suggest a close regulatory link between Rac1 and RAR1 at the transcriptional and possibly posttranscriptional levels.
Figure 6.
Os RAR1 Expression Is Regulated by Os Rac1.
(A) Correlation of mRNA expression levels of Os Rac1, Os RAR1, and Os SGT1. Overexpression of CA-OsRac1 strongly increased mRNA accumulation of RAR1 and SGT1 in rice cell cultures. Levels of mRNAs were measured by RT-PCR.
(B) Levels of mRNA (RT-PCR) and protein accumulation (protein gel blot [WB]) for Rac1, RAR1, and SGT1. DEX-inducible CA-OsRac1 (ICA) and an OsRac1-RNAi cell culture were examined by RT-PCR and immunoblotting after DEX induction and ethanol as a control.
DISCUSSION
RAR1 Is an Important Component in Innate Immunity in Rice
In barley, Arabidopsis, and tobacco, RAR1, together with HSP90 and SGT1, plays a key role in R gene–mediated resistance (Shirasu and Schulze-Lefert, 2003). Our results demonstrate that RAR1 is required for basal resistance against compatible races of rice blast and bacterial blight (Figures 1D to 1F) and that this resistance is mediated by Os Rac1 (Figure 2). Interestingly, Os RAR1 is not required by the three examined rice R genes for blast resistance. Recently, it was shown that Os RAR1 is not required for Pish-mediated blast resistance (Takahashi et al., 2007). Since RAR1 is not required for all R genes (Shirasu and Schulze-Lefert, 2003), these results in rice are not unexpected. We demonstrated that RAR1 is critical for Os Rac1–mediated enhancement of PAMP signaling (Figures 4A and 4B). Therefore, RAR1 activity in rice innate immunity may be on a broader scale than in those other species.
Our results are consistent with the involvement of RAR1 in basal resistance in Arabidopsis and mlo barley (Holt et al., 2005; Jarosch et al., 2005). Basal resistance is considered to be a weak response to virulent pathogens, and its molecular mechanism is not known. Thus, it is possible that basal resistance is induced by weak R proteins that recognize cognate effectors (Jones and Dangl, 2006). In this model, RAR1 function associated with basal resistance is also mediated by R proteins. More recently, RAR1 was shown to be a target of the P. syringae effector AvrB (Shang et al., 2006). Therefore, considering the observation that there are many PAMP receptors in plant cells and that RAR1 could potentially form complexes containing such receptors and other key signaling proteins, the extent of RAR1 involvement in various defense signaling pathways remains to be studied.
Our yeast two-hybrid analysis showed that Os RAR1 and HSP90 do not directly interact with Os Rac1, although Os RAR1 interacts directly with HSP90 (data not shown). Thus, it seems that Os Rac1 forms a complex with Os RAR1 and HSP90 by indirect interactions. We recently obtained evidence that Os Rac1 interacts with Sti1/Hop in vivo and in vitro (L. Chen and K. Shimamoto, unpublished data). Sti1/Hop is a cochaperone in the HSP90 complex with HSP90 and HSP70 and is highly conserved in eukaryotes (Pratt and Toft, 2003; Zhang et al., 2003). Thus, Os RAR1 and HSP90 are likely to form a complex with Os Rac1 through Sti1/Hop.
Results of RT-PCR and immunoblotting showed that the overexpression of Os Rac1 strongly increased Os RAR1 mRNA and protein accumulation (Figure 6), suggesting that Rac1 may function upstream of RAR1 in defense signaling and that the presence of a regulatory link between Os Rac1 and Os RAR1 in SE-induced signaling. Interestingly, treatment with GDA did not alter mRNA or protein accumulation of either Os RAR1 or Os SGT1 (data not shown). These results may suggest the presence of multiple regulatory pathways in which various components interact with each other at multiple levels of regulation.
HSP90 Is a Critical Regulator in Os Rac1–Mediated PAMP Signaling in Rice
Heat shock proteins are well known for regulating the maturation of protein complexes, for degrading damaged or misfolded peptides, and for involvement in the activity of many signal transduction proteins (Pratt and Toft, 2003; Rutherford, 2003). Previous studies have demonstrated that HSP90 plays a general role in R protein–mediated immunity in plants and that it acts physically close to R proteins (Hubert et al., 2003; Kanzaki et al., 2003; Lu et al., 2003; Takahashi et al., 2003; Liu et al., 2004). However, no specific role for HSP90 in protein complex formation or specific immune responses in plants has been identified.
