Abstract
Natural variation in gene expression (expression traits or e-traits) is increasingly used for the discovery of genes controlling traits. An important question is whether a particular e-trait is correlated with a phenotypic trait. Here, we examined the correlations between phenotypic traits and e-traits among 10 Arabidopsis thaliana accessions. We studied defense against Pseudomonas syringae pv tomato DC3000 (Pst), with a focus on resistance gene–mediated resistance triggered by the type III effector protein AvrRpt2. As phenotypic traits, we measured growth of the bacteria and extent of the hypersensitive response (HR) as measured by electrolyte leakage. Genetic variation among accessions affected growth of Pst both with (Pst avrRpt2) and without (Pst) the AvrRpt2 effector. Variation in HR was not correlated with variation in bacterial growth. We also collected gene expression profiles 6 h after mock and Pst avrRpt2 inoculation using a custom microarray. Clusters of genes whose expression levels are correlated with bacterial growth or electrolyte leakage were identified. Thus, we demonstrated that variation in gene expression profiles of Arabidopsis accessions collected at one time point under one experimental condition has the power to explain variation in phenotypic responses to pathogen attack.
INTRODUCTION
Genetic variation among wild-type populations of plants is an important source of information about biological traits. The information potential of these natural accessions has long been recognized and is increasingly exploited to uncover genetic loci controlling biological traits, for example, using quantitative trait loci (QTL) analysis (Perchepied et al., 2006). Similarly, gene expression can be used to dissect complex traits in combination with natural variation, for example, by expression QTL (eQTL) analyses in which the expression level of a gene is considered as a trait (expression trait or e-trait) (Schadt et al., 2003; Keurentjes et al., 2007). Gene expression profiling technology is usually used for study of e-traits and eQTLs because it allows simultaneous detection and scoring of numerous e-traits and, consequently, numerous eQTLs. For use of e-traits and eQTLs to be successful for discovery of genetic loci controlling biological traits, it is essential to understand (1) how genotypic variation is correlated with variation in gene expression profiles and (2) how variation in gene expression profiles is correlated with phenotypic variation. Recently, the correlation between genotypic variation and variation in gene expression profiles was studied in detail in seven Arabidopsis thaliana accessions (Kliebenstein et al., 2006). Here, we investigate the correlation between variation in gene expression profiles and phenotypic variation among 10 Arabidopsis accessions.
We investigated natural variation in the context of the plant response to pathogen attack. During early stages of infection, plants recognize molecules that are common among large groups of microbes, called microbe-associated molecular patterns (MAMPs), as signals of pathogen attack and turn on defense responses (Jones and Dangl, 2006). This MAMP-triggered defense is the cause of basal resistance, which is considered to be the first layer of inducible plant resistance. However, successful pathogens have acquired effectors that interfere with MAMP-triggered defense and overcome basal resistance. For example, the Pseudomonas syringae effectors AvrRpt2 and AvrRpm1 are known to perturb MAMP-triggered defense by modifying the Arabidopsis protein RIN4 (Kim et al., 2005). The next layer of plant defense is initiated by specific recognition by plant resistance (R) proteins of particular pathogen effectors or biochemical changes caused by effectors (Jones and Dangl, 2006). This recognition leads to rapid induction of defense responses and the hypersensitive response (HR). R gene–mediated resistance is typically very effective. Many defense responses are induced by both basal and R gene–mediated resistance, while others are specific to one or the other (Tao et al., 2003).
We focused on resistance mediated by the Arabidopsis R gene RPS2 against infection by the bacterial pathogen P. syringae pv tomato DC3000 (Pst) carrying the type III effector gene avrRpt2 (Pst avrRpt2). The interaction between the products of the avrRpt2 gene and the RPS2 resistance gene is one of the best-studied examples of R gene–mediated resistance (Axtell and Staskawicz, 2003; Mackey et al., 2003). It is clear that many genes are involved in the responses downstream of RPS2 (Tao et al., 2003). Thus, as long as the genotypes studied contain functional RPS2, this interaction can be considered a complex trait. Additionally, RPS2-mediated resistance is accompanied by dramatic changes in expression levels of many genes. This latter characteristic facilitates the identification of many potential candidate genes influencing the trait and therefore makes this an ideal system for studying natural variation in complex traits using gene expression profiling.
We analyzed gene expression profiles of plants infected with Pst avrRpt2 and sampled at a single time point. We found variation in these expression profiles among accessions. We also observed variation in phenotypes, such as growth of Pst avrRpt2, growth of Pst, and extent of the HR. Remarkably, for each of these phenotypes, we could identify subsets of gene expression profiles that were well correlated with phenotype. This finding indicates that the loci that control e-traits could also control biological traits and justifies the eQTL approach for discovery of QTLs that control important traits.
RESULTS
Phenotypic Characterization 1: Arabidopsis Accessions Show Variation in Resistance against Pst and Pst avrRpt2
We analyzed variation in growth, defined as the number of colony-forming units (cfu) of bacteria present in 1 cm2 of Arabidopsis leaf tissue of Pst avrRpt2 in 10 different Arabidopsis accessions. The accessions were chosen based on different geographic and climatic origins (see Supplemental Figure 1 online) and well represent known genetic variation among Arabidopsis accessions (Nordborg et al., 2005; Kliebenstein et al., 2006). As a control, we also assessed the growth of Pst avrRpt2 in the rps2 mutant. This mutant carries the null rps2-101C allele in the Columbia (Col-0) background (Mindrinos et al., 1994). Among the 45 possible pairwise comparisons among accessions, 39 showed significant differences in growth of Pst avrRpt2 (q < 0.05; Table 1). The rps2 mutant supported the highest growth of Pst avrRpt2 (Figure 1). Thus, there is considerable quantitative variation in growth of Pst avrRpt2 among these 10 accessions (Figure 1, Table 1). Broad-sense heritability indicates what proportion of the phenotypic variation (in this case, variation in the bacterial growth) can be explained by the genotypic variation (in this case, the accession difference). The remaining portion of the phenotypic variation could be caused by some other source of variation, such as uncontrolled environmental difference among replicated experiments. The broad-sense heritability of the resistance against Pst avrRpt2 1 and 2 d after inoculation was 58 and 70%, respectively (see Supplemental Table 1 online). Thus, a large part of the observed variation in growth of Pst avrRpt2 could be explained by genetic differences among Arabidopsis accessions.
Table 1.
Pairwise Comparisons between Genotypes
| Gene Expression Profilesa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial Growthb
|
Responsivenessc
|
|||||||||
| Genotype 1 | Genotype 2 | Pst | Pst avrRpt2 | Δcfu | ELd | Mocke | Pst avrRpt2e | Total | Mock | Pst avrRpt2 |
| Col-0 | Col-rps2 | NS | *** | *** | + | – | – | – | – | – |
| Col-0 | Cvi-1 | *** | NS | * | + | 137 | 110 | 78 | 39 | 30 |
| Col-0 | Est-1 | *** | *** | NS | + | 216 | 135 | 123 | 64 | 29 |
| Col-0 | Kas-1 | *** | ** | * | + | 267 | 131 | 146 | 101 | 19 |
| Col-0 | Kin-0 | * | *** | *** | + | 33 | 171 | 91 | 2 | 77 |
| Col-0 | Ler-0 | NS | NS | NS | + | 187 | 72 | 119 | 73 | 25 |
| Col-0 | Mt-0 | *** | *** | ** | + | 15 | 49 | 11 | 0 | 9 |
| Col-0 | Tsu-1 | NS | *** | *** | + | 36 | 66 | 43 | 6 | 27 |
| Col-0 | Van-0 | ** | *** | *** | + | – | – | – | – | – |
| Col-0 | Ws-2 | * | ** | NS | + | 11 | 141 | 101 | 4 | 74 |
| Col-rps2 | Cvi-1 | *** | *** | *** | + | – | – | – | – | – |
| Col-rps2 | Est-1 | *** | *** | *** | + | – | – | – | – | – |
| Col-rps2 | Kas-1 | *** | *** | *** | + | – | – | – | – | – |
| Col-rps2 | Kin-0 | * | *** | *** | − | – | – | – | – | – |
| Col-rps2 | Ler-0 | NS | *** | *** | + | – | – | – | – | – |
| Col-rps2 | Mt-0 | *** | *** | *** | + | – | – | – | – | – |
| Col-rps2 | Tsu-1 | NS | *** | *** | + | – | – | – | – | – |
| Col-rps2 | Van-0 | *** | *** | NS | − | – | – | – | – | – |
| Col-rps2 | Ws-2 | * | *** | *** | + | – | – | – | – | – |
| Cvi-1 | Est-1 | *** | *** | ** | − | 119 | 180 | 37 | 9 | 19 |
| Cvi-1 | Kas-1 | *** | ** | *** | + | 164 | 151 | 22 | 7 | 10 |
| Cvi-1 | Kin-0 | NS | *** | *** | + | 88 | 115 | 39 | 11 | 18 |
| Cvi-1 | Ler-0 | ** | NS | ** | − | 64 | 81 | 7 | 1 | 4 |
| Cvi-1 | Mt-0 | *** | *** | NS | − | 91 | 102 | 23 | 9 | 12 |
| Cvi-1 | Tsu-1 | * | ** | NS | − | 52 | 60 | 2 | 0 | 2 |
| Cvi-1 | Van-0 | *** | *** | *** | + | – | – | – | – | – |
| Cvi-1 | Ws-2 | * | ** | *** | + | 71 | 123 | 9 | 1 | 4 |
| Est-1 | Kas-1 | *** | *** | * | − | 54 | 77 | 7 | 2 | 3 |
| Est-1 | Kin-0 | *** | *** | ** | + | 201 | 230 | 110 | 38 | 50 |
| Est-1 | Ler-0 | *** | *** | NS | − | 83 | 160 | 30 | 4 | 20 |
| Est-1 | Mt-0 | *** | * | ** | + | 183 | 136 | 74 | 37 | 19 |
| Est-1 | Tsu-1 | *** | *** | *** | − | 160 | 164 | 64 | 22 | 27 |
| Est-1 | Van-0 | *** | *** | *** | + | – | – | – | – | – |
| Est-1 | Ws-2 | *** | *** | NS | + | 147 | 225 | 82 | 22 | 52 |
| Kas-1 | Kin-0 | *** | *** | *** | + | 251 | 237 | 98 | 38 | 32 |
| Kas-1 | Ler-0 | *** | *** | NS | − | 102 | 146 | 7 | 1 | 6 |
| Kas-1 | Mt-0 | NS | *** | *** | + | 256 | 136 | 89 | 59 | 19 |
| Kas-1 | Tsu-1 | *** | NS | *** | + | 213 | 160 | 42 | 13 | 16 |
| Kas-1 | Van-0 | *** | *** | *** | + | – | – | – | – | – |
| Kas-1 | Ws-2 | *** | *** | * | − | 222 | 232 | 51 | 18 | 25 |
| Kin-0 | Ler-0 | * | *** | *** | + | 154 | 121 | 43 | 23 | 12 |
| Kin-0 | Mt-0 | *** | *** | *** | + | 34 | 144 | 54 | 7 | 43 |
| Kin-0 | Tsu-1 | NS | *** | *** | + | 33 | 96 | 24 | 2 | 19 |
| Kin-0 | Van-0 | *** | NS | ** | − | – | – | – | – | – |
| Kin-0 | Ws-2 | NS | *** | ** | + | 45 | 143 | 49 | 7 | 37 |
| Ler-0 | Mt-0 | *** | *** | ** | + | 138 | 55 | 1 1 | 5 | 4 |
| Ler-0 | Tsu-1 | NS | *** | *** | − | 70 | 43 | 4 | 1 | 2 |
| Ler-0 | Van-0 | ** | *** | *** | + | – | – | – | – | – |
| Ler-0 | Ws-2 | NS | NS | NS | + | 87 | 64 | 7 | 0 | 3 |
| Mt-0 | Tsu-1 | *** | *** | ** | − | 21 | 46 | 0 | 0 | 0 |
| Mt-0 | Van-0 | * | *** | *** | + | – | – | – | – | – |
| Mt-0 | Ws-2 | *** | *** | * | + | 22 | 101 | 27 | 1 | 22 |
| Tsu-1 | Van-0 | *** | *** | *** | + | – | – | – | – | – |
| Tsu-1 | Ws-2 | NS | *** | *** | + | 31 | 57 | 7 | 0 | 4 |
| Van-0 | Ws-2 | *** | *** | *** | + | – | – | – | – | – |
–, Not measured.
