Abstract
The conserved eukaryotic protein SGT1 (for Suppressor of G2 allele of skp1) has characteristics of an HSP90 (for heat shock protein 90 kD) cochaperone and in plants regulates hormone responses and Resistance gene–triggered immunity. We affinity-purified SGT1-interacting proteins from Arabidopsis thaliana leaf extracts and identified by mass spectrometry cytosolic heat shock cognate 70 (HSC70) chaperones as the major stable SGT1 interactors. Arabidopsis SGT1a and SGT1b proteins associate with HSC70 in vivo and distribute with HSC70 in the cytosol and nucleus. An intact C-terminal SGT1-specific (SGS) domain that is required for all known SGT1b functions in immunity and development is needed for HSC70 interaction and for the nuclear accumulation of SGT1b. Interaction assays of transiently expressed proteins or their domains in Nicotiana benthamiana point to a role of SGT1 as a HSC70 cofactor. Expression of two HSC70 isoforms is upregulated by pathogen challenge, and while loss of function of individual cytosolic HSC70 genes has no defense phenotype, HSC70-1 overexpression disables resistance to virulent and avirulent pathogens. Moreover, mutations in SGT1b lead to a similar degree of heat shock tolerance as deregulation of HSC70-1. We conclude that an HSC70-SGT1 chaperone complex is important for multiple plant environmental responses and that the evolutionarily conserved SGS domain of SGT1 is a key determinant of the HSC70–SGT1 association.
INTRODUCTION
Molecular chaperones are essential for cell viability by ensuring the proper folding of nascent polypeptides, protein complex maturation, and the translocation of proteins through membranes. In eukaryotic cells, the conserved chaperones of the Hsp90 (for heat shock protein 90 kD) and DnaK/Hsc70 (for heat shock cognate 70 kD) families control cellular protein homeostasis through ATP-dependent cycles (Young et al., 2003). Hsc70 predominantly binds nonnative polypeptides and folding intermediates that are either newly synthesized or stress-induced (Erbse et al., 2004; Bukau et al., 2006). By contrast, Hsp90 chaperones bind proteins in their near-native state and mediate the maturation and activation of signaling complexes (Young et al., 2003). In yeast and mammalian cells, the activities of Hsp90 and Hsc70 are modulated by a complex network of cochaperones that define the balance of protein assembly and degradation by the ubiquitin-proteasome machinery (Esser et al., 2004; Bukau et al., 2006).
Sgt1 (for Suppressor of G2 allele of skp1) is a conserved, essential protein in eukaryotes that interacts with multiple protein complexes and has features of a cochaperone (Shirasu and Schulze-Lefert, 2003). The Sgt1 protein has three domains: an N-terminal tetratricopeptide repeat (TPR) domain that resembles the folds of Hop/Sti1 (for Hsp70/Hsp90 organizing protein); a central CHORD-Sgt1 (CS) domain that is similar to the Hsp20/α-crystallin domain of the human p23 cochaperone family; and a C-terminal Sgt1-specific (SGS) domain that is structurally less well defined although highly conserved relative to the other SGT1 domains (Dubacq et al., 2002; Garcia-Ranea et al., 2002; Lee et al., 2004). Yeast Sgt1p associates with Skp1p (for Suppressor of kinetochore protein) and Hsp90p through its TPR and CS domains, respectively, and is needed to assemble an active centromere binding factor3 kinetochore complex in cell cycle progression (Kitagawa et al., 1999; Bansal et al., 2004; Lingelbach and Kaplan, 2004; Catlett and Kaplan, 2006). Yeast Sgt1p is also required for activation of an SCF (for Skp1-Cul1-F box) ubiquitin ligase complex that mediates the ubiquitination of Sic2p, an inhibitor of Cdc28 kinase (Kitagawa et al., 1999). A Skp1p-independent but essential function of Sgt1p was identified in the activation of the yeast adenylyl cyclase (Cyr1p) protein through interaction between the Sgt1p SGS and Cyr1p leucine-rich repeat (LRR) domains (Dubacq et al., 2002). Thus, Sgt1 has a role in diverse signaling processes.
Plant SGT1 proteins associate with cytosolic HSP90s in vivo, consistent with a conserved cochaperone activity (Hubert et al., 2003; Takahashi et al., 2003; Liu et al., 2004). Also, in tobacco (Nicotiana benthamiana) and barley (Hordeum vulgare) extracts, SGT1 coimmunoprecipitated with the SCF structural subunits SKP1 and CUL1 and the COP9 signalosome that regulates the SCF ubiquitin-proteasome degradation system (Azevedo et al., 2002; Liu et al., 2002a). A link between SGT1 in plants and yeast (Kitagawa et al., 1999; Lyapina et al., 2001) with components of the ubiquitin-proteasome machinery suggests that SGT1 may assist in the controlled degradation of target proteins. Indeed, the loss of Arabidopsis thaliana SGT1b (one of two functional Arabidopsis SGT1 genes, SGT1a and SGT1b) compromised the functions of SCFTIR1 and SCFCOI1 that mediate the ubiquitination-dependent degradation of proteins in response to the phytohormones auxin and jasmonic acid, respectively (Gray et al., 2003).
Plant SGT1 interacts through its CS domain with another cytosolic HSP90 binding protein, RAR1 (for Required for Mla12 Resistance) (Hubert et al., 2003; Takahashi et al., 2003; Liu et al., 2004), and both RAR1 and SGT1 were identified as components of plant resistance mediated by intracellular nucleotide binding–leucine-rich repeat (NB-LRR) immune receptors (Shirasu et al., 1999; Austin et al., 2002; Azevedo et al., 2002; Liu et al., 2002b; Muskett et al., 2002; Tornero et al., 2002). A body of genetic and molecular evidence points to functions of plant SGT1 and RAR1 as cofactors in HSP90-mediated stabilization of preactivated NB-LRR protein complexes (Tornero et al., 2002; Hubert et al., 2003; Lu et al., 2003; Bieri et al., 2004; Liu et al., 2004; Azevedo et al., 2006). These receptors (also known as R proteins) are present in the cell in a constrained conformation and can be specifically activated by the action of pathogen-derived effectors (Shirasu and Schulze-Lefert, 2003). Pathogen recognition potentiates low-level basal defense that limits the growth of virulent pathogens and is often accompanied by localized programmed cell death (Chisholm et al., 2006). SGT1 can interact with the LRR domains of certain NB-LRR proteins and may assist in their proper folding (Bieri et al., 2004; Leister et al., 2005). There is no evidence for a direct association of RAR1 with NB-LRR proteins; therefore, RAR1 may operate at another level of immune receptor assembly or maintenance. While genetically additive contributions of SGT1b and RAR1 were observed in resistance mediated by the NB-LRR genes Arabidopsis RPP5 and barley MLA6 (Austin et al., 2002; Azevedo et al., 2002), an antagonistic relationship was found between SGT1b and the assembly roles of RAR1 and HSP90 in certain Arabidopsis NB-LRR conditioned responses (Holt et al., 2005). This likely reflects a fine balance between the assembly and degradative activities of the chaperone/cochaperone machineries in maintaining NB-LRR proteins poised for activation. Also, the Arabidopsis SGT1 homolog SGT1a may compensate for the loss of SGT1b in controlling the steady state levels of certain NB-LRR proteins, since SGT1a has intrinsic SGT1 activity but is expressed at a lower level than SGT1b (Azevedo et al., 2006). SGT1a and SGT1b have redundant essential roles in early embryo development, but only mutations in SGT1b compromise plant immunity or auxin signaling (Azevedo et al., 2006).
Therefore, SGT1 is necessary for plant development and disease resistance, but it is unclear how it operates molecularly and whether its activity as a HSP90 cofactor accounts entirely for its diverse cellular functions. We report here that affinity purification–tagged Arabidopsis SGT1 protein interacts stably with cytosolic/nuclear HSC70 chaperones in vivo. This interaction occurs with native SGT1 protein and requires an intact SGS domain for which no direct partners were known. Mutations in SGT1b and deregulation of HSC70-1, the predominant cytosolic HSC70 isoform in Arabidopsis, disable R protein–specified and basal disease resistance and lead to increased heat shock tolerance. We conclude that the SGT1–HSC70 association is important for the regulation of plant responses to biotic and abiotic stresses.
RESULTS
Functional Characterization of SGT1 Proteins Tagged by the StrepII Epitope
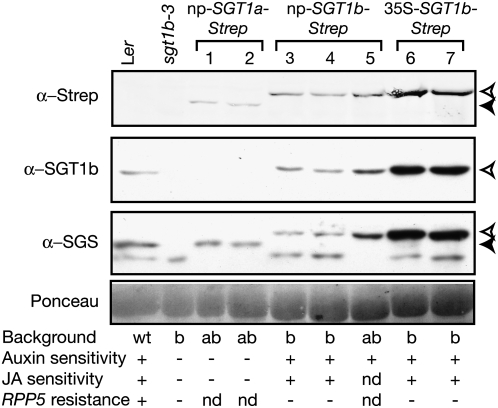
In order to search for biochemical interactors of the Arabidopsis SGT1 proteins, SGT1a and SGT1b were fused to a C-terminal StrepII (Strep) affinity purification tag under the control of the constitutive cauliflower mosaic virus 35S promoter or their respective native promoters. SGT1b constructs were transformed into the Landsberg erecta (Ler) sgt1b-3 null mutant (Austin et al., 2002), and SGT1a constructs were transformed into a Ler/Wassileskija (Ws-0) hybrid that was homozygous for sgt1b-3 (Ler) and heterozygous for sgt1a-1 (Ws-0 background) (Azevedo et al., 2006). Multiple transgenic lines were selected that expressed the SGT1a-Strep and SGT1b-Strep fusion proteins in the appropriate mutant backgrounds, as shown for representative lines in Figure 1. The functionality of the SGT1b-Strep fusion proteins was tested based on complementation of the known sgt1b-3 mutant defects. The SCF ubiquitin E3 ligase–dependent functions of SGT1b (root growth sensitivity to auxin and jasmonic acid) were fully complemented irrespective of the promoter used (Figure 1; see Supplemental Figure 1 online). RPP5 resistance to the oomycete pathogen Hyaloperonospora parasitica was not restored (Figure 1; see Supplemental Figure 1 online), because transgenic plants exhibited a delayed cell death response. The double homozygote mutant sgt1a-1 sgt1b-3 is embryo-lethal (Azevedo et al., 2006). Therefore, we crossed SGT1b-Strep transgenic plants into the Ler/Ws-0 hybrid that was homozygous for sgt1b-3 and heterozygous for sgt1a-1. Double homozygote sgt1a-1 sgt1b-3 mutants expressing SGT1b-Strep could be selected and were fully viable, indicating that SGT1b-Strep complements the lethality of sgt1b-3 sgt1a-1 (Figure 1). Therefore, SGT1b-Strep complemented three of the four known sgt1b mutant phenotypes in Arabidopsis. SGT1a-Strep expressed under its own promoter complemented the embryo lethality of sgt1b-3 sgt1a-1, which is the only sgt1a mutant phenotype known to date.