The close association of HSP90 with Os Rac1 (Figure 3) prompted an investigation into the possible involvement of HSP90 in Rac1-mediated enhancement of PAMP signaling in rice cell cultures. To examine the function of HSP90, we used GDA, which binds the N-terminal structural domain of HSP90 and inhibits ATPase activity by blocking its highly conserved ATP binding pocket, and it has little effect on prokaryotic pathogens (Stebbins et al., 1997; Picard, 2002). GDA experiments on rice cell cultures demonstrated the requirement of HSP90 for Rac1-mediated enhancement of PAMP signaling to induce PR genes (Figure 4C). These results could be explained by the dissociation of RAR1 and HSP90 from the Rac1 complex caused by GDA (Figure 5), since it has been demonstrated that GDA treatment alters the conformation and dissociation of cochaperones, such as p23, Hop, and HSP70, from the steroid receptor complex in mammalian cells (Whitesell et al., 1994; Bagatell et al., 2001; Waza et al., 2006). These findings are similar to those in recent studies of innate immunity in mammals (da Silva Correia et al., 2007; Mayor et al., 2007), which demonstrate that GDA causes inactivation of NLR-containing protein complexes and that HSP90 is required for stability of the signaling complex.
We demonstrated the critical role of HSP90 in Os Rac1–mediated enhancement of PAMP signaling; thus, we tested whether HSP90 is also required for SE-dependant ROS production. We examined H2O2 production after GDA treatment followed by SE treatment. However, GDA failed to suppress SE-induced H2O2 production in CA-OsRac1 cell cultures (data not shown). One possible interpretation of this result is that GDA itself increases ROS generation (Dikalov et al., 2002).
With regard to the possible function of HSP70 in innate immune responses in rice, we have no information at the moment. The rice genome contains 14 HSP70 genes that can be classified into three groups (N.P. Thao and K. Shimamoto, unpublished data), much like Arabidopsis, which also contains 14 genes for HSP70 (Sung et al., 2001). We attempted to knock down expression of nine genes in group 1 and three genes in group 2 by RNAi methods employing conserved coding sequences as targets (Miki et al., 2005). However, we were not able to obtain transgenic plants in which expression of HSP70 genes in groups 1 or 2 was clearly decreased (N.P. Thao and K. Shimamoto, unpublished data). A different approach would therefore be required to determine the function of HSP70, if any, in rice innate immunity.
A Network of Proteins Involved in Innate Immunity in Rice
CA-OsRac1 activates ROS production and HR in rice and confers resistance to rice blast and bacterial blight (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002). However, the molecular mechanisms of Os Rac1 immune response regulation are largely unknown. In this study, we present evidence suggesting that Os Rac1 associates with well-studied components of plant innate immunity, such as RAR1 and HSP90, and HSP70. We have previously demonstrated that Os Rac1 also associates with Os MAPK6 (Lieberherr et al., 2005). Furthermore, a WD repeat–containing receptor of activated C kinase homolog (RACK1/RWD), which is involved in hormone signaling and development (Chen et al., 2006), specifically binds CA-OsRac1 and is essential for basal resistance to rice blast and PAMP-triggered immunity in rice (A. Nakashima and K. Shimamoto, unpublished data). RACK1/RWD is likely to be a second link between Rac1 and RAR1 because RWD interacts directly with Os RAR1 in yeast two-hybrid and in vivo immunoprecipitation assays (A. Nakashima and K. Shimamoto, unpublished data), thus confirming the observation that RAR1 is part of the Rac1 complex. These results connect Rac/Rop GTPase with known players in plant defense pathways and reveal the general importance of Rac/Rop GTPase in plant innate immunity. The observations that overexpression of CA-OsRac1 increased transcript levels of Os RAR1 and Os SGT1 and that CA-OsRac1 complements the loss of Os RAR1 in basal resistance to rice blast (Figure 2) suggest that Rac1 is a central mediator of disease resistance in rice and that it coordinates the activity of other important factors, such as RAR1, HSP90, and the MAPK cascade. Recently, we have found a direct interaction between CA-OsRac1 and the N terminus of the NADPH oxidases Os RBOHs and St RBOHB (Wong et al., 2007). Thus, it is possible that Os RAR1 and HSP90 are required for full activation of the Os Rac1 complex, at least partially through regulation of NADPH oxidase.