NS, not significant; *, q < 0.05; **, q < 0.01; ***, q < 0.001.
“Responsiveness” indicates a comparison between the genotypes in the log ratio of expression values between Pst avrRpt2– and mock-infected samples for each gene. “Total” indicates the total number of the genes for which the log-ratio values are significantly different between the genotypes. Among the “Total” genes, “Mock” indicates the number of the genes for which the log-ratio difference was due to the difference in the “Mock” expression values, while “Pst avrRpt2” indicates the number of the genes of which the log-ratio difference was due to the difference in the Pst avrRpt2 expression values. Note that the sum of the numbers in “Mock” and “Pst avrRpt2” are smaller than that in “Total.” This is because in some genes, both “Mock” and “Pst avrRpt2” expression values, as well as the log ratios, were different between the accessions.
−, Not significant; +, significant, based on 95% confidence intervals of ΔΔconductivity.
These are the numbers of the genes in which the log-transformed expression values between the accessions are significantly different in mock- and Pst avrRpt2–infected samples, respectively.
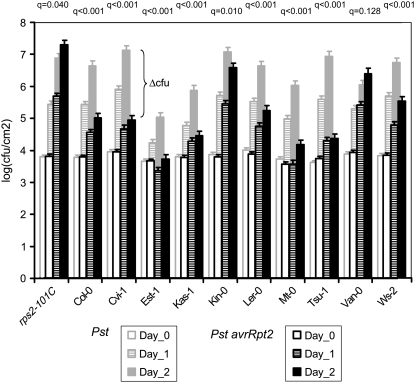
Figure 1.
The Growth of Pst and Pst avrRpt2 Is Highly Variable among Different Arabidopsis Accessions.
Titers of Pst (gray borders) and Pst avrRpt2 (black borders) were measured 0 (white), 1 (striped), and 2 (gray and black, respectively) d after inoculation. Significant variation among accessions was found for both bacterial strains at day one and day two (see text for details). The rps2 mutant was analyzed as a control. Values and se (error bars) of bacterial growth were calculated using analysis of variance (ANOVA) of the raw data. The figure also illustrates how Δcfu (the difference in growth of Pst and Pst avrRpt2) can be calculated. Results from t tests comparing growth of Pst avrRpt2 with growth of Pst per plant genotype are indicated by q values.
To determine whether the variation in growth of Pst avrRpt2 is due to variation in AvrRpt2-induced responses, such as R gene–mediated resistance and virulence effects of AvrRpt2, we also analyzed growth of Pst without avrRpt2. Again, a large part of the observed variation in growth of Pst could be attributed to genetic differences among Arabidopsis accessions, as the broad-sense heritabilities were 50 and 43% 1 and 2 d after inoculation, respectively (see Supplemental Table 1 online). However, due to larger variation within the accessions, the broad-sense heritability was lower than that of resistance against Pst avrRpt2.
To determine which accessions show AvrRpt2-induced resistance, we compared the growth of these two bacterial strains in each accession. Two days after inoculation, all accessions except Van-0 showed more growth of Pst than Pst avrRpt2 (q < 0.05), demonstrating that in these accessions AvrRpt2 triggers R gene–mediated resistance. By contrast, Pst avrRpt2 grew better than Pst in the rps2 mutant, demonstrating the contribution of AvrRpt2 to virulence in Col-0 plants that do not recognize AvrRpt2 (Kim et al., 2005). Van-0 supported growth of both strains to similar levels, indicating that in this accession AvrRpt2 does not trigger R gene–mediated resistance (Figure 1).
To investigate the differences in resistance to Pst avrRpt2 among accessions, we used pairwise comparisons to test for differences in growth of Pst, differences in growth of Pst avrRpt2, and differences in Δcfu, the log ratio of growth between Pst and Pst avrRpt2 (Figure 1, Table 1). We envision several classes of mechanisms that together explain resistance to Pst and Pst avrRpt2. Class I includes basal resistance mechanisms that become redundant when AvrRpt2-induced resistance is triggered. Differences in these mechanisms among accessions should affect Pst growth but not the growth of Pst avrRpt2. If Class I mechanisms are the only difference between two accessions, both Pst growth and Δcfu would be different, but growth of Pst avrRpt2 would not. Class II includes basal resistance mechanisms that do not become redundant when AvrRpt2-induced resistance is triggered. Differences in these mechanisms should affect the growth of Pst and Pst avrRpt2 equally. If Class II mechanisms are the only difference between accessions, growth of both Pst and Pst avrRpt2 would be different, but Δcfu would not. Class III includes AvrRpt2-specific mechanisms. Differences in these mechanisms should affect the growth of Pst avrRpt2 but not the growth of Pst. If Class III mechanisms are the only difference between accessions, both growth of Pst avrRpt2 and Δcfu would be different, but growth of Pst would not. If all three measurements (growth of Pst, growth of Pst avrRpt2, and Δcfu) are different between accessions, either (1) more than one class of mechanisms are different or (2) basal resistance and AvrRpt2-induced responses share signaling pathways to induce a common set of responses to different extents and the accessions differ in some of these shared pathways. We cannot distinguish these two possibilities. From the 36 possible pairwise comparisons among the nine accessions that showed AvrRpt2-induced resistance (Van-0 excluded), three pairwise comparisons showed differences only in Class I mechanisms (P [Pst avrRpt2] > 0.05, P [Pst] < 0.05, and P [Δcfu] < 0.05; e.g., Col-0 and Cvi-1), seven pairwise comparisons showed differences only in Class II mechanisms (P [Pst avrRpt2] < 0.05, P [Pst] < 0.05, and P [Δcfu] > 0.05; e.g., Col-0 and Est-1), and seven pairwise comparisons showed differences only in Class III mechanisms (P [Pst avrRpt2] < 0.05, P [Pst] > 0.05, and P [Δcfu] < 0.05; e.g., Col-0 and Tsu-1; Figure 1, Table 1). Additionally, 17 pairwise comparisons showed differences in growth of Pst, growth of Pst avrRpt2, and Δcfu (P [Pst avrRpt2] < 0.05, P [Pst] < 0.05, and P [Δcfu] < 0.05; e.g., Col-0 and Kas-1; Figure 1, Table 1). Two comparisons did not show any differences (e.g., Col-0 and Ler-0; Figure 1, Table 1). Thus, the variation in resistance to Pst avrRpt2 among the chosen accessions can be explained by variation in both basal resistance and AvrRpt2-induced responses.
Phenotypic Characterization 2: Arabidopsis Accessions Show Variation in Extent of the HR
Analysis of the growth of the two different bacterial strains on different accessions as discussed above is a collective measurement of plant defenses and bacterial responses. To study defense responses more specifically, we analyzed the HR triggered by recognition of Pst avrRpt2 using an electrolyte leakage assay (Figure 2; see Supplemental Figures 2 to 4 online). Based on 95% confidence intervals of the difference in electrolyte leakage between Pst avrRpt2– and Pst-inoculated plants (Δconductivity; Figures 2A and 2B), two accessions (Kin-0 and Van-0) did not show significant differences in the electrolyte leakage between Pst-inoculated and Pst avrRpt2–inoculated plants (Figure 2D; see Supplemental Figure 3 online). Thus, in these accessions, AvrRpt2 does not trigger HR. As expected, the same was true for the rps2 mutant (Figures 2B and 2D; see Supplemental Figure 3 online). We also asked whether the difference between electrolyte leakage by Pst-inoculated plants and Pst avrRpt2–inoculated plants varied between accessions (ΔΔconductivity) (Figure 2C, Table 1; see Supplemental Figure 4 online). Among the 28 pairs of accessions that did show AvrRpt2-induced HR (excluding Kin-0 and Van-0), 17 showed differences in induction of HR at some time points during the measured 24-h interval, demonstrating that extensive variation in both timing and extent of AvrRpt2-induced HR exists among accessions (Table 1). The broad-sense heritability values reach up to 65% after inoculation with Pst avrRpt2 (see Supplemental Table 1 online), indicating that genetic differences among Arabidopsis accessions can explain a large part of the variation in HR. The broad-sense heritability after inoculation of Pst was much lower (maximum 38%). This is probably because low electrolyte leakage that was caused by mechanisms other than HR in the assays with Pst was not as robust and, therefore, not as reproducible as high electrolyte leakage that was caused by HR in the assays with Pst avrRpt2.