Figure 1.
Stability and Functionality of StrepII-Tagged SGT1 Proteins in Transgenic Arabidopsis.
Total leaf protein extracts were prepared from 3-week-old plants and analyzed by SDS-PAGE and immunoblotting using anti-SGT1b and anti-SGS antibodies or the Strep-Tactin-AP conjugate, as indicated. The open and closed arrowheads mark SGT1b-Strep and SGT1a-Strep, respectively. 1/2, 3/4, and 6/7 indicate independent transgenic lines. Line 5 is derived from a cross between a Ler/Ws-0 hybrid, sgt1b-3/sgt1b-3 sgt1a-1/SGT1a, and plant 4. Equal loading of samples was checked by Ponceau red staining of the blot. Background refers to the SGT1 alleles: wt, wild-type SGT1a and SGT1b; a, homozygous sgt1a-1; b, homozygous sgt1b-3; ab, double sgt1a-1 sgt1b-3 homozygotes. + indicates plants that have wild-type sensitivity to 0.075 μM 2,4-D or 10 μM methyl jasmonate in root growth inhibition assays, and − indicates plants that are insensitive to these hormone concentrations. nd, not determined. RPP5 gene–mediated resistance assays were performed using H. parasitica avirulent isolate Noco2. For RPP5 resistance, + indicates wild-type hypersensitive response and − indicates trailing necrosis/sporulation. See Supplemental Figure 1 online for detailed phenotypic characterization.
In Planta Interaction of StrepII-Tagged Arabidopsis SGT1 with Cytosolic HSC70 Chaperones
We previously reported the identification of a 70-kD band that specifically copurified with StrepII- and tandem affinity purification–tagged Arabidopsis SGT1b expressed under the control of the 35S promoter in healthy plant tissues (Witte et al., 2004). This protein band was processed for mass spectrometric analysis. All 14 tryptic peptides belonged to cytosolic Arabidopsis HSC70 (predicted molecular mass of 71 kD; see Supplemental Table 1 online). The Arabidopsis genome encodes 14 HSC70 proteins of the DnaK superfamily, five of which (HSC70-1 to -5) are predicted to be cytosolic and/or nuclear due to the presence of predicted nuclear localization signals (Lin et al., 2001). Despite the high sequence similarity within the family (83 to 94% amino acid identity), identification of the HSC70 isoforms in the sample was achieved based on two informative tryptic fragments, C and G (see Supplemental Figure 2 online), whose identities were confirmed by quadrupole time-of-flight tandem mass spectrometry. From this analysis, HSC70-1 (CAB85987) and HSC70-3 (AAF14038) were unambiguously identified and could explain all 14 peptides detected by mass spectrometry. Therefore, HSC70-1 and HSC70-3 are two novel SGT1b-Strep interactors in healthy leaf tissue.
We tested whether the interaction with cytosolic HSC70s is specific to SGT1b compared with SGT1a or is influenced by pathogen challenge by performing a StrepII affinity purification of SGT1a-Strep and SGT1b-Strep expressed under the control of their respective native promoters (Figure 2). Leaf material was either nontreated or collected at 24 h after inoculation with the avirulent bacterial pathogen Pseudomonas syringae pv tomato (Pst) DC3000 expressing the effector AvrRpm1 (recognized by RPM1) (Figure 2A). As observed by silver staining and on an immunoblot probed with anti-HSC70 antiserum (which does not discriminate between the different isoforms), HSC70s could be purified with SGT1a-Strep or SGT1b-Strep expressed under the control of their native promoters (Figure 2B). These data show that cytosolic HSC70s do not discriminate strongly between SGT1a and SGT1b in their binding and that the observed SGT1–HSC70 interaction is not due to SGT1 overexpression. The amounts of purified HSC70 were globally proportional to the amounts of SGT1a or SGT1b purified. For example, SGT1a-Strep accumulated to higher levels in total extracts after pathogen challenge, and this was reflected in the levels of purified SGT1a and HSC70 proteins (Figure 2B). We reasoned that SGT1 and HSC70 most likely interact directly, since no other protein could be detected by silver staining in several independent purification experiments. This analysis also confirmed indirectly that the tagged SGT1 proteins are soluble and present in the cytosol and/or the nucleus, since they interact with cytosolic/nuclear HSC70s.
Figure 2.
Interactions of StrepII-Tagged SGT1 Proteins with Cytosolic HSC70 Chaperones in Arabidopsis Transgenic Lines.
(A) Total leaf protein extracts were prepared from Ler sgt1b-3 mutants transgenic for SGT1a-Strep or SGT1b-Strep expressed from their native promoters (np). A parallel experiment was performed with leaf tissue sampled at 24 h after infiltration with a bacterial suspension of Pst DC3000/avrRpm1 (107 colony-forming units [cfu]/mL). Protein extracts were analyzed by SDS-PAGE and immunoblotting with anti-HSC70 and anti-SGS antibodies. The open and closed arrowheads mark SGT1b-Strep and SGT1a-Strep, respectively. The closed circle indicates the SGT1b-GFP (for green fluorescent protein) fusion protein used as a negative control in the StrepII purification.
(B) Elution fractions from the purification were analyzed by SDS-PAGE and silver staining or immunoblotting with anti-HSC70 and anti-SGS antibodies. The asterisk marks 70-kD SGT1-interacting proteins. Molecular masses of protein markers are indicated at right in kilodaltons.
(C) SGT1b-Strep expressed from its native promoter was purified using the Strep-Tactin Macroprep resin and boiled off the resin. A parallel experiment was performed with leaf tissue sampled at 24 h after infiltration with Pst DC3000/avrRpm1 (107 cfu/mL). Extracts were analyzed by SDS-PAGE and Coomassie blue staining. The 70-kD bands were sampled, digested by trypsin, and analyzed by quadrupole time-of-flight tandem mass spectrometry. The relative abundance of the five cytosolic HSC70 isoforms was determined: −, not detected; (+), weak signal; +, clear signal; ++, strong signal.
We tested whether the spectrum of HSC70 isoforms copurified was affected by pathogen challenge. Unchallenged and pathogen-treated leaf samples were processed as above, and SGT1-Strep protein was collected using the Strep-Tactin-Macroprep resin, which allows higher recovery. Copurified HSC70 protein amounts were too low to perform the analysis with SGT1a-Strep. For SGT1b-Strep, HSC70-1 and HSC70-3 remained the principal interactors of SGT1b expressed under the control of its native promoter in both samples (Figure 2C; see Supplemental Figure 2 online). Weak but reproducible signals were also unambiguously identified as HSC70 isoforms 2 (CAB85986) and 4 (BAB02269) in the pathogen-treated samples only.
Expression of Cytosolic HSC70-2 and HSC70-4 Is Pathogen Inducible
Previous studies showed that HSC70-1 and HSC70-3 transcripts are the most abundant of the cytosolic isoforms in young healthy Arabidopsis tissue (Lin et al., 2001). In order to establish whether pathogen challenge modulates SGT1 affinity to individual HSC70 isoforms or simply reflects HSC70 abundance in the cell, we measured mRNA accumulation for the different cytosolic HSC70 isoforms by RT-PCR (Figure 3A). Ecotype Columbia (Col-0) plants were infiltrated with MgCl2 buffer, virulent Pst DC3000 containing an empty vector, or avirulent Pst DC3000 expressing either AvrRpm1 or AvrRps4 (recognized by RPS4). Expression of Pathogenesis-Related1 mRNA was also measured to assess the responsiveness of tissues and ensure that unchallenged plants were not stressed prior to infection. Standardization of cDNA samples used for RT-PCR was done by measuring the expression of a constitutive Tubulin gene. HSC70-1 and HSC70-3 expression did not change significantly after pathogen infection. By contrast, HSC70-2 and HSC70-4 expression was barely detectable in untreated samples and was weakly induced in plants infiltrated with MgCl2. HSC70-2 and HSC70-4 mRNA levels increased substantially following inoculation with avirulent Pst DC3000 strains, the response to AvrRpm1 being earlier than that to AvrRps4. A weaker induction of HSC70-2 mRNA was also observed in samples responding to virulent Pst DC3000. HSC70-5 mRNA was barely detectable but was slightly induced at 24 h after inoculation with Pst DC3000/AvrRpm1. SGT1a mRNA accumulated to high levels in avirulent pathogen-treated samples, whereas SGT1b expression was not strongly pathogen-responsive, consistent with earlier analysis of SGT1 promoter:β-glucuronidase (GUS) fusions (Azevedo et al., 2006). Samples taken from the same material used for RT-PCR analysis were processed for immunoblot analysis with anti-HSC70 antibody. The results (Figure 3B) reveal that HSC70 levels increased at late time points (6 to 24 h) in the incompatible interactions, consistent with the constitutive expression of the HSC70-1 and HSC70-3 isoforms and the pathogen-inducible expression of HSC70-2 and HSC70-4. Since the HSC70 isoforms interacting with SGT1b broadly mirror the HSC70 expression pattern, we conclude that SGT1b does not discriminate strongly in its interaction with different HSC70 isoforms present in the cytoplasm and/or nucleus.
Figure 3.
Expression of HSC70-2 and HSC70-4 Is Pathogen Inducible.
Four-week-old Col-0 plants were nontreated (NT), hand-inoculated with Pst DC3000 strains (107 cfu/mL) as indicated, or treated with 10 mM MgCl2 as a control (MgCl2). Samples were harvested at 0, 2, 6, and 24 h after inoculation (hpi).
(A) RNAs were isolated from leaves, and RT-PCR products were separated by agarose gel electrophoresis. Ethidium bromide–stained gels are shown.
(B) Total leaf protein extracts were analyzed by SDS-PAGE and immunoblotting with anti-HSC70 antibodies. Equal loading of samples was checked by Ponceau red staining of the blot.