With respect to the intracellular localization of the Os Rac1 complex, the results obtained in this study and previous studies (Ono et al., 2001) suggest that the primary location of Rac1 complex is likely to be at the plasma membrane. However, since recent studies indicate that some R proteins also function in the nucleus (Shen and Schulze-Lefert, 2007), whether the Rac1 complex could be also located in the nucleus or the cytoplasm remains to be studied.
SGT1 plays diverse roles in plants, whereas RAR1 has much more specialized resistance functions (Shirasu and Schulze-Lefert, 2003). SGT1 was not detected in immunoprecipitated RAR1 complexes in rice cell cultures under the conditions we used, whereas yeast two-hybrid analysis showed that rice SGT1 and RAR1 directly interact (data not shown). One explanation for this is that the interaction between Os SGT1 and Os RAR1 is transient in planta. It remains to be shown whether Os RAR1 and/or Os SGT1 associate with HSP90 and Os Rac1 and R proteins in a single complex or whether they simultaneously coregulate other complexes required for downstream signaling events. One current model derived from these observations is that a network of proteins including some known components of plant innate immunity, such as Os Rac1, RAR1, SGT1, HSP90, HSP70, Os MAPK6, and RBOH (NADPH oxidase), can form one or more protein complexes.
METHODS
Biological Materials
Oryza sativa japonica var Kinmaze was used as the wild type. The Os Rac1 mutants Os Rac1-G19V (CA-OsRac1), Os Rac1-T24N (DN-OsRac1), and Os Rac1-C212S (CS-OsRac1) were described previously (Kawasaki et al., 1999; Ono et al., 2001). Os Rac1 protein was tagged with the myc epitope at the N terminus, and the expression of each construct was under the control of maize (Zea mays) Ubiquitin promoters. To make an Os RAR1-RNAi construct, a cDNA fragment amplified by PCR using two primers, Os RAR1-F (5′-TCTGAGTGAGCCTAGGGTTTG-3′) and Os RAR1-R (5′-GACCGAAGTCTCCACACACA-3′), was inserted into pANDA developed as a vector for RNAi (Miki and Shimamoto, 2004). Agrobacterium tumefaciens–mediated transformation of rice calli was performed according to a published method (Hiei et al., 1994). Plants regenerated from transformed calli were selected by hygromycin resistance. Rice suspension cell cultures expressing Os RAR1-RNAi and CA-OsRac1/Os RAR1-RNAi were also produced.
Infection of Rice Plants with Blast Fungus and Bacterial Blight
Growth conditions for Magnaporthe grisea and methods for leaf punch infection have been described previously (Takahashi et al., 1999). For bacterial blight infection, leaves were inoculated by the clipping method (Ono et al., 2001) with the Japanese Xanthomonas oryzae pv oryzae race 1 (T7174), which is compatible with var Kinmaze. Disease lesions were measured 12 d after inoculation.
Rice Cell Cultures and Elicitor Treatment
Rice cell cultures expressing CA- and DN-OsRac1 were generated as described previously (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002). For analysis of gene expression, rice cell cultures were collected after treatment with 5 μg/mL of an SE prepared from rice blast fungus.
Gene Expression
Total RNA was extracted from cultured cells and seedlings using RNeasy plant RNA extraction kit (TAKARA). One μg RNA was digested with DNaseI (TaKaRa) and reverse transcribed with Super Script II (Invitrogen). PCR reactions were performed with specific primer sets: Os Rac1 (5′-AGATAGGGCCTATCTTGCTGATCATC-3′ and 5′-ACAAGCGCTTCCGCAAAAGT-3′), Actin (5′-ACAGGAGAGAGCAGGCAGAG-3′ and 5′-TGCAAAGGAGTGAGGCTTTT-3′), PBZ1 (5′-GGGCACCATCTACACCATGAA-3′ and 5′-GTCGCACACCGCCACC-3′), Chitinase1 (5′-TCTTAACATCACTGCAACTCAG-3′ and 5′-CTGCGAGCTCTGGACAC-3′), Ubiquitin (5′-CCAGGACAAGATGATCTGCC-3′ and 5′-AAGAAGCTGAAGCATCCAGC-3′), Os SGT1 (5′-ATGGATCCATATGGCAACCGCCGCCGCG-3′ and 5′-ATGCGGCCGCTTAGTACTCCCATTTCTTAAGC-3′), and Os RAR1 (5′-TGCAAAACTGGAAAGCACAC-3′ and 5′-GGAACTGCAGGCTTCTCAAC-3′).