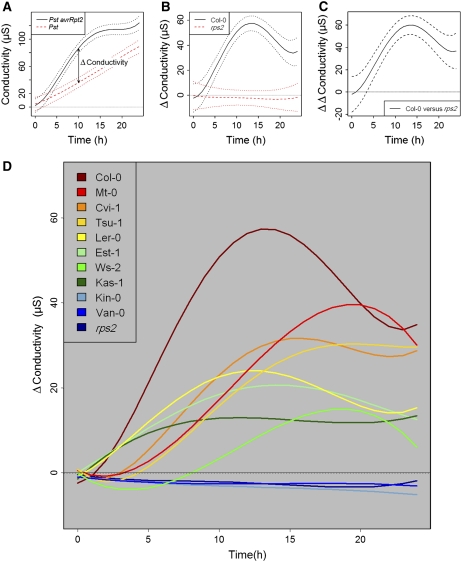
Figure 2.
Variation in HR among the Accessions Was Revealed by an Electrolyte Leakage Assay.
Conductivity changes of water containing leaf discs inoculated with Pst or Pst avrRpt2 were measured over a 24-h time period for all genotypes.
(A) Conductivity changes of Col-0 inoculated with Pst (red) or Pst avrRpt2 (black). Dashed lines indicate 95% confidence intervals. This graph also illustrates how the Δconductivity (the difference between conductivity after Pst avrRpt2 and Pst inoculation) can be calculated. See Supplemental Figure 2 online for similar graphs of all genotypes.
(B) Changes in the Δconductivity over time for Col-0 (black) and Col-rps2 (red). Dashed lines indicate 95% confidence intervals. The graph shows that for Col-0 after ∼3 h, 95% confidence intervals do not include a Δconductivity of zero. By contrast, 95% confidence intervals of Col-rps2 include a Δconductivity of zero during the complete 0- to 24-h time span, illustrating the absence of HR in Col-rps2 after inoculation with Pst avrRpt2. See Supplemental Figure 3 online for similar graphs of all genotypes.
(C) Changes in the ΔΔconductivity (the difference in the Δconductivity for two selected genotypes) over time for Col-0 and Col-rps2. After ∼3 h, the 95% confidence intervals do not include a ΔΔconductivity of zero, indicating that from this time point, Δconductivity of Col-0 and Col-rps2 differ significantly. See Supplemental Figure 4 online for similar graphs of all the pairwise comparisons.
(D) ΔConductivity of all genotypes without 95% confidence intervals.
As HR is considered an induced plant defense response, we tested for a correlation between HR and bacterial growth among accessions. We compared HR measured by conductivity after inoculation with Pst avrRpt2 from 0 to 24 h at 3-h intervals with titer of Pst avrRpt2 0, 1, and 2 d after inoculation. Only data from the eight accessions that showed AvrRpt2-induced HR (excluding Kin-0 and Van-0) were included. The bacterial titer 0 d after inoculation and conductivity 0 h after inoculation were included as negative controls: no correlations between HR and bacterial growth at zero time points were expected, and indeed no significant correlation was found. In fact, the broad-sense heritabilities of the bacterial titer on zero day and the conductivity at zero hour were <6% (see Supplemental Table 1 online). More interestingly, there was no significant correlation between conductivity at any time point and titer of Pst avrRpt2 at any day (maximum r2 = 0.40, df = 6, q = 0.44) or between Δcfu and Δconductivity (maximum r2 = 0.55, df = 6, q = 0.17) among accessions. Thus, variation in HR does not correlate with variation in resistance to Pst avrRpt2.
Variation in RPS2 Coding Sequence or Expression Cannot Fully Explain Variation in Phenotypic Responses among Arabidopsis Accessions
From the phenotypic analyses, we can conclude that there is considerable variation in AvrRpt2-induced responses among the accessions. As RPS2 is essential for AvrRpt2-induced R gene–mediated resistance, differences in the coding region of RPS2 and/or differences in RPS2 expression could potentially explain the observed differences. To address this, we sequenced the entire RPS2 coding region from all the accessions (Table 2) and analyzed expression of RPS2 in the nine accessions showing R gene–mediated resistance (excluding Van-0) 6 h after Pst avrRpt2 or mock inoculation using quantitative RT-PCR (qRT-PCR) (Figure 3).
Table 2.
Nucleotide and Amino Acid Polymorphisms in the RPS2 Sequence
| nta | 142 | 311 | 426 | 461 | 704 | 1092 | 1233 | 1245 | 1255 | 1311 | 1315 | 1326 | 1359 | 1374 | 1414 | 1438 | 1440 | 1458 | 1543 | 1544 | 1548 | 1569 | 1634 | 1698 | 1815 | 1923 | 1926 | 1931 | 1945 | 1946 | 1958 | 2097 | 2109 | 2334 | 2498 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Col-0 | A | A | A | C | G | A | C | A | T | C | C | C | T | T | C | T | G | G | G | T | C | C | C | G | A | C | T | A | G | G | C | A | A | C | G |
| rps2 | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Cvi-1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | C |
| Est-1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . |
| Kas-1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | C |
| Kin-0 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | C | T | . | . |
| Ler-0 | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . |
| Mt-0 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . |
| Tsu-1 | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | G | C |
| Van-0 | G | G | C | . | . | T | T | T | C | T | A | . | C | C | T | C | A | A | . | . | T | T | A | T | G | G | A | G | C | A | T | . | T | . | C |
| Ws-2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . |
| aaa | 48 | 104 | 154 | 235 | 419 | 439 | 472 | 515 | 515 | 545 | 644 | 649 | 653 | 699 | 703 | 778 | 833 | ||||||||||||||||||
| Col-0 | I | Y | S | W | S | H | H | V | V | S | E | G | A | E | E | N | R | ||||||||||||||||||
| V | C | Y | .b | P | N | Y | L | A | Y | G | Q | V | D | D | K | T |
nt, nucleotide position, starting from start codon; aa, amino acid position.
Stop codon.
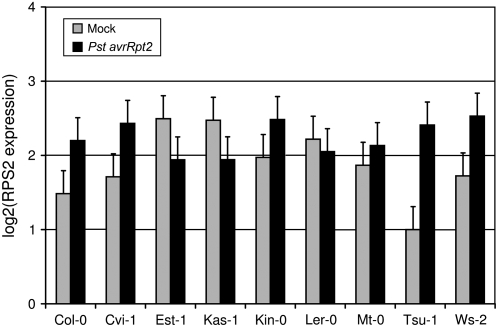
Figure 3.
The Expression Level of RPS2 Has Limited Variation among the Accessions.
RPS2 expression was measured using qRT-PCR and quantified using ΔCt (the difference in cycle number to reach a set threshold of product amount) between RPS2 and Actin2, 6 h after mock inoculation (gray) or inoculation with Pst avrRpt2 (black). Values and se of gene expression were obtained using ANOVA of the raw data.
Some of the observed phenotypic variation may be explained by variation in RPS2 sequence. For example, the RPS2 sequence of Van-0 is identical to that of Po-1. Like Van-0, Po-1 does not show RPS2-mediated resistance (Banerjee et al., 2001). Based on sequence analyses of RPS2, Mauricio et al. (2003) divided Arabidopsis accessions into two clades. One clade, which includes Po-1, consists mainly of susceptible accessions; the other clade consists of resistant accessions. Similarly, the RPS2 sequence of Kin-0 is identical to that of Yo-0. Like Kin-0, Yo-0 shows only marginal RPS2-mediated resistance (Mauricio et al., 2003). Both examples illustrate that RPS2 sequence may explain part of the observed variation, although it is also possible that these accessions share sequence identity at other loci that can explain this variation. However, accessions with the same RPS2 sequence, such as Est-1, Mt-0, and Ws-2, showed significant differences with respect to bacterial growth and HR. Thus, variation in RPS2 coding sequence cannot explain all the phenotypic variation observed.
RPS2 expression showed moderate variation among mock-treated accessions (a 2.8-fold difference between highest and lowest expression) and low variation among Pst avrRpt2–treated accessions (a 1.5-fold difference between highest and lowest expression; Figure 3). The broad-sense heritability of RPS2 expression was higher in mock-inoculated than in Pst avrRpt2–inoculated accessions (49 and 19%, respectively; see Supplemental Table 1 online). Overall, Pst avrRpt2 inoculation affects RPS2 expression with on average a 1.4-fold induction (df = 1, F = 6.5, P = 0.015 for the treatment factor in ANOVA). There is significant variation in the effect of Pst avrRpt2 on RPS2 expression among accessions (df = 8, F = 2.5, P = 0.027 for the genotype:treatment interaction in ANOVA). Comparing RPS2 expression after the Pst avrRpt2 treatment only, there were no significant differences among accessions. Comparing RPS2 expression after mock treatment only, Tsu-1 was significantly different from Est-1 (q = 0.041) and Kas-1 (q = 0.041). Thus, much of the variation in RPS2 expression among accessions is due to variation between Est-1 and Tsu-1 and Kas-1 and Tsu-1 after mock inoculation. Additionally, RPS2 expression 6 h after inoculation with Pst avrRpt2 was not significantly correlated with growth of Pst avrRpt2 at day one or day two (r2 = 0.41 and 0.40, respectively, df = 7 and q = 0.10 for both), and no significant correlations were found between expression of RPS2 after Pst avrRpt2 inoculation and HR at any time point (maximum r2 = 0.44, df = 6, q = 0.60). Thus, variation in RPS2 expression among accessions 6 h after inoculation with Pst avrRpt2 cannot explain the variation of growth of Pst avrRpt2 or the variation in HR.
