Abstract
Nitric oxide (NO) is a free radical product of cell metabolism that plays diverse and important roles in the regulation of cellular function. S-Nitrosylation is emerging as a specific and fundamental posttranslational protein modification for the transduction of NO bioactivity, but very little is known about its physiological functions in plants. We investigated the molecular mechanism for S-nitrosylation of peroxiredoxin II E (PrxII E) from Arabidopsis thaliana and found that this posttranslational modification inhibits the hydroperoxide-reducing peroxidase activity of PrxII E, thus revealing a novel regulatory mechanism for peroxiredoxins. Furthermore, we obtained biochemical and genetic evidence that PrxII E functions in detoxifying peroxynitrite (ONOO−), a potent oxidizing and nitrating species formed in a diffusion-limited reaction between NO and O2− that can interfere with Tyr kinase signaling through the nitration of Tyr residues. S-Nitrosylation also inhibits the ONOO− detoxification activity of PrxII E, causing a dramatic increase of ONOO−-dependent nitrotyrosine residue formation. The same increase was observed in a prxII E mutant line after exposure to ONOO−, indicating that the PrxII E modulation of ONOO− bioactivity is biologically relevant. We conclude that NO regulates the effects of its own radicals through the S-nitrosylation of crucial components of the antioxidant defense system that function as common triggers for reactive oxygen species– and NO-mediated signaling events.
INTRODUCTION
Nitric oxide (NO) is a well-known signaling molecule with a broad spectrum of regulatory functions involved in physiological and pathophysiological processes in both animals and plants (Wendehenne et al., 2004; Delledonne, 2005). Its broad chemistry involves an array of interrelated redox forms with distinct chemical reactivities, and numerous potential targets of NO activity exist in plants. As a regulator of development, NO promotes germination, leaf extension, and root growth and delays leaf senescence and fruit maturation (Neill et al., 2003). As a crucial component of the resistance mechanisms against pathogenic infections, it cooperates with reactive oxygen species (ROS) to trigger hypersensitive cell death in infected tissue and participates in the activation of several defense genes (Delledonne et al., 1998).
It is now recognized that NO and its related species can introduce posttranslational modifications of proteins using different routes. In addition to its capability to bind metal ions of heme groups, as reported in animals for guanylate cyclase activation (Denninger and Marletta, 1999), NO can perform important posttranslational protein modifications through S-nitrosylation and nitration, which are the major NO-dependent protein modifications currently investigated (Zaninotto et al., 2006). S-Nitrosylation refers to the incorporation of the NO moiety to a Cys sulfur atom to form a S-NO bond (Martinez-Ruiz and Lamas, 2004). S-Nitrosylation of protein Cys residues can be produced by different reactive nitrogen species, including nitrogen oxides formed by the reaction of NO and O2 (Martinez-Ruiz and Lamas, 2004), or it can result from the transfer of the nitroso group from low molecular mass thiols like glutathione or free Cys (Hess et al., 2005).
Like phosphorylation, S-nitrosylation is a precisely targeted and rapidly reversible posttranslational modification that is employed by the cells to flexibly and specifically respond to changes in their environment (Mannick and Schonhoff, 2004). Indeed, S-nitrosylated proteins can be easily denitrosylated, as the S-NO bond is labile in a reductive microenvironment, allowing cells to flexibly and precisely adapt protein function in response to environmental signals (Hess et al., 2005). The remarkable specificity of S-nitrosylation is conferred by different factors, such as the subcellular compartmentalization of NO sources and the target protein (Hess et al., 2005). Although many examples of protein S-nitrosylation and the consequent modifications in activity have been reported in animal systems (Stamler et al., 2001), knowledge of S-nitrosylation target proteins in plants is scarce. Recently, a proteomic approach led to the identification of 63 proteins from cell culture extracts treated with S-nitrosoglutathione (GSNO) and 52 proteins from leaves treated with NO, confirming the existence of targets for S-nitrosylation in Arabidopsis thaliana (Lindermayr et al., 2005). Nevertheless, very little is known about the extent and physiological function of S-nitrosylation in plants (Zaninotto et al., 2006).