Arabidopsis SGT1b Interacts with HSC70 in Vivo
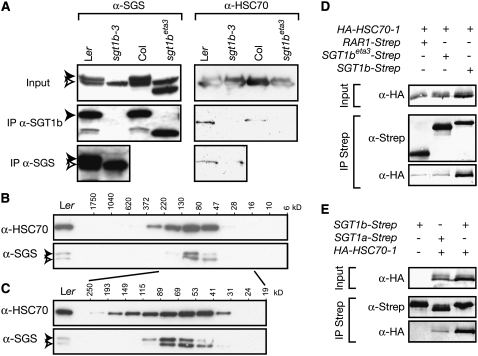
We considered that interaction between SGT1-Strep and HSC70s may be due to incorrect folding of SGT1, making it a HSC70 substrate (Erbse et al., 2004). Therefore, we tested whether native SGT1 proteins associate with HSC70 in wild-type tissues. This was examined by coimmunoprecipitation from plant soluble protein extracts using either anti-SGT1b antibodies that bind only SGT1b or anti-SGS antibodies that recognize both SGT1 isoforms (Austin et al., 2002) (Figure 4A). HSC70s could be coimmunoprecipitated in Ler and Col-0 total extracts with anti-SGT1b but not in extracts of the Ler sgt1b-3 mutant that lacks SGT1b protein (Austin et al., 2002). Anti-SGS antibody coimmunoprecipitated lower amounts of HSC70s from Ler sgt1b-3 extracts that contain SGT1a. These results indicate that native SGT1a and SGT1b interact with HSC70 in vivo, implying a physiologically relevant association.
Figure 4.
SGT1b Interacts with HSC70s in Vivo.
(A) Total leaf protein extracts of 3-week-old healthy plants were immunoprecipitated with anti-SGT1b or anti-SGS antibodies as indicated. Plant genotypes used were Ler, Ler sgt1b-3, Col-0, and Col-0 sgt1beta3. Total extracts and immunoprecipitates (IP) were analyzed by SDS-PAGE and immunoblotting with anti-HSC70 and anti-SGS antibodies.
(B) and (C) Wild-type Ler leaf protein extracts were separated by size-exclusion chromatography. Collected fractions were analyzed by SDS-PAGE and immunoblotting with anti-HSC70 and anti-SGS antibodies. Molecular mass ranges of the fractions are indicated in kilodaltons based on the column calibration.
(D) and (E) HA-tagged HSC70-1 and Strep-tagged SGT1 were coexpressed using Agrobacterium-mediated transient transformation of N. benthamiana leaves. Total protein extracts (Input) were subjected to affinity purification of the Strep-tagged protein using Strep-Tactin Sepharose and specifically eluted (IP Strep). Protein extracts were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody or the Strep-Tactin-AP conjugate, as indicated. SGT1beta3 indicates residues 1 to 322.
In order to characterize the interaction further, soluble protein extracts from healthy Ler plants were prepared as for the coimmunoprecipitations and analyzed by size-exclusion chromatography on a Superdex 200 column. Fractions were then collected and analyzed on an immunoblot (Figures 4B and 4C). SGT1a (39 kD) and SGT1b (40 kD) were collected in fractions containing proteins with apparent molecular mass ranges of 41 to 89 kD and 41 to 115 kD, respectively. This observation suggests that SGT1 proteins are either not globular or that their apparent mobility is affected by interaction with other protein partners. Fractions that contained SGT1 proteins (41 to 115 kD) also contained the major HSC70 pool, consistent with a SGT1–HSC70 association and direct interaction in a 1:1 stoichiometric ratio. Under these conditions, we did not detect higher molecular mass complexes containing SGT1.
The SGT1 SGS Domain Is Necessary and Sufficient for Interaction with HSC70
To better define the relationship between SGT1 and HSC70 proteins, we characterized the domains responsible for the SGT1–HSC70 interaction. First, we tested the effect of the enhancer of tir1-1 auxin resistance3 (eta3) mutant allele of Arabidopsis SGT1b on HSC70 interaction. The sgt1beta3 mutation causes a splicing error that results in a frame shift and a premature stop codon, leading to the synthesis of a truncated SGT1b protein that lacks the last 36 amino acids and therefore has a disrupted 94–amino acid C-terminal SGS domain (Gray et al., 2003). Although the mutant SGT1beta3 protein is more stable than wild-type SGT1b, sgt1beta3 phenotypes are indistinguishable from those of the Ler sgt1b-3 protein null mutant (Gray et al., 2003). Levels of HSC70 recovered in immunoprecipitates of sgt1beta3 extracts were reduced compared with those in the wild type in 7 of 11 repetitions, as shown in Figure 4A. This result indicates that an intact SGS domain is needed for efficient HSC70 binding in vivo.
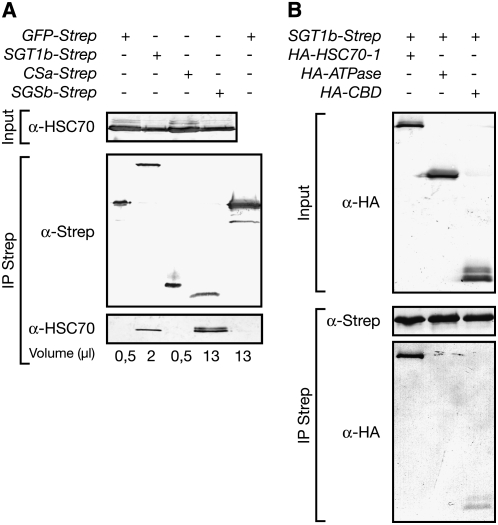
The association between SGT1 and HSC70s was confirmed and analyzed further using Agrobacterium tumefaciens–mediated transient coexpression of Arabidopsis SGT1 (Strep-tagged) and HSC70-1 (hemagglutinin [HA]-tagged) domains in Nicotiana benthamiana followed by detection of StrepII copurified protein on immunoblots (Figures 4D and 4E). In this analysis, SGT1b-Strep protein bound significant amounts of HSC70-1, whereas RAR1-Strep or SGT1beta3-Strep did not (Figure 4D). SGT1a accumulated to similar levels as SGT1b in N. benthamiana extracts but bound lower amounts of HSC70-1 protein (Figure 4E). In another N. benthamiana interaction assay, we assessed the efficiency with which endogenous HSC70 copurified with approximately equivalent amounts of transiently expressed Strep-tagged SGT1b or the individual CS and SGS domains (Figure 5A). The CS domain from SGT1a (the SGT1b CS domain was not expressed) and a GFP-Strep control protein failed to bind HSC70. By contrast, SGT1b-Strep and SGSb-Strep bound significant amounts of HSC70. These results indicate that the SGS domain is necessary and sufficient for SGT1 association with HSC70. The loss of function of SGT1beta3 is more likely due to the loss of interaction with HSC70-1, since HSP90-1 and RAR1 still interacted with SGT1beta3 tested in a yeast two-hybrid assay (see Supplemental Figure 3 online).
Figure 5.
Mapping of HSC70-1– and SGT1b-Interacting Domains in Transient Plant Expression Assays.
Strep-tagged SGT1 domains (A) and HA-tagged HSC70-1 domains with Strep-tagged SGT1 (B) were expressed using Agrobacterium-mediated transient transformation of N. benthamiana leaves. Total protein extracts (Input) were subjected to affinity purification of Strep-tagged protein using Strep-Tactin Sepharose and specifically eluted (IP Strep). Protein extracts were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody, the Strep-Tactin-AP conjugate, or the anti-HSC70 antibody to detect endogenous Nb HSC70, as indicated. Different volumes of the elution fraction were analyzed on the gel in order to normalize the amounts of purified Strep-tagged proteins. GFP-Strep was used as a non-HSC70-interacting protein in the purification process. ATPase, ATPase domain of HSC70-1 (residues 1 to 405); CBD; client binding domain of HSC70-1 (residues 374 to 543); CSa, SGT1a CS domain (residues 149 to 260); SGSb, SGT1b SGS domain (residues 268 to 357).
We then tested which of the HSC70-1 domains interacts most efficiently with SGT1b by coexpressing its HA-tagged ATPase domain (N-terminal; 45 kD) or client binding domain (C-terminal; 19 kD) with SGT1b-Strep in N. benthamiana. Interaction between SGT1b and HSC70-1 was again strong (Figure 5B). We detected no interaction between SGT1b and the ATPase domain of HSC70-1 and very weak or no interaction with the client binding domain of HSC70-1. These results argue against SGT1b being a substrate of HSC70 and suggest that effective SGT1b binding is probably only achieved with the complete HSC70-1 protein.
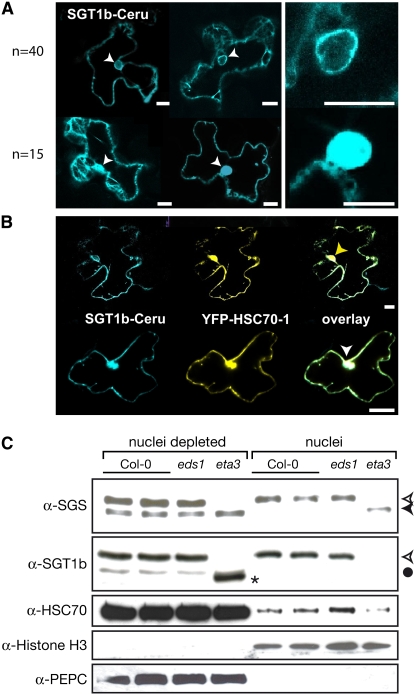
Subpools of SGT1b and HSC70 Localize to the Nucleus
We reasoned that colocalization of SGT1 and HSC70 in one or more compartments would be necessary for functional interaction. We first investigated the subcellular localizations of fluorescent protein–tagged HSC70-1 and SGT1b after biolistic transfection of N. benthamiana epidermal cells (Figures 6A and 6B). SGT1b fused to Cerulean (a derivative of cyan fluorescent protein) localized to the cytosol but could be seen in nuclei of ∼25% of 55 transformed cells examined (Figure 6A), suggesting movement of SGT1b between the cytosol and nucleus. In agreement with a predicted nuclear localization signal in cytosolic HSC70s (Sung and Guy, 2003), yellow fluorescent protein (YFP)–HSC70-1 was detected in the cytosol and nuclei of 100% of cells examined and colocalized with cytosolic and nuclear SGT1b-Cerulean (Figure 6B). We then tested the subcellular distribution of SGT1a and SGT1b and cytosolic HSC70 proteins in Arabidopsis by preparing nuclear extracts from leaves of Col-0, eta3, and the defense signaling mutant Col eds1-2. HSC70 and SGT1b signals were detected in both cytosolic and nucleus-enriched fractions of Col-0 and eds1-2 (Figure 6C). Notably, SGT1beta3 protein (detected by anti-SGT1b antibody) accumulated only in the nucleus-depleted fraction but stimulated the nuclear accumulation of SGT1a (detected by anti-SGS antibody) compared with wild-type and eds1-2 tissues (Figure 6C). These data show that subpools of SGT1 and HSC70 protein colocalize in the cytosol and nucleus and that the SGS domain of SGT1b, which is needed for HSC70-1 interaction, is also required for nuclear accumulation. While nuclear import of SGT1b appears to predominate in wild-type cells, SGT1a has the capacity to enter nuclei in the absence of functional SGT1b.