Inoculation with Pst avrRpt2 Strongly Affects the Variation in Gene Expression Profiles among Accessions
To investigate whether variation in gene expression profiles can be related to variation in phenotypic responses, we first measured the mRNA levels of 571 genes, 465 of which are pathogen responsive and 106 are stably expressed in Col-0, using the Arabidopsis miniarray (Pathoarray_464) (Sato et al., 2007). To assess how inoculation with Pst avrRpt2 changes variation among accessions, we measured gene expression in both mock-inoculated and Pst avrRpt2–inoculated plants 6 h after inoculation. In a previous study, gene expression profiles of Col-0 showed dramatic expression changes 6 h after inoculation with Pst avrRpt2 (Tao et al., 2003). We limited this analysis to the nine accessions that show AvrRpt2 induced resistance (Van-0 excluded) based on Δcfu and/or Δconductivity (Figures 1 and 2).
Noise reduction by removing genes that did not show gene expression levels higher than a negative control (a probe that does not match any Arabidopsis or Pst sequence) in any of the genotype treatment combinations removed 105 genes from the data set (P < 0.01), leaving 466 genes (see Supplemental Figure 5 online). Of these 466 genes, 436 were induced or repressed by Pst avrRpt2 treatment in at least one accession (q < 0.05), and more than one-third of these (167) were responsive in all accessions. As the majority of the genes on the miniarray were selected based on their induction or repression upon pathogen or viral attack, it is not surprising that most of these genes are responsive to Pst avrRpt2 treatment in at least one accession.
Subsequently, we analyzed the gene expression profiles by pairwise comparisons of accessions (Table 1). Of the 466 genes, 409 showed variation in at least one pair of Pst avrRpt2–inoculated accessions (q < 0.05). Thus, considerable variation exists among gene expression profiles after inoculation with Pst avrRpt2. However, some of this variation may not depend on the interaction with Pst avrRpt2. Indeed, 386 genes showed variation in at least one pair of mock-treated accessions (q < 0.05). To assess whether genes are differentially induced or repressed by treatment with Pst avrRpt2, we compared the log ratio of expression values in Pst avrRpt2–inoculated and mock-treated plants between accession pairs for each gene: 284 genes showed variation in the log ratio of Pst avrRpt2–inoculated and mock-treated plants in at least one pair of accessions (q < 0.05). Of these 284 genes that are differentially induced or repressed among accessions, 90 genes show similar basal expression levels but reach different expression levels after treatment with Pst avrRpt2, 49 genes show different basal expression levels but reach a similar expression level after treatment with Pst avrRpt2, and 139 show differences in both basal expression levels and expression levels after treatment with Pst avrRpt2. Thus, considerable variation exists among expression profiles of Pst avrRpt2–inoculated accessions, and much of this variation is caused by differential induction or repression of genes.
More detailed information on specific accession pairs is shown in Table 1. In some accession pairs, very few genes were differentially induced or repressed (e.g., Cvi-1 and Tsu-1: two genes), whereas other accession pairs showed extensive differences in gene induction or repression (e.g., Col-0 and Kas-1: 146 genes). As described above, in general more genes were differentially induced or repressed due to differences in Pst avrRpt2–inoculated plants than to differences in mock-inoculated plants (53% versus 27%). However, this also varied considerably among accession pairs. For example, between Col-0 and Kas-1, only 13% (19/146) of the differentially induced or repressed genes were different in Pst avrRpt2–inoculated plants only, while 69% were different in mock-inoculated plants only, the remaining genes being differentially expressed after both treatments. By contrast, between Col-0 and Ws-2, 73% (74/101) of the differentially induced or repressed genes were different in Pst avrRpt2–inoculated plants only, and just 4% were different in mock-inoculated plants only. Thus, when interested in gene expression changes related to resistance against Pst avrRpt2, it may be more worthwhile to compare, for example, Col-0 and Ws-2 than Col-0 and Kas-1.
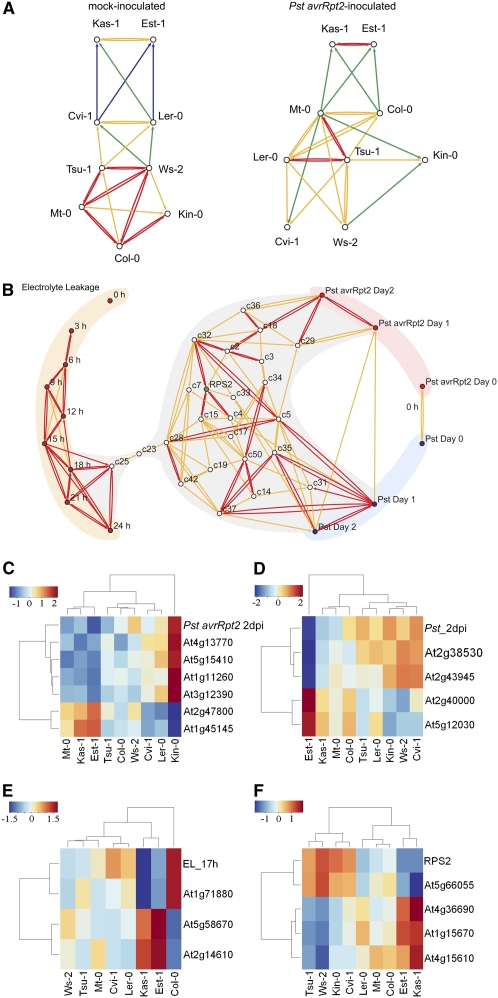
The variation in expression profiles among accessions was further explored using the algorithm locally linear embedding graph generator (LEGG). LEGG uses locally linear embedding (Roweis and Saul, 2000) to perform nonlinear dimensionality reduction and defines the relationships among expression profiles based on the type and the degree of similarities among them: Directed links are made to a particular expression profile from the most similar expression profiles, while directed links that correspond to redundant information are removed by the dimensionality reduction procedure. Thus, LEGG uses the same logic as local context finder (LCF) (Katagiri and Glazebrook, 2003), but it uses a different algorithm (see Methods). Unlike LCF, LEGG has an option to consider expression profiles with large negative correlations to share similarities, so exactly opposite expression profile patterns are considered the same. When a regulatory pathway results in upregulation of some genes and downregulation of others, both the upregulated and downregulated genes report the signal strength in the regulatory pathway. Assuming that differences among accessions are due to differences in signal strength, completely opposite expression patterns likely indicate the same regulatory pathway. Therefore, in this study, the negative correlation option of LEGG was used. LEGG analysis was performed with the expression profiles after mock and Pst avrRpt2 inoculation for the nine accessions (Figure 4A). From this analysis, several observations can be made. First, the expression profiles, which are indicated as nodes in the figure, were organized into two disconnected graphs: one contains the profiles of the mock-inoculated accessions, and the other contains the profiles of the Pst avrRpt2–inoculated accessions. As the miniarray is highly enriched with pathogen-responsive genes, this is not surprising. Second, many directed links between nodes for accessions change in response to treatment with Pst avrRpt2. Of the 27 directed links among mock-inoculated accessions and 27 directed links among Pst avrRpt2–inoculated accessions, only 14 overlap, and even in the overlapping directed links, the strength of the connections changes. Thus, treatment with Pst avrRpt2 changes the relationships among profiles of accessions considerably, illustrating large effects of the treatment on variation in gene expression profiles among accessions.
Figure 4.
Visualization of the Relationships among Expression Profiles.
(A) LEGG was applied to gene expression profiles of the accessions 6 h after mock inoculation or inoculation with Pst avrRpt2. The expression values of the 466 selected genes are the parameters of each profile. Expression profiles are indicated as nodes in the graph, and the relationships among the profiles that were determined by LEGG are depicted as directed links. Gene expression profiles are clearly affected by the different treatments, resulting in two disconnected graphs: mock-inoculated accessions (left) and accessions inoculated with Pst avrRpt2 (right). The directed links in these two graphs can be compared based on their presence, absence, or strength. Strength of connections measured by the r2 value is indicated by color: blue, 0.951 to 0.964; green, 0.964 to 0.974; orange, 0.974 to 0.984; and red, 0.984 to 0.990.
(B) LEGG was applied to expression profiles of the gene clusters and the phenotypic data profiles with the accessions as the parameters of each profile. Clusters of genes (white nodes) were identified based on correlated expression using the data 6 h after inoculation with Pst avrRpt2. The gene clusters that do not have links to the shown network are not included. Centered and scaled data for both gene expression and phenotypic data across the accessions was used as input. The analysis shows that electrolyte leakage (brown nodes) is only weakly connected to the major part of the gene expression graph, whereas bacterial growth (Pst, blue nodes; Pst avrRpt2, red nodes) is much more connected to the gene expression graph. RPS2 expression level (green node) is embedded in the center of the gene expression graph. Strength of the connections (r2): orange, 0.58 to 0.76; red, 0.76 to 1. The colors of the shaded background indicate the different types of data: red, growth of Pst avrRpt2; blue, growth of Pst; orange, electrolyte leakage of Pst avrRpt2–inoculated plants; gray, gene expression clusters.
(C) to (F) High correlations between expression profiles of member genes in selected gene clusters and phenotypic data across the accessions are illustrated by a heat map. Data were centered and scaled across the accessions before applying agglomerative hierarchical clustering with average linkage to generate the heat map. The selected gene expression clusters are the ones that are most correlated to the phenotypic data (see text, Table 3, Figure 4, and Supplemental Table 2 online for details).
(C) Titer of Pst avrRpt2 2 d after inoculation and expression of genes in cluster c29.
(D) Titer of Pst and expression of genes in cluster c35.
(E) Electrolyte leakage at 17 h after inoculation with Pst avrRpt2 and expression of genes in cluster c25.