Emerging evidence indicates that S-nitrosothiol turnover may be part of the regulation of S-nitrosylation in addition to contributing to the regulation of NO biosynthesis. In animals, S-nitrosylation of the antioxidant tripeptide glutathione that produces GSNO is proposed as a possible reservoir of NO bioactivity (Jia et al., 1996). An enzyme that metabolizes GSNO, namely GSNO reductase, has been shown to control intracellular levels of GSNO and S-nitrosylated proteins in yeast and mice (Liu et al., 2004). Recently, a protein possessing GSNO reductase activity was found in Arabidopsis (Sakamoto et al., 2002), and mutation of the corresponding gene At GSNOR1 has provided the first insight into its function in controlling cellular S-nitrosothiol levels under regular metabolic conditions as well as during plant–pathogen interactions (Feechan et al., 2005). Since the loss of At GSNOR1 function compromises plant defense responses to pathogenic infections (Feechan et al., 2005), it is likely that S-nitrosylated molecules are involved in modulating signaling pathways in plants.
Using a proteomic approach involving two-dimensional gel electrophoresis and mass spectrometry, we monitored changes in the S-nitrosylated proteome of Arabidopsis during the progression of the hypersensitive response (HR) (Romero-Puertas et al., 2008), a condition in which NO is well known to accumulate. Among the proteins identified, we focused our attention on peroxiredoxin II E (PrxII E), a member of the peroxiredoxin family consisting of peroxidases that reduce H2O2 and alkyl hydroperoxides to H2O and the corresponding alcohol using reducing equivalents from thioredoxin or glutaredoxins (Dietz, 2003; Horling et al., 2003). In this work, we investigated the molecular mechanism for the S-nitrosylation of PrxII E and its functional significance.
RESULTS
Analysis of S-Nitrosylated PrxII E
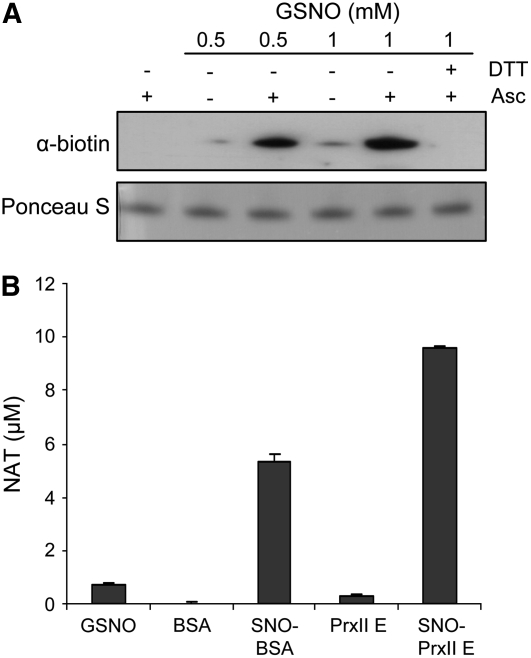
A recombinant form of PrxII E was expressed in Escherichia coli, purified, incubated with a molar excess of GSNO, and subjected to the biotin-switch assay, in which S-nitrosylated Cys residues are identified by the ascorbate-dependent release of NO followed by biotinylation of the free thiol. PrxII E was found to be S-nitrosylated in vitro, and the immunoreactivity of the protein was both GSNO- and ascorbate-dependent (Figure 1A). S-Nitrosylation of PrxII E was reversed by treatment with the reductant DTT (Figure 1A) and was unequivocally demonstrated by measuring the fluorescent compound 2,3-naphthyltriazole (NAT). NAT is stoichiometrically converted from 2,3-diaminonaphthalene (DAN) by NO released from S-nitrosylated proteins and thus provides a quantitative measure of S-nitrosothiol content (Gu et al., 2002). GSNO treatment of PrxII E resulted in significant S-nitrosothiol formation, as revealed by NAT formation (Figure 1B). GSNO itself as well as untreated proteins did not result in significant NAT formation (Figure 1B), thus ensuring that the detected fluorescence was specifically due to S-nitrosylated PrxII E.
Figure 1.
S-Nitrosylation of PrxII E.
(A) Immunodetection of PrxII E incubated with the indicated amounts of GSNO and then subjected to biotin-switch analysis. Where specified, the biotin-switch assay was performed in the presence of 1 mM ascorbate (Asc). As a negative control, GSNO-treated PrxII E was incubated with 10 mM DTT before performing the biotin switch. Protein loading was controlled by Ponceau red staining of polyvinylidene difluoride membrane after protein transfer.