Figure 6.
Subcellular Localization of SGT1b and HSC70 Proteins.
(A) and (B) Plasmids containing p35S:SGT1b-Cerulean (A) or p35S:SGT1b-Cerulean/35S:YFP-HSC70-1 (B) were delivered to N. benthamiana epidermal cells using a particle gun. Imaging with a confocal laser scanning microscope was done at 18 to 48 h after transformation. Bars = 20 μm.
(A) SGT1b enters the nucleus of some cells (n = 15 of 55 examined). The top panel shows two representative cells with SGT1b excluded from the nucleus, and the bottom panel shows two representative cells with SGT1b inside the nucleus. White arrowheads mark the nuclei. Enlarged views of the right nuclei are also shown.
(B) SGT1b colocalizes with HSC70-1. The top panel depicts the colocalization of both proteins outside the nucleus. The yellow arrowhead indicates the nuclear localization of YFP-HSC70-1 but not SGT1b-Cerulean. The bottom panel shows a representative cell with both proteins colocalized outside and within the nucleus. The white arrowhead indicates the nuclear localization of both YFP-HSC70-1 and SGT1b-Cerulean.
(C) Nucleus-depleted and -enriched protein extracts from Col-0, Col eds1-2, and sgt1beta3 leaves were analyzed by SDS-PAGE and immunoblotting with anti-SGS, anti-SGT1b, anti-HSC70, anti-histone H3 (nuclear marker), and anti-PEPC (for phosphoenolpyruvate carboxylase; cytosolic marker) antibodies, as indicated. Nuclear samples were 16 times more concentrated than nuclei-depleted preparations. The open and closed arrowheads indicate SGT1b and SGT1a signals, respectively. The asterisk marks the SGT1eta3 protein, and the closed circle marks a weak nonspecific signal with anti-SGT1b antibody.
HSC70-1 Overexpression Disables Plant Immune Responses
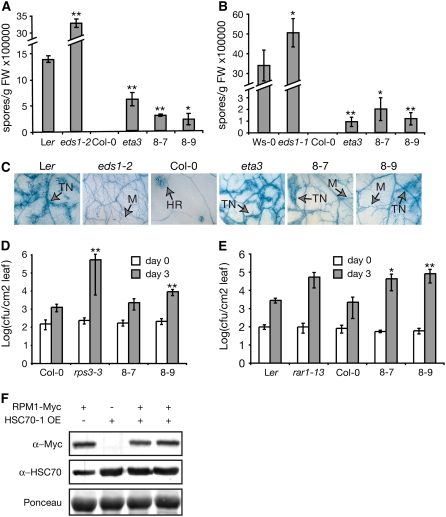
To explore whether cytosolic HSC70s are involved in plant processes known to require SGT1b, T-DNA insertion mutants of HSC70-1 to HSC70-3 were isolated (see Methods) and characterized for resistance to pathogens. One insertion found in HSP70-4 did not diminish transcript levels (see Supplemental Figure 4 online) and was not analyzed further. We did not detect alterations in the phenotypes of the hsc70-1, hsc70-2, and hsc70-3 mutants after infection with virulent or avirulent strains of H. parasitica and Pst (see Supplemental Figure 4 online). Gene silencing of the HSC70 gene family causes embryo lethality (Sung and Guy, 2003) and therefore was not an option to overcome likely functional redundancy within these highly conserved proteins. Also, no specific HSC70 inhibitors have been described (Brodsky and Chiosis, 2006). In animal and yeast cells, overexpression of HSC70 enhances tolerance to heat stress by increasing protein disaggregation but impairs normal HSC70 cellular function in protein maturation, degradation, and transport (Krobitsch and Lindquist, 2000; Brodsky and Chiosis, 2006). Therefore, we examined the effect of modulating HSC70 function on the plant immune response through overexpression. Several available Col-0 transgenic lines that express HSC70-1 under the control of the 35S promoter (Sung and Guy, 2003) were characterized, and lines 8-7 (a single T-DNA insertion; sevenfold overexpression) and 8-9 (multiple T-DNA insertions; fourfold overexpression) were selected for further study. These lines exhibited some developmental defects, such as a dwarf stature and a short root system, as described before (Sung and Guy, 2003), but they did not have altered photosynthetic efficiency (Sung and Guy, 2003), root architecture, or responsiveness to auxin (L.D. Noël, unpublished data). The HSC70-1 overexpression lines displayed a partial breakdown of resistance specified by four different Arabidopsis R genes tested (Figure 7). H. parasitica isolates Cala2 (recognized by RPP2) and Emwa1 (recognized by RPP4) were able to complete their life cycles in lines 8-7 and 8-9. Pathogen colonization was almost as extensive as that observed in the Col-0 sgt1beta3 mutant, measured by the production of conidiospores (Figures 7A and 7B) and trypan blue staining of leaves (Figure 7C). Partial loss of resistance was visualized by the appearance of trailing necrosis around pathogen hyphae, similar to that seen in Col sgt1beta3 (Figure 7C). RPM1-mediated recognition of Pst DC3000/avrRpm1 and RPS4 recognition of Pst DC3000/avrRps4 were also compromised (Figures 7D and 7E). Thus, HSC70-1 overexpression partially disables R gene–conditioned resistance to avirulent isolates of H. parasitica and Pst. Combining the sgt1beta3 mutation and HSC70-1 overexpression caused an additive loss of RPP4-mediated resistance (see Supplemental Figure 5 online), in support of a genetic interaction between these two components.
Figure 7.
HSC70-1 Overexpression Disables R Gene–Mediated Resistance to Pathogens.
Arabidopsis Col-0 transgenic lines 8-7 and 8-9 overexpressing HSC70-1 were inoculated with avirulent H. parasitica ([A] to [C]) or strains of Pst DC3000 ([D] and [E]). Two-week-old seedlings were inoculated with H. parasitica isolates Cala2 ([A] and [C]) or Emwa1 (B). Spores were counted at 6 d after inoculation ([A] and [B]), and leaves were stained with trypan blue (C) to visualize fungal structures and tissue necrosis. Ler and Ws-0 plants are susceptible to Cala2 and Emwa1, respectively. Ler eds1-2 and Ws-0 eds1-1 display enhanced susceptibility to Cala2 and Emwa1, respectively. FW, fresh weight; HR, hypersensitive response; M, mycelium; TN, trailing necrosis. Growth of Pst DC3000 strains expressing avrRpm1 (D) or avrRps4 (E) was measured at 0 and 3 d after vacuum infiltration of different lines, as indicated. Col-0 rps3-3 is an rpm1 mutant that has lost recognition of Pst DC3000/avrRpm1. The rar1-13 mutation (Ler ecotype) disables RPS4. Each data point was analyzed in triplicate, and error bars indicate se. Three independent experiments gave similar results. Statistically significant differences for values compared with the wild type were determined by Student's t test (* P < 0.05, ** P < 0.005). For (F), HSC70-1 overexpression (OE) does not affect RPM1-myc accumulation. Total leaf protein extracts were prepared from 3-week-old plants homozygous for RPM1-myc and HSC70-1 overexpression (line 8-7) transgenes and analyzed by SDS-PAGE and immunoblotting using anti-Myc and anti-HSC70 antibodies. Equal loading was checked by Ponceau red staining of the membrane.
In order to test the effect of HSC70-1 overexpression on R protein accumulation, line 8-7 was crossed with Col-0 plants expressing a functional RPM1-myc protein (Boyes et al., 1998), and plants homozygous for the RPM1-myc and HSC70-1 overexpression constructs were selected. As shown in Figure 7F, HSC70-1 overexpression did not alter RPM1-myc steady state levels. It is unlikely, therefore, that perturbation of the assembly or maintenance of preexisting R proteins is the cause of the resistance defects arising from HSC70-1 overexpression.
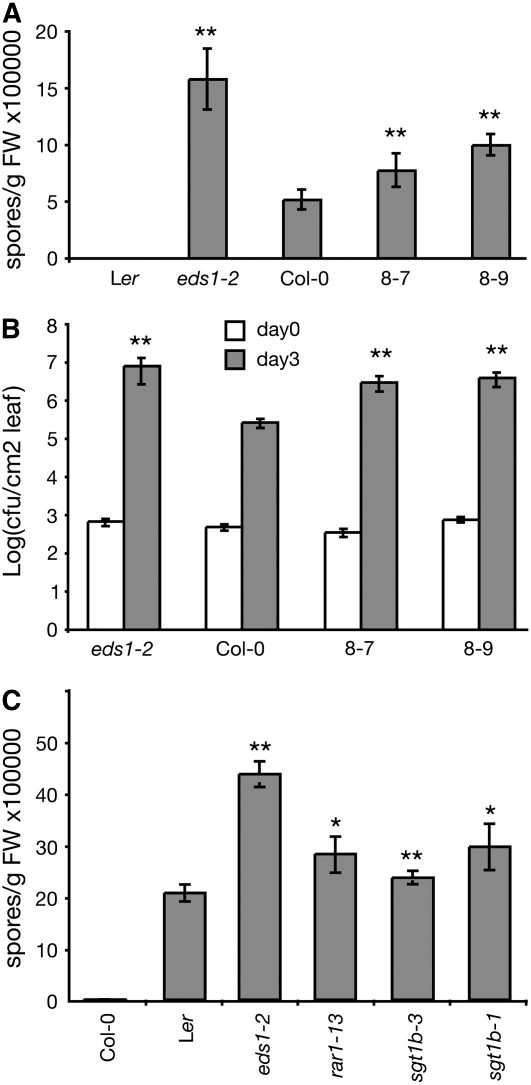
We considered whether the above phenotypes might reflect a defect in basal resistance that normally restricts the growth of virulent pathogen isolates. Growth of the virulent H. parasitica isolate Noco2 (Figure 8A) and Pst DC3000 (Figure 8B) was greater in lines 8-7 and 8-9 than in wild-type Col-0 but not as extreme as in the basal defense mutant eds1-2. This observation prompted us to investigate the contribution of SGT1b to basal defense. Growth of the virulent H. parasitica isolate Cala2 was higher in Ler sgt1b-1 and sgt1b-3 null mutants compared with the Ler wild type (Figure 8C). We conclude that SGT1b and HSC70-1 modulate both basal and R protein–specified immune responses.
Figure 8.
HSC70 and SGT1 Modulate Plant Basal Resistance.