Protein Analysis
Rice suspension cell cultures expressing CA-OsRac1, DN-OsRac1, and C212S-OsRac1 were ground in cold extraction buffer (137 mM NaCl, 8.1 mM Na2HPO4 anhydrous, 1.47 mM Na2HPO4, pH 7.4, 10% sucrose, and complete protein inhibitor tablets [Roche]). Cell debris was removed by centrifugation at 12,000g for 25 min. Protein concentrations were determined using Bradford protein assay with BSA as the standard. Proteins were separated in 10 or 12.5% SDS polyacrylamide gels and blotted onto nitrocellulose membranes (Millipore) with a semidry electroblotting-Trans blot SD cell (Bio-Rad).
For immunodetection, membranes were incubated for 1 h with primary antibodies against the myc epitope (mouse monoclonal [Invitrogen] or rabbit polyclonal [Santa Cruz Biotechnology]). Hv RAR1 (Takahashi et al., 2003), Hv HSP90 (Takahashi et al., 2003), SGT1 (Takahashi et al., 2003), and HSP70 (Stressgen Biotechnologies), followed by anti-mouse/rat/rabbit/IgG were conjugated to horseradish peroxidase (Sigma-Aldrich). Specific protein bands were visualized with the ECL chemiluminescent Western blotting detection reagent (GE Healthcare) and Hyperfilm ECL (GE Healthcare).
For immunoprecipitations, 1 g extract (in a 1-mL volume) was incubated with protein A or G Sepharose 4 Fast Flow beads (GE Healthcare) at 4°C for 3 h. Supernatants were collected and combined with 20 μL anti-myc antibody and rotated end-over-end at 4°C for 1 h. Fifty microliters of protein A or G were added, and the incubation was continued overnight. Immunocomplexes were washed three times with 1 mL ice-cold washing buffer (extraction buffer plus 150 mM NaCl and 0.5% Triton X-100), resuspended in 20 μL SDS-PAGE sample buffer, heated to 90°C for 3 min, and separated by SDS-PAGE as described.
Quantification of H2O2
Quantification of SE-induced H2O2 production was performed with modifications described by Suharsono et al. (2002). Briefly, 0.4 g of suspension cultured cells were precultured in a fresh R2S medium at 30°C for 16 h. Cells were transferred to 2 mL of fresh medium containing SE (10 μg/mL). Aliquots were collected following incubation and filtered through a 0.22-μm filter. The filtered aliquot (100 μL) was mixed with 1 mL xylenol orange buffer [0.25 mM FeSO4, 0.25 mM (NH4)2SO4, 25 mM H2SO4, 10 mM sorbitol, and 12.5 mM xylenol orange] and incubated for 2 h at room temperature. Absorbance was measured in a spectrophotometer (Beckman) at 650 nm, and H2O2 levels were determined based on a standard curve made from known concentrations of H2O2 dissolved in the R2S medium.
For GDA treatment, GDA was diluted from a 10 mM stock in DMSO into fresh R2S medium in the preculture step (described above) to a concentration of 10 μM. DMSO alone was added into the R2S medium as a control.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AB029508 (Os Rac1), AK111881 (Os RAR1), AAF18438 (Os SGT1), D38170 (PBZ1), and D16221 (Os Chitinase1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Basal Resistance to Rice Blast and Bacterial Blight Infection in Os RAR1 RNAi T0 Plants.
Supplemental Figure 2. Os RAR1 Transcript Levels Are Consistent with Protein Levels in Os RAR1-RNAi Rice Cultured Cell Lines.