(B) Specific fluorescence assay for the detection of S-nitrosylation by the conversion of DAN to the fluorescent compound NAT. BSA, reported for comparison, and PrxII E were incubated with 1 mM GSNO and then analyzed for NO release by measuring the fluorescence intensity of the reaction product. BSA and PrxII E not incubated with GSNO as well as GSNO itself resulted in insignificant S-nitrosothiol readings in the assay. The data represent means ± sd of three independent experiments.
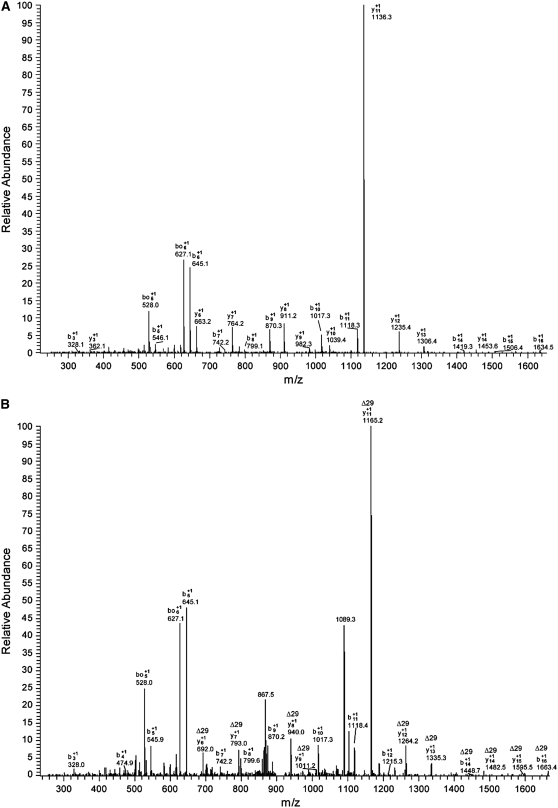
To determine PrxII E Cys residue(s) as target(s) of S-nitrosylation, we analyzed by electrospray ionization–tandem mass spectrometry (ESI-MS/MS) the protein treated with two different NO donors, GSNO and diethylamine-NONOate (DEA-NONOate). Trypsin digestion of the untreated protein released a peptide of m/z 890.73, which was identified by MS/MS to contain the residue Cys-121 of PrxII E. In both GSNO- and DEA-NONOate–treated samples, a corresponding tryptic peptide of m/z 905.90 (mass shift of 29) was observed at a similar retention time, and MS/MS analysis showed many of the fragmentation ions (containing Cys-121) shifted by 29. However, the MS/MS spectra obtained from treated proteins showed a low intensity, probably due to the labile nature of S-nitroso Cys (Cys-SNO), and could be further confounded with an abundant tryptic peptide (positions 79 to 95, with m/z 904.98) that eluted just before the modified peptide of interest. To get spectra of better quality and to confirm the MS/MS fragmentation patterns of the m/z 905.90 precursor ion obtained from tryptic digestions of treated PrxII E, we performed a second experiment in which untreated PrxII E was first digested with trypsin and AspN (predicted to cut between positions 79 and 95 but not between 108 and 124), then subjected to GSNO or DEA-NONOate treatment, and finally immediately added to the reaction mixture for MS analysis. This approach allowed us to reduce the time between NO donor treatment and MS analysis from ∼4 h to 15 min, increasing the yield of the labile SNO modification. Additionally, the abundance of the contaminating peptide (positions 79 to 95) was slightly reduced. In this way, improved MS/MS spectra of the SNO-modified peptide were obtained, showing the characteristic mass shift of Cys-121–containing fragment ions (Figure 2). It should be noted that other modifications of Cys-121 were also observed (i.e., sulfinic acid formation in treated and control samples and sulfonic acid formation in treated samples only). Nevertheless, MS analysis demonstrated that SNO modification occurs at this specific site of PrxII E.
Figure 2.
Tandem Mass Spectra of the Peptide TILFAVPGAFTPTCSQK from PrxII E.
The fragmentation is dominated by cleavages before the Pro residues at positions 7 and 12, resulting in intense b6 and y11+1 ions; this is typical of ion-trap MS. The MS/MS fragmentation was performed on the unmodified peptide (precursor m/z 890.70 [A]) and on the SNO-modified peptide (precursor m/z 905.90 [B]). Fragmentation of ions showing the diagnostic mass shift of +29 is indicated.