Different Arabidopsis lines were inoculated with virulent isolates of H. parasitica Noco2 (A) and Cala2 (C) or Pst DC3000 (B). Pathogen growth was measured as described for Figure 7. 8-7 and 8-9 are two transgenic lines (Col-0) overexpressing HSC70-1. Ler eds1-2 and rar1-13 have compromised basal resistance. Ler sgt1b-1 and sgt1b-3 are two independent sgt1b null mutants. Statistically significant differences for values compared with the wild type were determined by Student's t test (* P < 0.05, ** P < 0.005). FW, fresh weight.
Mutations in SGT1b and Deregulation of HSC70-1 Lead to Heat Shock Tolerance
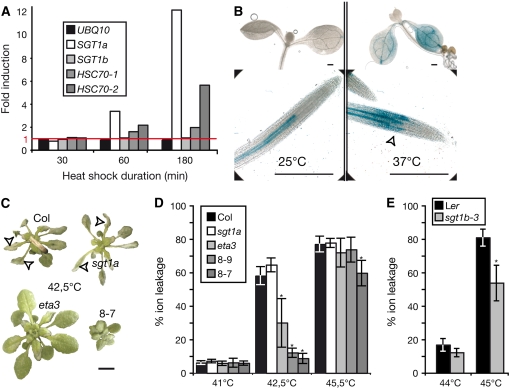
We explored whether the interaction between SGT1 and HSC70 cytosolic isoforms has broader biological significance by examining the requirement for SGT1 in the Arabidopsis heat shock response, one of several abiotic stresses that recruit HSC70 chaperone functions (Sung and Guy, 2003). Data from a gene expression microarray experiment that traced the heat shock response of 18-d-old Arabidopsis plantlets grown in liquid at 25°C and then incubated at 38°C for 30, 60, or 180 min (L. Nover and P. von Koskull-Doring, http://www.Arabidopsis.org/servlets/TairObject?type=expression_setandid=1007967124) were examined. After 60 and 180 min at 38°C, SGT1a mRNA accumulated to 3- and 12-fold higher levels, respectively, than in control samples incubated at 25°C, whereas SGT1b and UBQ10 expression remained unchanged (Figure 9A). HSP70-1 was moderately and HSC70-2 was strongly induced by heat shock treatment, as described previously (Lin et al., 2001). There was also a dramatic increase in the activity of the SGT1a promoter followed by a GUS reporter gene (Azevedo et al., 2006) after heat shock. In seedlings grown at 25°C, detectable pSGT1a:GUS activity was restricted to two cell lineages of the root pericycle (Figure 9B). After 3 h of incubation at 37°C, GUS activity was detected in cotyledon tissues and was expressed in outer cell layers of the root (Figure 9B). The strong induction of mRNAs for SGT1a and the comparative low responsiveness of SGT1b after heat shock broadly mirror their modes of expression in response to pathogens (Figures 2 and 3) (Azevedo et al., 2006). We then tested the heat shock tolerance of plants by immersing 4-week-old plants grown at 25°C in a water bath at 41 to 48°C for 10 min, as described (Sung and Guy, 2003). Three days after treatment, extensive tissue collapse was observed in leaves of wild-type Col-0 and in the sgt1a mutant after treatment at 42.5°C (Figure 9C). By contrast, sgt1beta3 mutant plants displayed reduced tissue damage resembling that of the thermotolerant HSC70-1–overexpressing lines 8-7 and 8-9 (Sung and Guy, 2003). The extent of cell collapse in these lines was quantified in five plants by measuring the percentage of total ions that leaked from leaves after heat shock. One hundred percent refers to the total ion content in the plant sample after it had been microwaved to release all ions. sgt1beta3 exhibited a similar degree of heat shock tolerance as the HSC70-1–overexpressing lines 8-7 and 8-9 (Figure 9D). Ler sgt1b-3 mutant plants were also more tolerant of heat shock than was the Ler wild type (Figure 9E). We conclude that SGT1 and HSC70 proteins have some overlapping functions in modulating responses to biotic and abiotic stresses. A correlation between the loss of HSC70 interaction of the sgt1beta3 mutant protein and the increased thermotolerance of sgt1beta3 plants further implies that heat shock sensitivity in the wild type involves the interaction of HSC70 with SGT1.
Figure 9.
Arabidopsis sgt1b Mutants Exhibit Enhanced Tolerance to Heat Shock.
(A) SGT1a, SGT1b, HSC70-1, HSC70-2, and UBQ10 gene expression after heat shock (38°C) relative to control conditions (25°C) was determined by meta-analysis of Arabidopsis gene expression microarray hybridization experiments. Samples were harvested after 30, 60, or 180 min. The red line indicates the induction level of genes whose expression is not affected by heat shock at 38°C. UBQ10, ubiquitin constitutive control.
(B) SGT1a promoter activity. Transgenic plants expressing the GUS reporter gene under the control of the SGT1a promoter were grown for 7 d at 25°C (left panel) and placed at 37°C (right panel) for 3 h before staining for GUS activity (shown in blue). Bar = 0.4 mm. The open arrowhead marks the site of intense GUS activity in the root after heat shock. One representative transgenic line is shown (AB1-1; Azevedo et al., 2006).
(C) Appearance of 5-week-old plants at 3 d after a 10-min incubation at 42.5°C. Open arrowheads indicate tissue collapse. Bar = 1 cm.
(D) and (E) The extent of cell collapse was measured in different lines by ion leakage at 3 d after a 10-min incubation at 41 to 45.5°C (D) or at 44 and 45°C (E). Ion leakage was normalized to the total ion content in the plant sample after it had been microwaved to release all ions. The percentage of total ions leaking from leaf tissue after heat shock reflects the proportion of cells that have lost cell integrity. Values are means from five individual plants. Error bars indicate sd. Statistically significant differences for values compared with the wild type were determined by Student's t test (* P < 0.005).
DISCUSSION
We present evidence that two functional isoforms of Arabidopsis SGT1, SGT1a and SGT1b, form stable interactions with cytosolic/nuclear HSC70 chaperones in vivo and that these interactions depend on the SGS domain of SGT1. Several observations point to SGT1 behavior as a HSC70 cofactor rather than a substrate. First, SGT1 is not bound strongly by the HSC70 client binding domain. Second, interaction between SGT1beta3 and HSC70 is reduced, arguing against potentially misfolded SGT1 serving as an HSC70 client. Third, the 36 residues that are missing in SGT1beta3 form part of a hydrophilic domain with helices that are not optimally recognized as a substrate by HSC70 chaperones (Rudiger et al., 1997; Lee et al., 2004). Resistance responses in Arabidopsis to virulent and avirulent pathogen isolates are compromised both by mutations in SGT1b and by deregulation of HSC70-1 expression. This newly identified connection between SGT1 and HSC70 activities correlates with an enhanced heat shock tolerance of sgt1b mutants, resembling the HSC70-1 overexpression phenotype. Together with the colocalization of SGT1 and HSC70 proteins in the cytosol and nuclei, these data lead us to propose that the interaction between SGT1 and HSC70 chaperones is important for plant defense against pathogens and some other stress responses that are controlled by HSC70 chaperones. Our findings are reinforced by a global study of the yeast Hsp90 interaction network that identified Sgt1 as an Hsc70 (Ssa1, Ssa2) interactor (Zhao et al., 2005). Also, coimmunoprecipitation of human Sgt1 and Hsp70 was recently reported after transfection of cultured cells and in ELISAs of recombinant proteins (Spiechowicz et al., 2007). The analysis by Spiechowicz et al. (2007) and results from our size-exclusion and N. benthamiana interaction assays (Figures 4 and 5) support direct chaperone–cochaperone binding. However, we did not observe any specific interaction between recombinant SGT1 and HSC70-1 in vitro or in yeast two-hybrid assays; therefore, we cannot exclude the possibility that their association is mediated or assisted by another component.
Cytosolic HSC70s and SGT1 Function in Plant Postinvasive Defense
Consistent with a role of cytosolic HSC70s in plant resistance, transcripts of the HSC70-2 and HSC70-4 isoforms were responsive to bacterial pathogen infiltration (Figure 3A). Induction of HSC70-2 and HSC70-4 mRNAs preceded the appearance of cell death (data not shown) and SGT1a mRNA accumulation (Figure 3A) and exhibited slightly faster kinetics than was reported for Arabidopsis HSP90-1, which is preferentially recruited in RPS2 resistance (Takahashi et al., 2003). Therefore, the observed boost in cytosolic HSC70 levels (Figure 3B) is unlikely to be a consequence of plant cell death. There was no impairment of the response of the Arabidopsis sgt1beta3 mutant or the HSC70-1–overexpressing lines to a host nonadapted fungal pathogen, Blumeria graminis f. sp. hordei (V. Lipka, personal communication). Resistance to this pathogen in wild-type Arabidopsis is expressed prior to the invasion of cells and is not associated with programmed plant cell death (Lipka et al., 2005). Also, virus-induced gene silencing of SGT1 in N. benthamiana showed that it is necessary for multiple cell death–associated disease resistance programs (Peart et al., 2002). Therefore, the activities of both cytosolic/nuclear HSC70s and SGT1 appear to be most important for plant resistance to pathogens once they have invaded host cells.
We did not observe alterations in resistance phenotypes associated with the depletion of specific cytosolic HSC70 mRNAs, probably due to high levels of functional redundancy. Consistent with this, combining the hsc70-1 and hsc70-3 mutations produced seedling lethality (data not shown). However, overexpression of Arabidopsis HSC70-1 disabled immune responses (Figure 7) and enhanced tolerance to heat shock (Figure 9). Given that sgt1b mutants displayed similar immune response (Figures 7 and 8) and heat stress phenotypes (Figure 9) as HSC70-1 overexpression and that an sgt1b mutation enhanced the disease susceptibility of an HSC70-1 overexpression line (see Supplemental Figure 5 online), SGT1 could behave as an important modulator of HSC70, balancing its various activities and interactions within the chaperone–cochaperone network.
Arabidopsis SGT1b did not discriminate in association with particular cytosolic HSC70 isoforms in leaves (Figure 2C), consistent with the presence of stable SGT1-HSC70 complexes in healthy and pathogen-treated tissues. However, we observed stronger binding of SGT1b than SGT1a to HSC70-1 in N. benthamiana interaction assays (Figure 4E). SGT1a was also reported to bind to HSP90 less strongly than SGT1b (Hubert et al., 2003). In healthy Arabidopsis leaves, SGT1b protein accumulates to approximately fourfold higher amounts than SGT1a, although SGT1a is strongly pathogen-induced (Figure 3) and can compensate for the loss of SGT1b when expressed at high levels (Azevedo et al., 2006). The lower abundance of SGT1a coupled with the reduced affinity for HSC70 may account for the preferential genetic recruitment of SGT1b in immunity and auxin or jasmonic acid sensing (Austin et al., 2002; Gray et al., 2003). Also, differences in the subcellular distribution of SGT1a and SGT1b observed here (Figure 6C) may influence their competence as cochaperones. Notably, SGT1b import to nuclei predominated over SGT1a in wild-type tissues. The finding that SGT1a accumulated in nuclear preparations of mutant plants expressing the nonfunctional SGT1eta3 protein suggests that SGT1 nuclear accumulation is an important aspect of SGT1 activity and possibly also HSC70 interaction.