Supplementary Material
Acknowledgments
We thank Mika Nobuhara, Yuko Tamaki, and Masako Hamane for technical assistance. This research was supported by Grants-in-Aid from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project IP4001) and the Japan Society for Promotion of Science (13G0023) to K.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ko Shimamoto (simamoto@bs.naist.jp)
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Betsuyaku, S., Peart, J., Takahashi, A., Noel, L., Sadanandom, A., Casais, C., Parker, J., and Shirasu, K. (2006). Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295 2073–2076. [DOI] [PubMed] [Google Scholar]
- Bagatell, R., Khan, O., Paine-Murrieta, G., Taylor, C.W., Akinaga, S., and Whitesell, L. (2001). Destabilization of steroid receptors by heat shock protein 90-binding drugs: A ligand-independent approach to hormonal therapy of breast cancer. Clin. Cancer Res. 7 2076–2084. [PubMed] [Google Scholar]
- Bieri, S., Mauch, S., Shen, Q.H., Peart, J., Devoto, A., Casais, C., Ceron, F., Schulze, S., Steinbiss, H.H., Shirasu, K., and Schulze-Lefert, P. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett, M.G., and Kaplan, K.B. (2006). Sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J. Biol. Chem. 281 33739–33748. [DOI] [PubMed] [Google Scholar]
- Chen, J.G., Ullah, H., Temple, B., Liang, J., Guo, J., Alonso, J.M., Ecker, J.R., and Jones, A.M. (2006). RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J. Exp. Bot. 57 2697–2708. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Cooper, B., Clarke, J.D., Budworth, P., Kreps, J., Hutchison, D., Park, S., Guimil, S., Dunn, M., Luginbühl, P., Ellero, C., Goff, S.A., and Glazebrook, J. (2003). A network of rice genes associated with stress response and seed development. Proc. Natl. Acad. Sci. USA 100 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Correia, J., Miranda, Y., Leonard, N., and Ulevitch, R. (2007). SGT1 is essential for Nod1 activation. Proc. Natl. Acad. Sci. USA 104 6764–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikalov, S., Landmesser, U., and Harrison, D.G. (2002). Geldanamycin leads to superoxide formation by enzymatic and non-enzymatic redox cycling. Implications for studies of Hsp90 and endothelial cell nitric-oxide synthase. J. Biol. Chem. 277 25480–25485. [DOI] [PubMed] [Google Scholar]
- Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., Yang, F., Chu, Z., Wang, G.L., White, F.F., and Yin, Z. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435 1122–1125. [DOI] [PubMed] [Google Scholar]
- Gu, Y., Wang, Z., and Yang, Z. (2004). ROP/RAC GTPase: An old new master regulator for plant signaling. Curr. Opin. Plant Biol. 7 527–536. [DOI] [PubMed] [Google Scholar]
- Hahn, J.S. (2005). Regulation of Nod1 by Hsp90 chaperone complex. FEBS Lett. 579 4513–4519. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Holt III, B.F., Belkhadir, Y., and Dangl, J.L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309 929–932. [DOI] [PubMed] [Google Scholar]
- Hubert, D.A., Tornero, P., Belkhadir, Y., Krishna, P., Takahashi, A., Shirasu, K., and Dangl, J.L. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch, B., Collins, N.C., Zellerhoff, N., and Schaffrath, U. (2005). RAR1, ROR1, and the actin cytoskeleton contribute to basal resistance to Magnaporthe grisea in barley. Mol. Plant Microbe Interact. 18 397–404. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kanzaki, H., Saitoh, H., Ito, A., Fujisawa, S., Kamoun, S., Katou, S., Yoshioka, H., and Terauchi, R. (2003). Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 4 383–391. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T., Koita, H., Nakatsubo, T., Hasegawa, K., Wakabayashi, K., Takahashi, H., Umemura, K., Umezawa, T., and Shimamoto, K. (2006). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 103 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4 21–33. [DOI] [PubMed] [Google Scholar]
- Koga, J., Yamauchi, T., Shimura, M., Ogawa, N., Oshima, K., Umemura, K., Kikuchi, M., and Ogasawara, N. (1998). Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J. Biol. Chem. 273 31985–31991. [DOI] [PubMed] [Google Scholar]
- Lieberherr, D., Thao, N.P., Nakashima, A., Umemura, K., Kawasaki, T., and Shimamoto, K. (2005). A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice 1. w. Plant Physiol. 138 1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Burch-Smith, T., Schiff, M., Feng, S., and Dinesh-Kumar, S.P. (2004). Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279 2101–2108. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S.P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30 415–429. [DOI] [PubMed] [Google Scholar]
- Lu, R., Malcuit, I., Moffett, P., Ruiz, M.T., Peart, J., Wu, A.J., Rathjen, J.P., Bendahmane, A., Day, L., and Baulcombe, D.C. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder, W., Yoshioka, K., and Klessig, D.F. (2005). Involvement of the small GTPase Rac in the defense responses of tobacco to pathogens. Mol. Plant Microbe Interact. 18 116–124. [DOI] [PubMed] [Google Scholar]