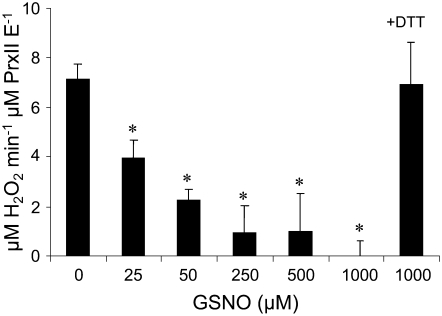
We then investigated the functional effect of S-nitrosylation on PrxII E peroxidase activity. GSNO was found to severely inhibit this activity in a concentration-dependent manner (Figure 3). This effect could be reversed by the thiol-specific reductant DTT, indicating that GSNO affects PrxII E activity through S-nitrosylation of the Cys residue.
Figure 3.
Effects of S-Nitrosylation on the Peroxidase Activity of PrxII E.
PrxII E was preincubated with the indicated amounts of GSNO for 30 min at room temperature and then subjected to the ferrithiocyanate assay using H2O2 as a peroxide substrate. At the end of the reaction, the remaining H2O2 was visualized as a red-colored complex quantified at 480 nm. Addition of 8 mM DTT at the end of the experiment restored peroxidase activity. The asterisks indicate values statistically different from those of the nontreated protein (0 μM GSNO; Student's t test, P < 0.05). Data shown are means ± sd of three independent experiments.
S-Nitrosylation Inhibits the Peroxynitrite Detoxification Activity of PrxII E
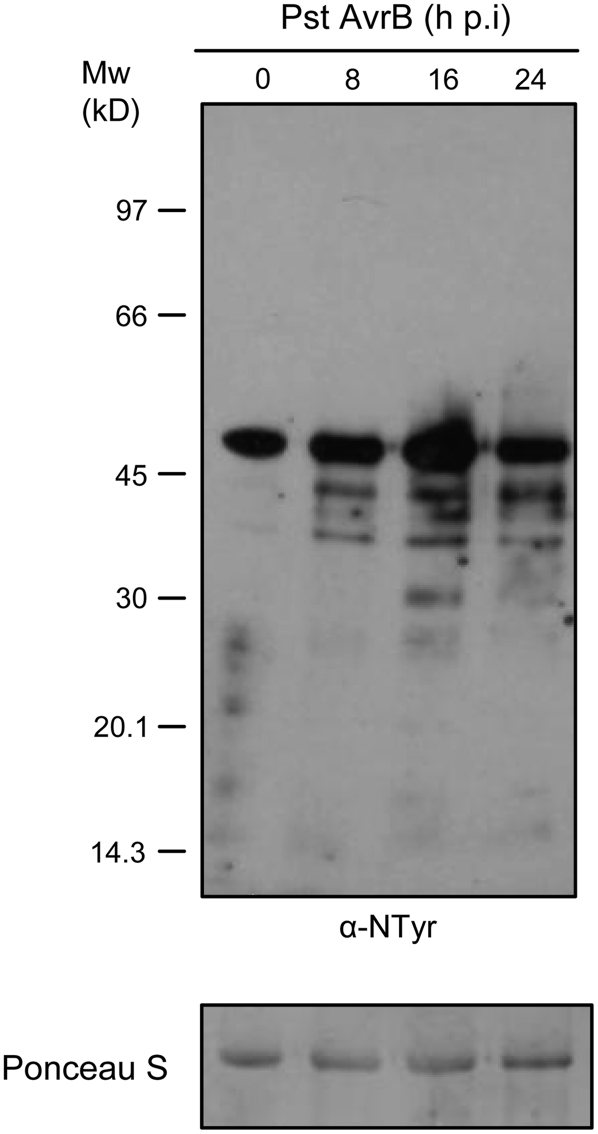
Recently, it was shown that some peroxiredoxin isoforms possess a peroxynitrite reductase activity (Bryk et al., 2000) that detoxifies peroxynitrite (ONOO−), a reactive nitrogen species generated by the reaction of NO with O2− (Koppenol et al., 1992). Since one of the earliest events in the HR is the rapid accumulation of ROS and NO through the activation of enzyme systems similar to neutrophil NADPH oxidase and NO synthase, ONOO− is formed during the disease resistance response (Alamillo and Garcia-Olmedo, 2001; Saito et al., 2006). Because ONOO− causes oxidative damage and protein modifications such as Tyr nitration (Radi, 2004), we examined protein nitration during the progression of the HR. Protein gel blot analysis from one-dimensional protein separation using an anti-nitrotyrosine antibody revealed a progressive increase in the number of nitrated proteins at 8 and 16 h after infection of Arabidopsis plants with Pseudomonas syringae pv tomato carrying the avirulence gene AvrB (Pst AvrB), indicating that nitrotyrosine formation is a biologically relevant protein modification (Figure 4).