HSC70 and SGT1b Modulate Basal Resistance to Virulent Pathogens
An unexpected finding was the partial disabling of basal resistance to H. parasitica isolates in sgt1b null mutants and the HSC70-1 overexpression lines (Figure 8). This form of low-level resistance restricts pathogen colonization in the absence of cell death (Chisholm et al., 2006). Since in yeast and plants, SGT1 affects the functions of several unrelated protein complexes (Muskett and Parker, 2003; Shirasu and Schulze-Lefert, 2003), it is conceivable that SGT1 controls several molecularly independent steps in plant immunity. HSP90 and SGT1 are needed for the steady state accumulation of at least one NB-LRR receptor, Rx (Azevedo et al., 2006), but it is not known whether this is universal for NB-LRR proteins. An absence of obvious effects of HSC70-1 overexpression on RPM1-myc accumulation (Figure 7F) supports HSC70 action at a site other than NB-LRR protein stabilization. There were no differences in SGT1a and SGT1b protein accumulation between HSC70-1 overexpression lines and the wild type (data not shown), arguing against the depletion of SGT1 being an explanation for the loss of resistance. Nevertheless, HSC70 may cooperate with SGT1 (and HSP90) to regulate the levels of numerous low-affinity immune receptors that together could constitute basal resistance to invasive virulent pathogens (Holt et al., 2005).
A broader biological relevance of an SGT1-HSC70 complex is suggested by the finding that exposure of plants to heat shock elicits strong induction of SGT1a expression (Figures 9A and 9B), as found in pathogen responses (Azevedo et al., 2006), and enhanced heat shock tolerance in sgt1b leaves, as seen in the HSC70-overexpressing lines (Figures 9C to 9E). We think that these related phenotypes reflect a functional connection between SGT1 and cytosolic/nuclear HSC70 chaperones. However, HSC70-1 overexpression does not mirror all sgt1b loss-of-function phenotypes, since it did not compromise sensitivity to auxin by the root growth inhibition assay (L.D. Noël, unpublished data).
Does SGT1 Bridge HSP90-HSC70 Chaperone Activities?
An association between Sgt1 and Hsp90 in multiple systems and our finding that Arabidopsis SGT1 and HSC70 interact in vivo prompt the question of whether SGT1 serves to coordinate HSP90-HSC70 chaperone functions. Although Hsp90 and Hsc70 proteins have distinct biochemical roles, there is a substantial degree of cooperativity between these two chaperones in regulating protein complex assembly and subcellular trafficking (Pratt et al., 2004). Such coordinated activities are in large part mediated by cochaperones. For example, the TPR protein Hop (known as Sti1 in yeast) binds Hsc70 and Hsp90 and ensures optimal substrate channeling within a single multiple-chaperone complex (Hernandez et al., 2002). Another cochaperone, CHIP (for C terminus of Hsc70-interacting protein) directs Hsp90/Hsc70 chaperone complexes to the proteasome by attaching a ubiquitin signal onto the chaperone-bound client protein (Connell et al., 2001; Esser et al., 2004). Analysis in yeast and human showed that the Hsp90–Sgt1 association is transient (Kitagawa et al., 1999; Lee et al., 2004; Lingelbach and Kaplan, 2004; Catlett and Kaplan, 2006), and this is likely also to be the case in plants, since only a very small proportion of the total SGT1 pool was bound to HSP90 in vivo (Hubert et al., 2003). We failed to identify HSP90 in SGT1-Strep affinity purification experiments. We also could not detect RAR1 or ASK1 (the major Arabidopsis SKP1 protein in leaves), probably due to the transient and unstable nature of these associations, as reported for interactions between Sgt1p and Skp1p in yeast (Kitagawa et al., 1999; Lyapina et al., 2001). In yeast, human, and plant cells, Sgt1 interaction with the Hsp90 ATPase domain is mediated principally through its central CS (p23-like) domain, although Sgt1 binding does not influence the ATP hydrolysis rate of Hsp90, in contrast with other cochaperones such as Hop/Sti1 and Hip/p60 (Takahashi et al., 2003; Lee et al., 2004; Catlett and Kaplan, 2006). Thus, the Sgt1 CS domain is both necessary and sufficient for Hsp90 binding and should not be affected directly by SGS modifications (Takahashi et al., 2003; Lee et al., 2004). Consistent with this model, the SGT1a CS domain failed to bind HSC70 in N. benthamiana interaction assays (Figure 5A) and the SGT1eta3 mutant protein retained interaction with RAR1 and HSP90 in a yeast two-hybrid assay (see Supplemental Figure 3 online).
Yeast Sgt1p appears to behave as an adaptor for client proteins and in this capacity is able to link Skp1 to Hsp90 (Catlett and Kaplan, 2006). Hop/Sti1 is one of a number of TPR domain cochaperones that bind HSP90 at its C-terminal EEVD domain and acts as a coupling factor between Hsp70 and Hsp90 to fold proteins such as the glucocorticoid receptor or to assemble protein complexes (Hernandez et al., 2002; Pratt et al., 2004). Hsp70 promotes the initial steps of glucocorticoid receptor folding and transfers its substrate to Hsp90 to complete maturation. Yeast Sgt1p can form ternary complexes with Hsp90 and Hop (Catlett and Kaplan, 2006), and this might influence substrate channeling between the two chaperones as well as their binding of other cofactors. Our finding that the plant SGT1 interaction with cytosolic/nuclear HSC70s requires its SGS domain could allow cooperative binding of HSP90 through the CS domain and an association with SKP1 and the core SCF ubiquitin E3 ligase system through the SGT1 TPR motif. However, the role of the TPR domain in plant SGT1 remains unclear, since it was dispensable for Arabidopsis SGT1 activity in R protein–triggered immunity and auxin sensing (Azevedo et al., 2006). If the association between SGT1 and SKP1 is solely through the TPRs (Catlett and Kaplan, 2006), this would argue against the SGT1–SKP1 interaction being important for SGT1 function in the plant immune and hormone responses. Another possibility is that SGT1 in plants connects with particular SCF E3 ligase complexes through an interaction between its SGS domain and the LRRs of certain F box proteins, such as TIR1 in SCFTIR1 and COI1 in SCFCOI1. In support of this, yeast two-hybrid interaction analysis by Dubacq et al. (2002) showed that yeast Sgt1p has a propensity to associate with LRRs or WD-40 repeats that are highly represented in F box and other signaling proteins. Also, interactions were observed between SGT1 and the LRR domains of barley MLA1 in yeast two-hybrid assays (Bieri et al., 2004) and pepper (Capsicum annuum) BS2 in transient N. benthamiana expression assays (Leister et al., 2005). Notably, the SGT1–MLA association was observed with TPR deletants and with a truncated SGT1 protein containing only the SGS domain (Bieri et al., 2004). We propose, therefore, that SGT1 may bridge the HSP90-HSC70 chaperone machinery with a selected number of domains during protein complex maturation and/or activation. The stable interaction of SGT1 with cytosolic HSC70 observed in our study may reflect a default state that is then directed toward assembly or disassembly functions through transient interaction with HSP90 and the activities of other cochaperones.
Significance of the SGT1 SGS Domain
The SGS domain is necessary and sufficient for the binding of SGT1 to HSC70 in N. benthamiana transient interaction assays. Several SGS domain mutant alleles (e.g., in sgt1-5) were identified in yeast Sgt1 that block the G1/S transition in cell cycle progression and are affected in the SCF-dependent degradation of Cln1p/Cln2p cyclins and Sic1p Cdk inhibitor (Kitagawa et al., 1999; Bansal et al., 2004). These contrast with mutations in the yeast Sgt1p TPR domain (such as those encoded by sgt1-3) that lose interaction with the kinetochore component Skp1p and are blocked in the G2/M transition of the cell cycle. Thus, in yeast, the SGS domain mediates distinct SGT1 functions, consistent with its genetic requirement for adenylyl cyclase Cyr1p activation (Dubacq et al., 2002). To our knowledge, HSC70 chaperones are the only direct SGS interactors to date. The SGS domain is the most highly conserved in SGT1 (59% identity between human SGT1 and Arabidopsis SGT1b) but is unfolded with a limited degree of helical structure (Lee et al., 2004). Due to the high conservation of the SGS domain in eukaryotes and the ability of an SGT1b TPR domain deletant to complement the sgt1b-1 mutation (Azevedo et al., 2006), we propose that sgt1beta3 may represent a plant sgt1 G1 allele. This raises the question of whether yeast sgt1 G1 phenotypes could in part be due to a loss of SGT1–HSP70 interaction. To date, HSP70 has been implicated in cell cycle regulation as a high-copy suppressor of the G2 cell cycle arrest induced by human immunodeficiency virus (Iordanskiy et al., 2004). A role of HSC70 in the cell cycle may be worth exploring.
METHODS
Plant Materials, Growth Conditions, and Pathology Assays
Wild-type Arabidopsis thaliana accessions used were Col-0, Ler, and Ws-0. The Ler sgt1b-3 and sgt1b-1(Austin et al., 2002), Ler eds1-2 (Falk et al., 1999), Ler rar1-13 (Muskett et al., 2002), Ws-0 eds1-1 (Parker et al., 1996), Col-0 rps3-3 (Bisgrove et al., 1994), Col-0 sgt1beta3 (Gray et al., 2003), Col-0 pad4-1 (Glazebrook et al., 1997), and Ws-0 sgt1a-1 (Hubert et al., 2003) mutants have been described. Lines overexpressing SGT1b-Strep (Ler sgt1b-3 pXCS-SGT1b-Strep) were described previously (Witte et al., 2004). Homozygous Col-0 plants overexpressing HSC70-1 (Sung and Guy, 2003) were used: lines 8-7 (single insertion) and 8-9 (multiple insertions). Col-0 plants expressing functional RPM1-myc protein (Boyes et al., 1998) were crossed with HSC70-1–overexpressing line 8-7, and plants homozygous for RPM1-myc and HSC70-1 transgenes were selected in the progeny. Col-0 T-DNA insertion mutants in HSC70-1 (SALK_135531), HSC70-2 (SALK_085076), HSC70-3 (GK_758E01), HSC70-4 (SALK_029571), and SGT1a (GK_266H09) were ordered from the SALK and GABI-KAT databanks, and the insertions were verified by PCR and made homozygous. HSC70 gene expression was measured in these lines by RT-PCR. Two independent transgenic lines expressing the GUS gene reporter under the control of the SGT1a promoter have been described (Azevedo et al., 2006).