(F) RPS2 expression and expression of genes in cluster c15. Note that some genes shown are strongly negatively correlated. The color scale is depicted in the top left part of each panel.
In summary, there is extensive variation in gene expression profiles among accessions after inoculation with Pst avrRpt2, and on average 49% of this variation can be explained by genetic differences among accessions (see Supplemental Table 1 online). Although some of the variation is present after mock inoculation, most of it is specifically caused by differential induction or repression after treatment with Pst avrRpt2. When comparing pairs of accessions, substantial deviations from this general trend can be found, with some accession pairs mainly showing differences after mock inoculation, whereas other accession pairs mainly show differences after inoculation with Pst avrRpt2. These results are indicative of significant phenotypic plasticity among accessions. This is illustrated in the LEGG analysis: accessions that are strongly connected after mock inoculation are not strongly connected after inoculation with Pst avrRpt2 and vice versa.
HR, Bacterial Growth, and RPS2 Expression Show Distinct Correlations with the Expression of Clusters of Genes
Clearly, there is significant variation among accessions in both phenotypic responses, as measured by variation in bacterial growth and HR and in treatment-dependent gene expression. Some of this variation (or lack thereof) may share the same genetic origin. For example, between Cvi-1 and Tsu-1, there were only two genes that showed differential induction or repression, and Cvi-1 and Tsu-1 did not show any variation in phenotypic responses, whereas between Col-0 and Kas-1, many genes were differentially induced or repressed, and these accessions showed significant variation in both bacterial growth and electrolyte leakage phenotypes. However, global results from gene expression profiling, such as the number of responsive genes or average responsiveness, do not correlate well with phenotypic data such as bacterial growth and HR (data not shown). To explore covariation between gene expression profiles and phenotypic data in more detail, we analyzed the correlation between phenotypic data and the gene expression profiles.
Separation of signal from noise is a problem with analyses of many possible predictors (i.e., genes). To address this problem, we used two sequential methods. First, we ran an ANOVA on the Pst avrRpt2–inoculated data set only and selected genes (360) that showed a significant genotype effect, thus selecting genes that show significant variation in expression among Pst avrRpt2–inoculated accessions. Second, we clustered genes using hierarchical clustering (see Methods), thus drastically reducing the number of comparisons between gene expression and phenotypic data and thereby increasing statistical power. This resulted in 28 clusters containing three or more genes (see Supplemental Table 2 online). Genes within a cluster showed correlated expression profiles over the different accessions and thus likely share upstream signaling factors, for example, regulation by the same transcription factor. Additionally, clustering filters out possible effects of sequence variation that affect binding of cRNA to the probes of the miniarray: It is likely that the measured expression differences of some genes are actually (partially) due to different probe binding properties of the mRNA because of sequence differences among Arabidopsis accessions. However, it is highly unlikely that similar patterns of probe efficiency variation across the accessions occur in more than one gene. Selection of clusters with multiple genes therefore eliminates this type of false signal. Thus, clustering has statistical, biological, and methodological advantages.
After clustering the genes, we analyzed the correlation between the expression of each cluster and phenotypic data (both bacterial growth and HR) and RPS2 expression. For bacterial growth, we used titers of the two bacterial strains on different days; for electrolyte leakage, we used the electrolyte leakage of Pst avrRpt2–inoculated plants in 3-h intervals; and for RPS2 expression, we used the data obtained after inoculation with Pst avrRpt2. Most phenotypic data correlated with the expression of at least one gene cluster, suggesting that genes in these clusters, or other genes not present on the miniarray but controlled by the same regulator, may influence the correlated phenotypic responses (Table 3, Figures 4C to 4F). For example, five clusters were correlated with growth of Pst on both day one and day two, and five other clusters were correlated with growth of Pst on day two only (q < 0.05). These correlations make biological sense if these genes induced or repressed by Pst avrRpt2 are also induced or repressed by Pst. Indeed, previous analyses of gene expression profile changes of Col-0 after inoculation with Pst and Pst avrRpt2 indicated that many genes are responsive to both treatments, albeit to a different extent and on a different time scale (Tao et al., 2003). Other clusters correlated with AvrRpt2-induced responses: Two clusters were correlated with growth of Pst avrRpt2 on day two, and one cluster was correlated with electrolyte leakage between 15 and 24 h after Pst avrRpt2 inoculation (q < 0.05). Additionally, 10 clusters were correlated with RPS2 expression (q < 0.05). Gene clusters that show correlated expression with RPS2 may indicate genes that are closely linked with RPS2 in the signaling network.
Table 3.
Correlations between Gene Expression Patterns and Phenotypic Data
| Growth of Pst
|
Growth of Pst avrRpt2
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster No. | Genes per Cluster | Day 1
|
Day 2
|
Day 1
|
Day 2
|
Electrolyte Leakage (18 h)
|
RPS2 Expression
|
||||||
| r2 | q | r2 | q | r2 | q | r2 | q | r2 | q | r2 | q | ||
| c2 | 10 | 0.45 | 0.09 | 0.41 | 0.12 | 0.47 | 0.13 | 0.56 | 0.09 | 0.01 | 0.86 | 0.71 | 0.03 |
| c3 | 3 | 0.25 | 0.21 | 0.20 | 0.27 | 0.25 | 0.29 | 0.33 | 0.16 | 0.00 | 0.93 | 0.46 | 0.08 |
| c4 | 3 | 0.71 | 0.02 | 0.61 | 0.04 | 0.45 | 0.13 | 0.52 | 0.10 | 0.42 | 0.29 | 0.79 | 0.01 |
| c5 | 30 | 0.77 | 0.02 | 0.72 | 0.04 | 0.58 | 0.13 | 0.59 | 0.09 | 0.25 | 0.41 | 0.65 | 0.03 |
| c7 | 5 | 0.43 | 0.10 | 0.41 | 0.12 | 0.21 | 0.29 | 0.37 | 0.16 | 0.48 | 0.26 | 0.58 | 0.05 |
| c8 | 4 | 0.04 | 0.64 | 0.04 | 0.64 | 0.15 | 0.35 | 0.08 | 0.50 | 0.23 | 0.41 | 0.06 | 0.55 |
| c10 | 3 | 0.06 | 0.59 | 0.08 | 0.52 | 0.28 | 0.28 | 0.32 | 0.16 | 0.24 | 0.41 | 0.20 | 0.25 |
| c14 | 4 | 0.38 | 0.12 | 0.30 | 0.20 | 0.06 | 0.54 | 0.05 | 0.57 | 0.16 | 0.43 | 0.37 | 0.11 |
| c15 | 4 | 0.57 | 0.05 | 0.52 | 0.07 | 0.21 | 0.29 | 0.24 | 0.24 | 0.23 | 0.41 | 0.84 | 0.01 |
| c16 | 3 | 0.46 | 0.09 | 0.37 | 0.14 | 0.19 | 0.31 | 0.11 | 0.44 | 0.16 | 0.43 | 0.46 | 0.08 |
| c17 | 4 | 0.30 | 0.17 | 0.26 | 0.23 | 0.09 | 0.48 | 0.15 | 0.36 | 0.18 | 0.43 | 0.33 | 0.13 |
| c18 | 4 | 0.31 | 0.17 | 0.31 | 0.20 | 0.54 | 0.13 | 0.77 | 0.03 | 0.03 | 0.75 | 0.51 | 0.06 |
| c19 | 5 | 0.35 | 0.15 | 0.29 | 0.20 | 0.22 | 0.29 | 0.35 | 0.16 | 0.36 | 0.32 | 0.44 | 0.09 |
| c21 | 3 | 0.00 | 0.92 | 0.02 | 0.74 | 0.00 | 0.91 | 0.04 | 0.59 | 0.14 | 0.43 | 0.04 | 0.61 |
| c23 | 3 | 0.23 | 0.23 | 0.24 | 0.24 | 0.15 | 0.35 | 0.29 | 0.19 | 0.58 | 0.26 | 0.30 | 0.14 |
| c25 | 3 | 0.55 | 0.05 | 0.60 | 0.04 | 0.45 | 0.13 | 0.47 | 0.12 | 0.87 | 0.02 | 0.33 | 0.13 |
| c28 | 3 | 0.63 | 0.05 | 0.63 | 0.04 | 0.48 | 0.13 | 0.60 | 0.09 | 0.46 | 0.26 | 0.61 | 0.04 |
| c29 | 6 | 0.57 | 0.05 | 0.61 | 0.04 | 0.75 | 0.07 | 0.78 | 0.03 | 0.48 | 0.26 | 0.39 | 0.11 |
| c31 | 4 | 0.76 | 0.02 | 0.66 | 0.04 | 0.48 | 0.13 | 0.36 | 0.16 | 0.19 | 0.43 | 0.63 | 0.04 |
| c32 | 6 | 0.57 | 0.05 | 0.58 | 0.05 | 0.45 | 0.13 | 0.55 | 0.09 | 0.23 | 0.41 | 0.80 | 0.01 |
| c33 | 3 | 0.56 | 0.05 | 0.63 | 0.04 | 0.41 | 0.16 | 0.41 | 0.15 | 0.14 | 0.43 | 0.69 | 0.03 |
| c34 | 4 | 0.32 | 0.17 | 0.23 | 0.24 | 0.23 | 0.29 | 0.40 | 0.15 | 0.12 | 0.44 | 0.39 | 0.11 |
| c35 | 4 | 0.82 | 0.01 | 0.80 | 0.03 | 0.55 | 0.13 | 0.50 | 0.11 | 0.17 | 0.43 | 0.69 | 0.03 |
| c36 | 4 | 0.19 | 0.28 | 0.22 | 0.25 | 0.39 | 0.16 | 0.62 | 0.09 | 0.15 | 0.43 | 0.45 | 0.08 |
| c37 | 5 | 0.81 | 0.01 | 0.75 | 0.04 | 0.36 | 0.19 | 0.34 | 0.16 | 0.52 | 0.26 | 0.54 | 0.06 |
| c39 | 3 | 0.04 | 0.63 | 0.01 | 0.78 | 0.09 | 0.48 | 0.08 | 0.50 | 0.40 | 0.29 | 0.04 | 0.61 |
| c42 | 5 | 0.46 | 0.09 | 0.44 | 0.11 | 0.27 | 0.28 | 0.39 | 0.15 | 0.49 | 0.26 | 0.37 | 0.11 |
| c50 | 4 | 0.58 | 0.05 | 0.45 | 0.11 | 0.23 | 0.29 | 0.28 | 0.19 | 0.31 | 0.38 | 0.54 | 0.06 |
| Electrolyte leakage (21 h) | |||||||||||||
| c29ma | 3 | 0.80 | 0.04 | ||||||||||
Significant correlations (q < 0.05) are in bold.