- Matsuzawa, S.I., and Reed, J.C. (2001). Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7 915–926. [DOI] [PubMed] [Google Scholar]
- Mayor, A., Martinon, F., De Smedt, T., Petrilli, V., and Tschopp, J. (2007). A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat. Immunol. 8 497–503. [DOI] [PubMed] [Google Scholar]
- Miki, D., Itoh, R., and Shimamoto, K. (2005). RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 138 1903–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45 490–495. [DOI] [PubMed] [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, E., Wong, H.L., Kawasaki, T., Hasegawa, M., Kodama, O., and Shimamoto, K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard, D. (2002). Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 59 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, W.B., and Toft, D.O. (2003). Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228 111–133. [DOI] [PubMed] [Google Scholar]
- Rutherford, S.L. (2003). Between genotype and phenotype: Protein chaperones and evolvability. Nat. Rev. Genet. 4 263–274. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert, P. (2004). Plant immunity: The origami of receptor activation. Curr. Biol. 14 R22–R24. [PubMed] [Google Scholar]
- Shang, Y., Li, X., Cui, H., He, P., Thilmony, R., Chintamanani, S., Zwiesler-Vollick, J., Gopalan, S., Tang, X., and Zhou, J.M. (2006). RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 103 19200–19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q.H., and Schulze-Lefert, P. (2007). Rumble in the nuclear jungle: Compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 26 4293–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.W., Zhou, F., Azevedo, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99 355–366. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., and Schulze-Lefert, P. (2003). Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci. 8 252–258. [DOI] [PubMed] [Google Scholar]
- Stebbins, C.E., Russo, A.A., Schneider, C., Rosen, N., Hartl, F.U., and Pavletich, N.P. (1997). Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell 89 239–250. [DOI] [PubMed] [Google Scholar]
- Suharsono, U., Fujisawa, Y., Kawasaki, T., Iwasaki, Y., Satoh, H., and Shimamoto, K. (2002). The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 99 13307–13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, Y.S., Vierling, E., and Guy, C.L. (2001). Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 126 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Agrawal, G.K., Yamazaki, M., Onosato, K., Miyao, A., Kawasaki, T., Shimamoto, K., and Hirochika, H. (2007). Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell 9 2940–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Casais, C., Ichimura, K., and Shirasu, K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Kawasaki, T., Henmi, K., Shii, K., Kodama, O., Satoh, H., and Shimamoto, K. (1999). Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 17 535–545. [DOI] [PubMed] [Google Scholar]
- Tornero, P., Merritt, P., Sadanandom, A., Shirasu, K., Innes, R.W., and Dangl, J.L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura, K., Ogawa, N., Yamauchi, T., Iwata, M., Shimura, M., and Koga, J. (2000). Cerebroside elicitors found in diverse phytopathogens activate defense responses in rice plants. Plant Cell Physiol. 41 676–683. [DOI] [PubMed] [Google Scholar]
- Wang, Y.S., Pi, L.Y., Chen, X., Chakrabarty, P.K., Jiang, J., De Leon, A.L., Liu, G.Z., Li, L., Benny, U., Oard, J., Ranal, P.C., and Song, W.Y. (2006). Rice XA21 binding protein 3 is a ubiquitin ligase required for full XA21-mediated disease resistance. Plant Cell 18 3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waza, M., Adachi, H., Katsuno, M., Minamiyama, M., Tanaka, F., Doyu, M., and Sobue, G. (2006). Modulation of Hsp90 function in neurodegenerative disorders: a molecular-targeted therapy against disease-causing protein. J. Mol. Med. 84 635–646. [DOI] [PubMed] [Google Scholar]
- Whitesell, L., Mimnaugh, E.G., De Costa, B., Myers, C.E., and Neckers, L.M. (1994). Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA 91 8324–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, H.L., Pinontoan, R., Hayashi, K., Tabata, R., Yaeno, T., Hasegawa, K., Kojima, C., Yoshioka, H., Iba, K., Kawasaki, T., and Shimamoto, K. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, H.L., Sakamoto, T., Kawasaki, T., Umemura, K., and Shimamoto, K. (2004). Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 135 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Qi, M., and Mei, C. (2004). Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40 909–919. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Quick, M.K., Kanelakis, K.C., Gijzen, M., and Krishna, P. (2003). Characterization of a plant homolog of hop, a cochaperone of hsp90. Plant Physiol. 131 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C., and Felix, G. (2005). Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 8 353–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.