For pathogenicity tests, plants were grown in soil under a 10-h photoperiod at 22°C with light intensity of 180 to 200 μE·m−2·s−1 and 65% humidity. Hyaloperonospora parasitica and Pseudomonas syringae pv tomato DC3000 isolates were cultured and prepared for inoculation as described (Aarts et al., 1998; Muskett et al., 2002). Two-week-old seedlings were sprayed with 4 × 104 H. parasitica conidiospores/mL in distilled water. Infection phenotypes were scored at 6 to 7 d after infection by lactophenol trypan blue staining and counting of conidiospores as described (Muskett et al., 2002). Bacterial growth tests were performed by vacuum infiltration of a bacterial suspension (1 × 105 cfu/mL) into leaves of 5- to 6-week-old plants. Each data point was analyzed at least in triplicate.
Arabidopsis Hormone and Heat Shock Response Assays
For root growth inhibition assays, seedlings were grown in sterile conditions on vertically oriented Murashige and Skoog (MS/10) medium containing 0.5% sucrose in a white light growth chamber under a 16-h photoperiod at 24/21°C (light/dark). Five-day-old seedlings were transferred to MS/10 medium containing different concentrations of methyl jasmonate (Duchefa) or 2,4-D (Sigma-Aldrich). Root length was measured at 3 d after transfer. To induce a heat shock response, 7-d-old plantlets growing on MS/10 medium at 25°C were incubated at 38°C for 3 h or kept at 25°C (control condition) and stained for GUS activity for 1 to 2 h as described (Jefferson, 1987). Heat shock tolerance was tested as described (Sung and Guy, 2003). Values are means of at least three biological replicates.
Generation of Strep-Tagged SGT1 Variants
In order to express SGT1a-Strep and SGT1b-Strep from their own promoters, SGT1 coding regions with their 1.3-kb upstream promoter sequences (Tor et al., 2002) were amplified from Col-0 genomic DNA. Primer sequence information is available upon request. Amplicons were cloned into pENTR/D-TOPO (Invitrogen) and transferred into pXCG-Strep by LR recombination (Invitrogen), giving pXCG-SGT1a-Strep and pXCG-SGT1b-Strep, respectively. A pXCSG-Strep derivative, pXCG-Strep (Witte et al., 2004), without the 35S promoter, was obtained by AscI/XhoI digestion of pXCSG-Strep, fill-in, and religation. To express SGT1 CS and SGS domains and the truncated SGT1beta3 protein with a C-terminal StrepII tag under the control of the 35S promoter, the corresponding SGT1 sequences were PCR-amplified from Ler cDNA clones and cloned into pENTR/D-TOPO (Invitrogen). Amplicons were transferred by LR recombination into pXCSG-Strep, giving pXCSG-CSa-Strep, pXCSG-SGSb-Strep, and pXCSG-ETA3-Strep.
Generation of HA-Tagged HSC70-1 Derivatives
Amplicons corresponding to the full-length ATPase domain and the client binding domain (CBD) of HSC70-1 were PCR-amplified from a Col-0 cDNA library and cloned into pENTR/D-TOPO (Invitrogen). The amplicons were transferred by LR recombination into pJ2B-3HA-GW (N. Medina-Escobar and J.E. Parker, unpublished data), giving pJ2B-3HA-HSC70FL, pJ2B-3HA-HSC70ATPase, and pJ2B-3HA-HSC70CBD.
RT-PCR Analysis
Four-week-old Col-0 plants were hand-inoculated with Pst DC3000 strains (107 cfu/mL) in 10 mM MgCl2 or with 10 mM MgCl2 alone as a control. RNA was isolated from leaves using Tri reagent (Sigma-Aldrich) for use as RT-PCR template. One microgram of total RNA was subjected to reverse transcription using SuperScript II (Invitrogen). Primer specificity was confirmed by direct sequencing of RT-PCR amplicons. The number of cycles used was determined empirically to be within the linear amplification phase. RT-PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
Plant Transformations
Agrobacterium tumefaciens strain GV3101∷pMP90RK was used for transient protein expression in Nicotiana benthamiana using pXC/pXN binary derivatives, as described (Witte et al., 2004). Stable transformation of Arabidopsis was done by flower dipping as described (Clough and Bent, 1998). Transformants were selected by spraying soil-grown 7-d-old seedlings with 0.1% Basta (Aventis). Resistant T1 transgenic lines expressing detectable amounts of the fusion proteins were self-pollinated to produce T2 seeds. Single insertion lines were selected by segregation analysis of the resistance in T2 populations on MS medium containing 10 μg/mL phosphinotricin (Duchefa).
StrepII Affinity Purification and Immunoprecipitation
StrepII affinity purifications were performed as described (Witte et al., 2004). For immunoprecipitation experiments, 1 g of leaf tissue was homogenized in 2 mL of W buffer (50 mM Tris, pH 8, 150 mM NaCl, 0.05% Triton X-100, and 2 mM DTT) supplemented with plant protease inhibitor cocktail (Sigma-Aldrich P9599; diluted 1:200). After centrifugation, 500 μL of the cleared protein extract was incubated with 10 μL of antibody by end-over-end rotation at 4°C for 2 h. The following antibodies were used: rabbit anti-SGS (raised against the SGT1a SGS domain; S. Betsuyaku, A. Takahashi, K. Shirasu, and J.E. Parker, unpublished data) and rabbit anti-SGT1b (Austin et al., 2002). Washed Protein A–Sepharose Fast Flow (Amersham Biosciences) was added to samples and incubated for 1 h. Beads were washed three times with 1 mL of W buffer and finally resuspended in 25 μL of Laemmli buffer.
SDS-PAGE and Immunoblotting
Proteins were separated by SDS-PAGE on 10 or 12% gels and transferred onto nitrocellulose or polyvinylidene difluoride membranes. Immunoblots with the Strep-Tactin-AP conjugate (catalog number IBA 2-1503-001) was performed as described (Witte et al., 2004). The following antibodies were used: monoclonal mouse anti-spinach HSP70 (Stressgen SPA-817); rat anti-HA antibody (Roche 1 867 423); mouse anti-c-Myc (clone 9E10; Santa Cruz Biotechnology); anti-SGS antibody (as above); rabbit anti-SGT1b antibody (Austin et al., 2002); rabbit anti-PEPC (Rockland); rabbit anti-histone H3 (Abcam) antibodies; goat anti-rabbit IgG horseradish peroxidase (HRP) conjugate (Santa Cruz Biotechnology sc-2004); goat anti-mouse IgG HRP conjugate (Santa Cruz Biotechnology sc-2005); and goat anti-rat IgG HRP conjugate (Santa Cruz Biotechnology sc-2006). AP and HRP activities were detected with p-nitroblue tetrazolium and enhanced chemiluminescence (SuperSignal West Femto chemiluminescent substrate; Pierce), respectively. Silver staining was performed as described (Shevchenko et al., 1996).
Preparation of Arabidopsis Nuclear Extracts
Nuclei-enriched/depleted fractions were prepared from leaves of 4-week-old-seedlings as described (Feys et al., 2005).
Yeast Two-Hybrid Analysis
Sequences encoding RAR1, HSP90-1, HSP90ATPase, SGT1b, SGT1beta3, and HSC70-1 were recombined into Gateway-converted versions of the vectors pGADT7 and pGBKT7 (Clontech) (T. Lahaye, unpublished data). Plasmids were transformed into yeast strains AH109 and Y187 (Clontech). Diploids containing both bait and prey constructs were constructed by mating. Expression of the fusion proteins was checked by protein gel blot analysis using c-Myc and HA antibodies. Dilution series of the obtained diploids were plated on synthetic defined selective medium to screen for interaction according to the manufacturer and grown for 3 d at 28°C.
Construction of Cerulean and YFP Fusions
To generate fluorescent protein destination vectors for Gateway (GW) cloning technology (Invitrogen), PCR-amplified Cerulean (Rizzo et al., 2004) and eYFP (Shah et al., 2002) and a Gateway recombination cassette were inserted into the pXCS-HisHA vector (Witte et al., 2004), resulting in p35S-GW-Cerulean-nos and p35S-eYFP-GW-nos, respectively. The cDNA sequences of SGT1b (Witte et al., 2004) and HSC70-1 cloned into pENTR/D-TOPO (Invitrogen) were recombined into p35S-GW-Cerulean-nos and p35S-eYFP-GW-nos, respectively.
Transient Single-Cell Gene Expression Assays
Detached leaves from 5- to 6-week-old N. benthamiana plants cultivated as described (Witte et al., 2004) were used for transient gene expression in epidermal cells mediated by particle bombardment (Bhat et al., 2005). For colocalization, equimolar amounts of plasmids were coated onto the gold particles. After bombardment, leaves were incubated for 18 to 48 h at room temperature prior to microscopic analysis.
Confocal Laser Scanning Fluorescence Microscopy
Transient intracellular fluorescence was observed by confocal laser scanning microscopy using a Leica SP2 AOBS inverted confocal microscope (Leica Microsystems) equipped with argon ion (458-, 476-, 488-, 496-, and 514-nm laser lines) lasers. Additionally, a 405-nm diode laser (BDL 405 SMC; Becker and Hickl) was also installed into the Leica SP2 AOBS system. SGT1b-Cerulean was excited either with the 405-nm diode laser or the 458-nm argon laser line, while eYFP was excited with the 514-nm argon laser line. In cells coexpressing both SGT1b-Cerulean and eYFP-HSC70-1, the imaging was done in sequential mode. Cerulean fluorescence was detected using the Leica AOBS system and a custom 485- to 505-nm band-pass emission filter, while eYFP fluorescence was detected using the Leica AOBS system and a custom 515- to 560-nm band-pass emission filter. A 63× HCX PLAN-APO water-immersible objective lens (numerical aperture = 1.2) was used for imaging.
Mass Spectrometry
Tryptic digestion of proteins separated by SDS-PAGE was performed using a protocol based on that of Shevchenko et al. (2000). Digests were desalted for electrospray mass spectrometry with a C18 reverse-phase resin (ZipTip; Millipore). A Micromass Q-Tof-2 mass spectrometer operating with nanospray and the Masslynx software (version 3.5) were used for analysis.