For gene expression clusters from mock-inoculated accessions, only the cluster that showed significant correlation is shown for a selected electrolyte leakage time point.
As the genes on the small-scale microarray were selected for broad representation of diverse expression patterns defined in Col-0 and for easy measurement of expression levels, rather than representation of biological processes, the selected genes are unlikely to adequately represent biological processes of potential interest. Furthermore, many clusters are so small that they are not appropriate for statistical analysis of biological processes associated with gene clusters. For these reasons, we did not attempt to associate biological processes with the clusters. However, these results do demonstrate that gene expression profiling at a single time point after Pst avrRpt2 infection contains information that can be correlated to several phenotypic responses. Additionally, these gene expression profiles likely contain information about the structure of the underlying signaling network. The correlation analyses also gives us some insight into how the signaling network connects different phenotypic responses, as several clusters show correlation with multiple phenotypic responses. For example, cluster 18 is specifically correlated with growth of Pst avrRpt2 and cluster 35 is specifically correlated with growth of Pst. Other clusters are correlated with more than one phenotypic response, such as cluster 29, which is correlated with growth of both bacterial strains, and cluster 25, which is correlated with HR and with growth of Pst. Thus, clusters correlating with several phenotypic responses may indicate overlaps in the signaling networks influencing the different phenotypes. Interestingly, of the 10 clusters correlated with growth of Pst, seven overlapped with clusters correlated with RPS2 expression, suggesting a role of RPS2 in basal resistance or coregulation of RPS2 with basal resistance. It was reported that an inducer of the basal defense response, flg22, induces RPS2 expression (Zipfel et al., 2004) and that a mutation in RPS2 does not affect the growth of a hrcC mutant of Pst (Katagiri and Sato, 2007). These facts indicate that RPS2 is coregulated with basal resistance but does not play a crucial role in basal resistance against Pst.
As a negative control, we also investigated whether any patterns were significantly correlated with bacterial titer at day zero, right after inoculation. No patterns correlated with bacterial titer at day zero for either bacterial strain (q ≥ 0.62). These results strengthen our conclusion that gene expression profiles contain information that is relevant to the phenotypic responses.
We performed the same analyses using gene expression profiles after mock inoculation. ANOVA resulted in the selection of 349 genes that showed significant variation in expression levels among accessions after mock treatment. Gene clustering resulted in 15 clusters of three or more genes. None of these clusters showed significant correlation with titer of either Pst or Pst avrRpt2 at any time point, but one cluster correlated with electrolyte leakage at 21 and 24 h (Table 3). Clearly, few differences in phenotypic responses can be explained by differences in expression profiles after mock inoculation. Thus, it appears that differences in basal gene expression levels do not strongly affect the phenotypic variation. By contrast, certain expression changes after Pst avrRpt2 infection are strongly correlated with phenotypic variation. This implies that an eQTL approach using uninfected plants has a very limited chance of detecting loci involved in resistance.
To visualize the relationships between gene expression patterns, HR, and bacterial growth, we used LEGG to embed the phenotypic data in a gene expression graph. As LEGG uses dimensionality reduction, not all significant correlations between gene expression clusters are represented by direct links in the LEGG analyses. Still, the LEGG analysis of combined phenotypic and gene expression data reflects and integrates many of the results from the statistical analyses (Figure 4B). First, Figure 4B shows many more links between the gene expression graph and bacterial growth than between the gene expression graph and HR. The gene cluster connected with HR is not strongly connected to the rest of the network. Thus, these results confirm the lack of strong correlation between HR and bacterial growth and show that this lack of correlation is reflected in separate parts of the gene expression graph that are associated with either phenotypic response. RPS2 expression level is embedded in the center of the graph and shows no direct connections with phenotypic data. Again, this fits with the observation that RPS2 expression is not correlated with the phenotypic data. Thus, it appears that variation in signaling downstream or independent of RPS2 is causing much of the observed variation in phenotypic responses to Pst avrRpt2 among accessions.
DISCUSSION
Phenotypic Characterization and Gene Expression Profiling of Arabidopsis Accessions Show Distinct and Shared Mechanisms of Resistance against Pst and Pst avrRpt2
We have demonstrated that variation exists in RPS2-mediated resistance among Arabidopsis accessions (Figure 1, Table 1). Such variation has been reported previously and as far as the accessions used overlap, our data confirm previous results (Caicedo et al., 1999; Mauricio et al., 2003). By analyzing variation in Δcfu and variation in growth of Pst and Pst avrRpt2, we can separate variation in resistance to Pst between accession pairs into four distinct classes. Some of the variation in resistance to Pst and Pst avrRpt2 among accessions suggests that the molecular mechanisms underlying basal and R gene–mediated resistance in Arabidopsis have shared features. The strong interconnectedness of gene expression clusters that are associated with growth of the two bacterial strains (Figure 4B) also support overlapping molecular mechanisms. Obviously, this overlap is not complete, with some variation specifically affecting one bacterial strain but not the other. Thus, our results with natural accessions of Arabidopsis corroborate the conclusion that basal and R gene–mediated resistance in Arabidopsis have both distinct and shared features (Tao et al., 2003).
Phenotypic Characterization and Gene Expression Profiling of Arabidopsis Accessions Show Uncoupling of HR and Resistance
In line with our finding that there is variation in the level of resistance among accessions that show AvrRpt2-induced resistance, there is also variation in the HR among accessions, as measured by electrolyte leakage (Figure 2, Table 1). However, there is no significant correlation between the variation in bacterial growth and HR. As HR-associated cell death may not be an appropriate defense to the hemibiotrophic Pst, the discrepancy between variation in cell death and bacterial growth may not be unexpected. However, the lack of correlation indicates that there is either relatively little variation between signaling events affecting both HR and bacterial growth among the chosen accessions or that signaling events affecting HR and signaling events affecting bacterial growth are not tightly connected. Indeed several Arabidopsis mutants show AvrRpt2-induced resistance with little or no HR (Clough et al., 2000; Jurkowski et al., 2004), and several Pseudomonas effector proteins induce R gene–mediated resistance without HR (Gassmann et al., 1999; Gassmann, 2005). Gene expression profiling reflected the lack of correlation between HR and resistance, with different clusters correlating to HR and to resistance (Table 3). Moreover, the gene clusters connected to HR are not well integrated into the gene expression graph that is connected to resistance (Figure 4B).
Gene Expression Profiling Identifies Robust yet Variable Responses to Pst avrRpt2
From these results, it is clear that infection with Pst avrRpt2 results in dramatic changes in gene expression, and these changes correlate with phenotypic responses induced by Pst avrRpt2. Interestingly, approximately one-third of the Pst avrRpt2–responsive genes are responsive in all accessions (see Supplemental Figure 5 online). Salicylic acid is a plant hormone that plays a key role in resistance against Pst avrRpt2 and other biotrophic pathogens. A recent study of the effect of salicylic acid on gene expression in seven accessions, all of which were included in our study, found that very few genes showed conserved induction (van Leeuwen et al., 2007). Thus, it appears that infection with a pathogen results in a much more robust change in gene expression profile than treatment with a single hormone, suggesting a high level of redundancy in signaling pathways affecting gene expression profiles and resistance.
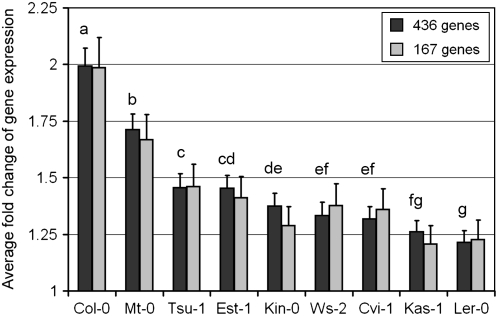
Even though many genes are responsive in all accessions, there is considerable variation in the extent of this response (Table 1). For example, when considering the average response of all 436 responsive genes, Ler-0 and Kas-1 are least responsive to Pst avrRpt2 (average absolute fold changes of 1.2 and 1.3; Figure 5), whereas Col-0 is most responsive (average absolute fold change of 2.0). Limiting the analyses to the 167 genes that are responsive in all accessions does not change these results dramatically (Figure 5). Thus, it is not only the number of responsive genes but also the extent of induction or repression that varies among accessions. In the previously mentioned study including the same accessions except for Kas-1, Ler-0, and Ws-2, van Leeuwen et al. (2007) demonstrated that Cvi-1 is hyporesponsive to salicylic acid, whereas Mt-0 is hyperresponsive to salicylic acid with respect to induction and repression of genes. Our data on those seven accessions show that Cvi-1 was least responsive, whereas Mt-0 showed the second highest responsiveness. Our observations after treatment with Pst avrRpt2 therefore resemble the observations after treatment with salicylic acid. Thus, even though there may be a high level of redundancy in signaling pathways, this redundancy seems only partial such that salicylic acid signaling has a strong effect.
Figure 5.
Average Fold Change of Gene Expression in Response to Pst avrRpt2.
The average fold change was determined using both induction and repression at 6 h after inoculation. Thus, a gene whose expression is twofold induced in response to Pst avrRpt2 and a gene whose expression is twofold reduced in response to Pst avrRpt2 both have a twofold response. The average fold change of these two genes would thus be 2. The average fold change is shown using all 436 genes that are responsive in at least one accession (black bars) or the 167 genes that are responsive in all accessions (gray bars). Letters above the black bars indicate significant differences among accessions after multiple t tests comparing average responsiveness among accession pairs using false discovery rate correction. The t tests were performed using the data of all 436 responsive genes.
From Correlation to Causal Effect
In this article, we have demonstrated that variation in gene expression patterns and phenotypic responses among natural accessions is correlated. This correlation indicates that variation in phenotypic plasticity is reflected in variation in gene expression patterns and that these gene expression patterns contain predictive information on phenotypic responses. However, correlations do not necessarily signify causal effects. To demonstrate causal effects, we need to identify the trans-acting genetic loci that affect clusters of genes and assess their effects on phenotypic variation. Identification of these loci can be achieved using a segregating population from a cross between two parental accessions in an eQTL approach. The information from expression profiles can aid us in selecting the parental accessions. For example, when interested in identifying trans-acting genetic loci that specifically affect R gene–mediated resistance, it may be more worthwhile to select Col-0 and Ws-2 as parental lines rather than Col-0 and Kas-1, even though Col-0 and Kas-1 show more significant phenotypic differences. This is because between Col-0 and Kas-1, differential induction or repression of genes is mainly caused by differences in expression profiles after mock inoculation, whereas differential induction or repression of genes between Col-0 and Ws-2 is mainly caused by differences in expression profiles after inoculation with Pst avrRpt2. Thus, we can quickly screen many candidate parental lines and select the most promising combinations using the results presented here. We are currently undertaking an eQTL approach to identify regulatory loci affecting clusters of genes and assess the effect of these loci on Arabidopsis resistance against Pst avrRpt2.
METHODS
Plants and Bacteria
Arabidopsis thaliana accessions were chosen to represent diverse geographic origins. Seeds of Arabidopsis accessions Col-0 (ABRC stock number CS22625), Cvi-1 (CS8580), Est-1 (CS22629), Kas-1 (CS22638), Kin-0 (22,654), Ler-0 (CS20), Mt-0 (CS22642), Tsu-1 (CS1640), Van-0 (CS22627), Ws-2 (CS2360), and the rps2 mutant rps2-101C (Col-0 background) (Mindrinos et al., 1994) were suspended in 0.1% agar for a minimum of 4 d at 4°C. Cold-treated seeds were transferred to 12 × 12 × 5-cm (l × w × h) pots filled with twice-autoclaved, sterile soil (either BM-2 soil from Berger Peat Moss or LG3 germinating mix from Sun Gro Horticulture). Plants were grown in a controlled-environment chamber at 22°C, with 80% RH and a 10/14 h (R gene expression and gene expression profiling) or 12/12 h (all other experiments) light/dark cycle. Plants used were either 5 (R gene expression and gene expression profiling) or 4 (all other experiments) weeks old. We noticed that Col-0 plants used in this study (CS22625) reproducibly showed higher bacterial growth than Col-0 plants that were used for the genome sequencing (CS60000). We do not know genetic difference underlying this phenotypic difference.
Pseudomonas syringae pv tomato DC3000 strains containing either avrRpt2 (Pst avrRpt2) or the empty pLAFR3 plasmid vector (Pst) were grown for 2 d on King's B plates containing appropriate antibiotics and subsequently were grown overnight in liquid King's B, again containing appropriate antibiotics (25 μg/mL rifampicin and 10 μg/mL tertracycline). Inoculum was prepared and hand-infiltrated as described by Katagiri et al. (2002), at indicated doses, using a needleless syringe.
Bacterial Growth Analysis
Zero, one, and two days after inoculation with either Pst avrRpt2 or Pst OD600 = 0.001 (1 × 106 cfu/mL), two leaf discs (total surface 0.57 cm2) were punched from a single leaf using a new plant for each time point. Leaf discs were pulverized in 400 μL 5 mM MgSO4, and a dilution series of the suspension was made. Of each dilution, 10 μL was streaked on King's B plates containing appropriate antibiotics. After 2 d, colonies were counted from the dilution that resulted in 15 to 150 colonies per streak. From this data, the log10 of the number of cfu per cm2 leaf surface was calculated. The experiment was repeated three times, with eight replicates per genotype per treatment per experiment.
Results were analyzed separately by day using a fixed-effects linear model: log2(Cijk) ∼ Gi + Tj + G:Tij + Rk + ɛijk, where C = cfu/cm2, G = genotype; T = treatment; R = replicate; ɛ = residual; i = 1,…,11; j = 1,2; and k = 1,…,4.
The results from the linear model were used for two-tailed t tests using Benjamini and Hochberg false discovery rate (BH-FDR) multiple tests correction per hypothesis tested (Benjamini and Hochberg, 1995). Results are shown using q values and the BH-FDR–corrected P values. All the statistical analyses in this study were performed in the R environment (R Development Core Team, 2007) with R packages for linear mixed-effects models (R package versions 0.99875-7 and 3.1-83).
Electrolyte Leakage Analysis
The use of an electrolyte leakage assay as a measure of HR has been described previously (Goodman and Novacky, 1994). In short, per sample, three leaves per plant were inoculated with Pst or Pst avrRpt2 in 5 mM MgSO4 at an OD600 of 0.1 (1 × 108 cfu/mL). One hour after inoculation, two leaf discs per leaf, six per plant, were transferred to a Petri dish containing 25 mL of purified water and placed on a shaker for 2 h. After this washing step, the leaf discs were transferred to glass tubes containing 6 mL of purified water. The conductivity of the samples was determined using a portable conductivity meter (VWR Scientific) every 3 h for 24 h. This experiment was repeated on five experimental days, with four to six replicates per genotype per treatment (either inoculated with Pst or Pst avrRpt2) per day.
Results were analyzed by fitting a polynomial linear model through the electrolyte leakage curves of individual plants and using a mixed-effect linear model on the coefficients of these curves: Cijk ∼0 + G:Tij + G:T:(Tm + Tm2+Tm3+Tm4)ij + (1+ Tm + Tm2+Tm3+Tm4)|Pijk + 1|Rl + ɛijk, where C = conductivity; G = genotype (fixed effect); T = treatment (fixed effect); Tm = time (fixed effect); P = plant (random effect); R = replicate (random effect); ɛ = residual; i = 0,…,11; j = 1,2; k = 1,..,336; and l = 1,…,5. To avoid convergence problems, the coefficients of the (1+ Tm + Tm2+Tm3+Tm4)|Pijk random effect were assumed to be independent and time was centered and scaled to range from −1 to 1.
RPS2 Sequencing
Six fragments were PCR amplified from DNA isolated as described by Weigel and Glazebrook (2002) (see Supplemental Table 3 online for primer sequences). Amplification products were sequenced using standard procedures. Per accession, the concatenated sequence of the six fragments was compared with public sequences using BLASTN 2.2.15 (Altschul et al., 1997). DNA and protein alignments were performed using DNASTAR MegAlign 6.1. For those accessions for which RPS2 sequence information was (partially) available at www.ncbi.nlm.nih.gov, our sequence data matched these sequences.
RPS2 Expression Analysis
RPS2 expression was analyzed using qRT-PCR as by Sato et al. (2007) from mRNA collected for gene expression profiling (see below) with Actin2 (At3g18780) as internal reference and using three technical replicates per reaction. Primer sequences are shown in Supplemental Table 4 online. Ct values of technical replicates were averaged, and from this ΔCt (RPS2-Actin2) was calculated. The following fixed-effects linear model was used to analyze the data: ΔCtijk ∼ Gi + Tj + G:Tij + Rk + ɛijk (model 3), where ΔCt = ΔCt; G = genotype; T = treatment; R = RNA replicate; ɛ = residual; i = 1,..,9; j = 1,2; and k = 1,2,3.
The results from the linear model were used for two-tailed t tests with BH-FDR multiple tests correction per hypothesis tested.
Gene Expression Profiling
Gene expression profiling was performed using a small-scale microarray containing 571 gene-specific probes (miniarray, Pathoarray_464; Sato et al., 2007). Per sample, plant mRNA was isolated from five leaves of a single plant; leaves were mock (water) inoculated or inoculated with Pst avrRpt2 in water at OD600 = 0.05 (5 × 107 cfu/mL) 6 h prior to harvest. For each of three replicate experiments, one plant per accession per treatment was used. Data from individual miniarrays were normalized as described by Sato et al. (2007) using the expression of 106 stably expressed genes. To reduce noise, gene expression values were compared with values from a negative control probe to which no Arabidopsis or Pseudomonas mRNA should bind, using a t test on the replicates of each genotype treatment combination, without multiple tests correction. Only genes that were significantly different (P < 0.01) from the values of the negative control in at least one of the genotype treatment combinations were used for further analyses. For each gene, the following fixed-effects linear model was used to determine which genes show significant effects for genotype:treatment interaction: log2(Eijk) ∼ Gi + Tj + G:Tij + Rk + ɛijk, where E = expression; G = genotype; T = treatment; R = replicate; ɛ = residual; i = 1,..,9; j = 1,2; and k = 1,2,3.
The results from the linear model were used to determine which genes show different treatment-dependent expression per genotype pair using two-tailed t tests with BH-FDR multiple tests correction. For selection of genes varying among accessions per treatment, the raw data set was split into two sets, each containing the raw data of one treatment. Per treatment data set, the following fixed-effects linear model was used to determine which genes show significant genotype effects: log2(Eik) ∼ Gi + Rk + ɛik. Results from this statistical analysis were corrected for multiple tests using BH-FDR.