Gel-Filtration Analysis
A 25-μL Ler protein sample prepared as for immunoprecipitation was loaded on a Superdex 200 HR 10/30 column (Amersham Biosciences) at 0.2 mL/min flow with W buffer without Triton X-100. Then, 0.5- and 1-mL fractions were sampled, precipitated using 10% trichloracetic acid, and analyzed by SDS-PAGE and immunoblotting. Column calibration was performed using the gel filtration low molecular weight and high molecular weight calibration kits (Amersham Biosciences).
Meta-Analysis of Arabidopsis Gene Expression Microarray Data
A microarray dataset describing the response of Arabidopsis plantlets to heat shock at 38°C was downloaded from The Arabidopsis Information Resource site (for detailed experimental setup and results, see http://www.Arabidopsis.org/servlets/TairObject?type=expression_setandid=1007967124) and processed using Genespring software (Silicon Genetics). Normalization per gene and per chip of the log2 values was performed, allowing comparison of the two independent replicates available. Normalized gene expression levels of duplicates were averaged and used to determine levels of induction relative to control growth conditions (25°C).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At4g16860 (RPP5); At3g48090 (EDS1); At3g52430 (PAD4); At5g51700 (RAR1); At4g23570 (SGT1a); At4g11260 (SGT1b); At5g02500 (HSC70-1); At5g02490 (HSC70-2); At3g09440 (HSC70-3); At3g12580 (HSC70-4); At1g16030 (HSC70-5); At2g14610 (PR1); At5g44340 (TUB4); At3g07040 (RPM1); At4g05320 (UBQ10); At2g39940 (COI1); and At5g52640 (HSP90-1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Functionality Tests of the StrepII-Tagged SGT1 Proteins in Transgenic Arabidopsis Described in Figure 1.
Supplemental Figure 2. Mass Fingerprinting of Two Peptides Discriminating between Cytosolic HSC70 Isoforms.
Supplemental Figure 3. Interaction Studies between SGT1 and RAR1 or HSP90-1 Using the Clontech Matchmaker GAL4 Yeast Two-Hybrid System.
Supplemental Figure 4. Individual Loss of Arabidopsis Isoforms 1, 2, and 3 Does Not Affect Plant Immune Responses.
Supplemental Figure 5. SGT1b Loss of Function and HSC70-1 Overexpression Have Additive Effects on R Gene–Mediated Resistance to H. parasitica.
Supplemental Table 1. Tryptic Fragments Observed by Mass Spectrometry of 70-kD Proteins Copurified with Overexpressed SGT1b-Strep.
Supplementary Material
Acknowledgments
We thank Volker Lipka for testing Arabidopsis nonhost resistance responses and the SALK Institute and Bernd Weisshaar (GABI-KAT collection) for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. We are grateful to Jacqueline Bautor and Serge Chiarenza for technical assistance and to Jorg Höhfeld (University of Bonn) and Raphael Guérois (Commissariat à l'Energie Atomique) for discussions on HSC70. This work was supported by the Max Planck Society and the Laboratoire de Biologie du Développement des Plantes, by Deutsche Forschungsgemeinschaft Grant SFB 635, by the Alexander von Humboldt Foundation, the Centre National de la Recherche Scientifique, and the Commissariat à l'Energie Atomique, and by Alexander von Humboldt and European Molecular Biology Organization long-term fellowships to L.D.N. and C.-P.W., respectively.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Laurent D. Noël (laurent.noel@cea.fr).
Online version contains Web-only data.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Betsuyaku, S., Peart, J., Takahashi, A., Noël, L., Sadanandom, A., Casais, C., Parker, J., and Shirasu, K. (2006). Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295 2073–2076. [DOI] [PubMed] [Google Scholar]
- Bansal, P.K., Abdulle, R., and Kitagawa, K. (2004). Sgt1 associates with Hsp90: An initial step of assembly of the core kinetochore complex. Mol. Cell. Biol. 24 8069–8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, R.A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., and Panstruga, R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 102 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri, S., Mauch, S., Shen, Q.H., Peart, J., Devoto, A., Casais, C., Ceron, F., Schulze, S., Steinbiss, H.H., Shirasu, K., and Schulze-Lefert, P. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove, S.R., Simonich, M.T., Smith, N.M., Sattler, A., and Innes, R.W. (1994). A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C., Nam, J., and Dangl, J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., and Chiosis, G. (2006). Hsp70 molecular chaperones: Emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 6 1215–1225. [DOI] [PubMed] [Google Scholar]
- Bukau, B., Weissman, J., and Horwich, A. (2006). Molecular chaperones and protein quality control. Cell 125 443–451. [DOI] [PubMed] [Google Scholar]
- Catlett, M.G., and Kaplan, K.B. (2006). Sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J. Biol. Chem. 281 33739–33748. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Connell, P., Ballinger, C.A., Jiang, J., Wu, Y., Thompson, L.J., Höhfeld, J., and Patterson, C. (2001). The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3 93–96. [DOI] [PubMed] [Google Scholar]
- Dubacq, C., Guerois, R., Courbeyrette, R., Kitagawa, K., and Mann, C. (2002). Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1 568–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse, A., Mayer, M.P., and Bukau, B. (2004). Mechanism of substrate recognition by Hsp70 chaperones. Biochem. Soc. Trans. 32 617–621. [DOI] [PubMed] [Google Scholar]
- Esser, C., Alberti, S., and Hohfeld, J. (2004). Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta 1695 171–188. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Wiermer, M., Bhat, R.A., Moisan, L.J., Medina-Escobar, N., Neu, C., Cabral, A., and Parker, J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ranea, J.A., Mirey, G., Camonis, J., and Valencia, A. (2002). p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529 162–167. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E.E., Crute, I.R., Holub, E.B., Hammerschmidt, R., and Ausubel, F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Muskett, P.R., Chuang, H.W., and Parker, J.E. (2003). Arabidopsis SGT1b is required for SCFTIR1-mediated auxin response. Plant Cell 15 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, M.P., Sullivan, W.P., and Toft, D.O. (2002). The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J. Biol. Chem. 277 38294–38304. [DOI] [PubMed] [Google Scholar]
- Holt, B.F., III, Belkhadir, Y., and Dangl, J.L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309 929–932. [DOI] [PubMed] [Google Scholar]
- Hubert, D.A., Tornero, P., Belkhadir, Y., Krishna, P., Takahashi, A., Shirasu, K., and Dangl, J.L. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanskiy, S., Zhao, Y., Dubrovsky, L., Iordanskaya, T., Chen, M., Liang, D., and Bukrinsky, M. (2004). Heat shock protein 70 protects cells from cell cycle arrest and apoptosis induced by human immunodeficiency virus type 1 viral protein R. J. Virol. 78 9697–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Kitagawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Heiter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a subunit of the SCF ubiquitin ligase complex. Mol. Cell 4 21–33. [DOI] [PubMed] [Google Scholar]
- Krobitsch, S., and Lindquist, S. (2000). Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA 97 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.T., Jacob, J., Michowski, W., Nowotny, M., Kuznicki, J., and Chazin, W.J. (2004). Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 279 16511–16517. [DOI] [PubMed] [Google Scholar]
- Leister, R.T., Dahlbeck, D., Day, B., Li, Y., Chesnokova, O., and Staskawicz, B.J. (2005). Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.L., Wang, J.S., Liu, H.C., Chen, R.W., Meyer, Y., Barakat, A., and Delseny, M. (2001). Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingelbach, L.B., and Kaplan, K.B. (2004). The interaction between Sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol. Cell. Biol. 24 8938–8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.W., and Dinesh-Kumar, S.P. (2002. a). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.L., Burch-Smith, T., Schiff, M., Feng, S.H., and Dinesh-Kumar, S.P. (2004). Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and RAR1 to modulate an innate immune response in plants. J. Biol. Chem. 279 2101–2108. [DOI] [PubMed] [Google Scholar]
- Liu, Y.L., Schiff, M., Marathe, R., and Dinesh-Kumar, S.P. (2002. b). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30 415–429. [DOI] [PubMed] [Google Scholar]
- Lu, R., Malcuit, I., Moffett, P., Ruiz, M.T., Peart, J., Wu, A.J., Rathjen, J.P., Bendahmane, A., Day, L., and Baulcombe, D.C. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C.S., Wolf, D.A., Wei, N., and Deshaies, R.J. (2001). Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 292 1382–1385. [DOI] [PubMed] [Google Scholar]
- Muskett, P., and Parker, J. (2003). Role of SGT1 in the regulation of plant R gene signalling. Microbes Infect. 5 969–976. [DOI] [PubMed] [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D.G., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, W.B., Galigniana, M.D., Morishima, Y., and Murphy, P.J. (2004). Role of molecular chaperones in steroid receptor action. Essays Biochem. 40 41–58. [DOI] [PubMed] [Google Scholar]
- Rizzo, M.A., Springer, G.H., Granada, B., and Piston, D.W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22 445–449. [DOI] [PubMed] [Google Scholar]
- Rudiger, S., Germeroth, L., Schneider-Mergener, J., and Bukau, B. (1997). Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, K., Russinova, E., Gadella, T.W.J., Willemse, J., and de Vries, S.C. (2002). The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev. 16 1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Chernushevich, I., Wilm, M., and Mann, M. (2000). De novo peptide sequencing by nanoelectrospray tandem mass spectrometry using triple quadrupole and quadrupole/time-of-flight instruments. Methods Mol. Biol. 146 1–16. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68 850–858. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.W., Zhou, F., Azevedo, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99 355–366. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., and Schulze-Lefert, P. (2003). Complex formation, promiscuity and multi-functionality: Protein interactions in disease-resistance pathways. Trends Plant Sci. 8 252–258. [DOI] [PubMed] [Google Scholar]
- Spiechowicz, M., Zylicz, A., Bieganowski, P., Kuznicki, J., and Filipek, A. (2007). Hsp70 is a new target of Sgt1—An interaction modulated by S100A6. Biochem. Biophys. Res. Commun. 357 1148–1153. [DOI] [PubMed] [Google Scholar]
- Sung, D.Y., and Guy, C.L. (2003). Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol. 132 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Casais, C., Ichimura, K., and Shirasu, K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tor, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Turk, F., Can, C., Dangl, J.L., and Holub, E.B. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Merritt, P., Sadanandom, A., Shirasu, K., Innes, R.W., and Dangl, J.L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte, C.P., Noël, L.D., Gielbert, J., Parker, J.E., and Romeis, T. (2004). Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55 135–147. [DOI] [PubMed] [Google Scholar]
- Young, J.C., Barral, J.M., and Ulrich Hartl, F. (2003). More than folding: Localized functions of cytosolic chaperones. Trends Biochem. Sci. 28 541–547. [DOI] [PubMed] [Google Scholar]
- Zhao, R., et al. (2005). Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120 715–727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.