Correlation Analysis Using LEGG
Nonlinear dimensionality reduction was performed using locally linear embedding (LLE) (Roweis and Saul, 2000). LCF (Katagiri and Glazebrook, 2003) and LEGG use the same mathematical principle to generate graphs based on the results of LLE, but they use different algorithms. Each profile was described by m parameters. Thus, each profile can be described as m-dimensional vector. LEGG uses the cosine correlation coefficient (uncentered Pearson correlation coefficient) as the similarity metric. First, for each profile vector (seed vector, 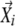 ), the n nearest neighboring profile vectors were identified (n, the number of neighboring vectors to explore;
), the n nearest neighboring profile vectors were identified (n, the number of neighboring vectors to explore; 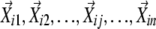 ). Second, the coefficients in the linear combination of the neighboring vectors that have the highest similarities to the seed vector were determined, that is, maximize
). Second, the coefficients in the linear combination of the neighboring vectors that have the highest similarities to the seed vector were determined, that is, maximize 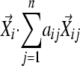 for each i when
for each i when 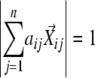 . This can be achieved by solving a set of simultaneous first-order equations for each i,
. This can be achieved by solving a set of simultaneous first-order equations for each i, 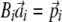 , where Bi is an n by n matrix of
, where Bi is an n by n matrix of 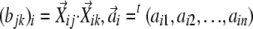 , and
, and 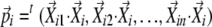 . The coefficients
. The coefficients 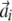 can be determined by multiplying both sides of the equations with
can be determined by multiplying both sides of the equations with 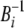 from the left and then scale for
from the left and then scale for 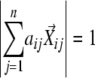 . In practice, the neighboring vectors
. In practice, the neighboring vectors 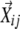 were included into Bi one by one in the order of similarity to
were included into Bi one by one in the order of similarity to 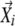 , and if addition of a particular
, and if addition of a particular 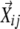 made the determinant of matrix Bi equal to 0, this particular
made the determinant of matrix Bi equal to 0, this particular 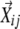 was omitted from being included in Bi; thus, the dimensionality is reduced. In LCF, the coefficients
was omitted from being included in Bi; thus, the dimensionality is reduced. In LCF, the coefficients 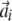 were determined by an optimization algorithm, but in LEGG, they were determined explicitly as described above. This algorithm difference made LEGG more accurate and faster than LCF. Another difference between LEGG and LCF is that any aij was limited to positive in LCF, but LEGG allows choices between limiting to positive and not limiting. When aij is not limited to positive in LEGG (this was the case in this study), by determining the neighboring vectors using the absolute value of the cosine correlation coefficient, LEGG considers the opposite expression patterns as similar patterns. Third, directed links were made from the neighboring profiles where the absolute values of the associated aij were larger than a threshold value to the seed profile. The directed graph generated in this way was visualized using Pajek (http://vlado.fmf.uni-lj.si/pub/networks/pajek/) (Batagelj and Mrvar, 1998). The Perl script for LEGG is freely available from F.K. for noncommercial research.
were determined by an optimization algorithm, but in LEGG, they were determined explicitly as described above. This algorithm difference made LEGG more accurate and faster than LCF. Another difference between LEGG and LCF is that any aij was limited to positive in LCF, but LEGG allows choices between limiting to positive and not limiting. When aij is not limited to positive in LEGG (this was the case in this study), by determining the neighboring vectors using the absolute value of the cosine correlation coefficient, LEGG considers the opposite expression patterns as similar patterns. Third, directed links were made from the neighboring profiles where the absolute values of the associated aij were larger than a threshold value to the seed profile. The directed graph generated in this way was visualized using Pajek (http://vlado.fmf.uni-lj.si/pub/networks/pajek/) (Batagelj and Mrvar, 1998). The Perl script for LEGG is freely available from F.K. for noncommercial research.
Clustering of Genes Based on Expression Profiles of Different Accessions
The squared Pearson correlation coefficient was used in average linkage, agglomerative hierarchical clustering of the expression value profiles for each gene across the accessions. The clustering procedure was terminated at a predetermined critical correlation coefficient value (P < 0.001, r2 > 0.81). From the resulting clusters, those that contained three or more genes were selected. The expression pattern of each cluster was calculated by taking the means of the centered and scaled expression values of the genes in a cluster. If genes clustered together due to negative correlation, the signs of the expression values of the negatively correlating genes were changed before taking the mean.
Accession Numbers
Expression profile depositions to the Gene Expression Omnibus are as follows: GSE8298, GSM205842, GSM205843, GSM205844, GSM205845, GSM205846, GSM205847, GSM205848, GSM205849, GSM205850, GSM205851, GSM205852, GSM205853, GSM205854, GSM205855, GSM205856, GSM205857, GSM205858, GSM205859, GSM205860, GSM205861, GSM205862, GSM205863, GSM205864, GSM205865, GSM205866, GSM205867, GSM205868, GSM205869, GSM205870, GSM205871, GSM205872, GSM205873, GSM205874, GSM205875, GSM205876, GSM205877, GSM205878, GSM205879, GSM205880, GSM205881, GSM205882, GSM205883, GSM205884, GSM205885, GSM205886, GSM205887, GSM205888, GSM205889, GSM205890, GSM205891, GSM205892, GSM205893, GSM205894, and GSM205895. The GenBank accession numbers for RPS2 sequences are EF560559 to EF560565.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Geographic Origins of Selected Arabidopsis Accessions.
Supplemental Figure 2. Conductivity for All Plant Genotypes.
Supplemental Figure 3. ΔConductivity for All Plant Genotypes.
Supplemental Figure 4. ΔΔConductivity for All Pairwise Comparisons among All Plant Genotypes.
Supplemental Figure 5. Heat Map of Gene Expression Profiles of Nine Selected Accessions.
Supplemental Table 1. Broad-Sense Heritabilities of the Measured Values.
Supplemental Table 2. Individual Genes in the Clusters Used in Figure 4B and Table 3.
Supplemental Table 3. Sequences of the PCR Primers Used in RPS2 Sequencing.
Supplemental Table 4. Primer Sequences Used in the qRT-PCR Experiments.
Supplementary Material
Acknowledgments
We thank all the undergraduate students who helped in this project, in particular Katie Wolf and Heather Cohen. We thank Ken Vernick for sharing his GenePix 4000B scanner and the Minnesota Supercomputing Institute for GenePix software. We thank all the members of the Katagiri and Glazebrook labs, especially Jane Glazebrook, and Jetske de Boer and Robert Stupar for their critical comments on the manuscript. R.M.P.V.P. was supported in part by the Netherlands Organization for Scientific Research under a Talent Fellowship. M.S. is a recipient of a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists. This project was supported by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (Grant 2004-35301-14525 to F.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Fumiaki Katagiri (katagiri@umn.edu).
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377. [DOI] [PubMed] [Google Scholar]
- Banerjee, D., Zhang, X., and Bent, A.F. (2001). The leucine-rich repeat domain can determine effective interaction between RPS2 and other host factors in arabidopsis RPS2-mediated disease resistance. Genetics 158 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagelj, V., and Mrvar, A. (1998). Pajek - Program for large network analysis. Connections 21 47–57. [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. [Google Scholar]
- Caicedo, A.L., Schaal, B.A., and Kunkel, B.N. (1999). Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., Fengler, K.A., Yu, I.C., Lippok, B., Smith, R.K., Jr., and Bent, A.F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann, W. (2005). Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol. Plant Microbe Interact. 18 1054–1060. [DOI] [PubMed] [Google Scholar]
- Gassmann, W., Hinsch, M.E., and Staskawicz, B.J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20 265–277. [DOI] [PubMed] [Google Scholar]
- Goodman, R.N., and Novacky, A.J. (1994). The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon. (St. Paul, MN: APS Press).
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Jurkowski, G.I., Smith, R.K., Jr., Yu, I.C., Ham, J.H., Sharma, S.B., Klessig, D.F., Fengler, K.A., and Bent, A.F. (2004). Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol. Plant Microbe Interact. 17 511–520. [DOI] [PubMed] [Google Scholar]
- Katagiri, F., and Glazebrook, J. (2003). Local Context Finder (LCF) reveals multidimensional relationships among mRNA expression profiles of Arabidopsis responding to pathogen infection. Proc. Natl. Acad. Sci. USA 100 10842–10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, F., and Sato, M. (2007). Expression profiles as detailed snapshots of biological states. Transgenic Res. 16 399–403. [DOI] [PubMed] [Google Scholar]
- Katagiri, F., Thilmony, R., and He, S.Y. (2002). The Arabidopsis thaliana–Pseudomonas syringae interaction. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0039, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Keurentjes, J.J., Fu, J., Terpstra, I.R., Garcia, J.M., van den Ackerveken, G., Snoek, L.B., Peeters, A.J., Vreugdenhil, D., Koornneef, M., and Jansen, R.C. (2007). Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc. Natl. Acad. Sci. USA 104 1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.G., da Cunha, L., McFall, A.J., Belkhadir, Y., DebRoy, S., Dangl, J.L., and Mackey, D. (2005). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121 749–759. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., West, M.A., van Leeuwen, H., Kim, K., Doerge, R.W., Michelmore, R.W., and St Clair, D.A. (2006). Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics 172 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Mauricio, R., Stahl, E.A., Korves, T., Tian, D., Kreitman, M., and Bergelson, J. (2003). Natural selection for polymorphism in the disease resistance gene Rps2 of Arabidopsis thaliana. Genetics 163 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.L., and Ausubel, F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78 1089–1099. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchepied, L., Kroj, T., Tronchet, M., Loudet, O., and Roby, D. (2006). Natural variation in partial resistance to Pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana. PLoS ONE 1 e123.17205127 [Google Scholar]
- R Development Core Team (2007). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing).
- Roweis, S.T., and Saul, L.K. (2000). Nonlinear dimensionality reduction by locally linear embedding. Science 290 2323–2326. [DOI] [PubMed] [Google Scholar]
- Sato, M., Mitra, R.M., Coller, J., Wang, D., Spivey, N.W., Dewdney, J., Denoux, C., Glazebrook, J., and Katagiri, F. (2007). A high-performance, small-scale microarray for expression profiling of many samples in Arabidopsis-pathogen studies. Plant J. 49 565–577. [DOI] [PubMed] [Google Scholar]
- Schadt, E.E., et al. (2003). Genetics of gene expression surveyed in maize, mouse and man. Nature 422 297–302. [DOI] [PubMed] [Google Scholar]
- Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H.S., Han, B., Zhu, T., Zou, G., and Katagiri, F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen, H., Kliebenstein, D.J., West, M.A., Kim, K., van Poecke, R., Katagiri, F., Michelmore, R.W., Doerge, R.W., and St Clair, D.A. (2007). Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell 19 2099–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.