Figure 4.
Tyr Nitration during the Progression of the HR in Arabidopsis Plants.
Protein extracts from wild-type Arabidopsis leaves at the indicated times after infiltration with 108 colony-forming units/mL Pst AvrB were subjected to protein gel blot analysis with an anti-nitrotyrosine antibody. The polyvinylidene difluoride membrane was stained with Ponceau red after protein transfer, and a portion of the stained blot corresponding to ribulose-1,5-bis-phosphate carboxylase/oxygenase is shown as a loading control. This experiment was repeated three times with similar results. hpi, hours after infiltration.
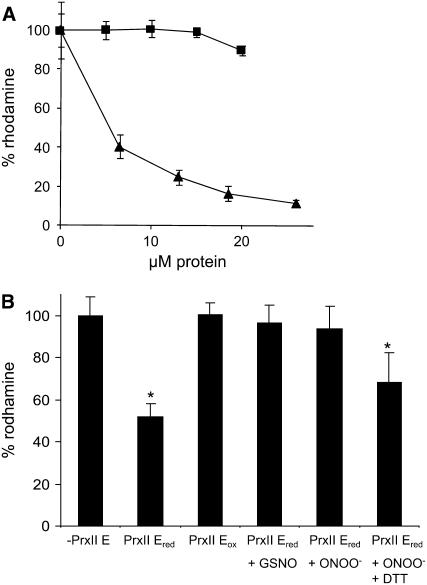
Therefore, we tested PrxII E for its ONOO− detoxification activity in vitro. We observed that reduced PrxII E was able to avoid the ONOO− -mediated oxidation of dihydrorhodamine 123 to rhodamine, demonstrating that PrxII E effectively detoxified ONOO− in a concentration-dependent manner (Figure 5A). On the contrary, BSA used as negative control did not show any protective effect. GSNO-mediated S-nitrosylation of PrxII E resulted in a significant reduction of this antioxidant activity (Figure 5B). Moreover, pretreatment of the protein with ONOO− abolished the PrxII E protection of dihydrorhodamine 123 from oxidation by ONOO−, but the protective effect could be recovered at ∼70% by incubation of the protein with DTT (Figure 5B). This indicates that PrxII E possesses peroxynitrite reductase activity: it can be reductively regenerated to the active form for a second catalytic cycle and does not just chemically decompose ONOO− in a one-to-one stoichiometry. It is likely that PrxII E–recovered peroxynitrite reductase activity can be augmented in vivo by specific interaction(s) with a still unknown reductase system, as demonstrated for other peroxiredoxins (Dubuisson et al., 2004).
Figure 5.
ONOO− Detoxification Activity of PrxII E.
(A) Indicated amounts of reduced PrxII E (triangles) and BSA (squares) were assessed for protection against the oxidation of dihydrorhodamine 123 to rhodamine caused by ONOO−. Each data set represents the average ± sd of at least three independent experiments.
(B) Dihydrorhodamine oxidation to rhodamine caused by ONOO− in the absence (−PrxII E) or presence of 10 μM reduced (PrxII Ered) or oxidized (PrxII Eox) PrxII E. Where specified, reduced PrxII E was pretreated with 1 mM GSNO or a stoichiometric amount of ONOO−. The 100% value was defined as the amount of rhodamine formed in the absence of protein (−PrxII E). Asterisks indicate values statistically different from the control value (−PrxII E; Student's t test, P < 0.05). Values shown are means ± sd of nine independent experiments.
We next generated transgenic Arabidopsis plants overexpressing PrxII E (35S-AtPrxII E) and obtained a line carrying a tDNA insertion that disrupted the prxII E gene (KD-AtPrxII E [for knockdown-AtPrxII E]) from the SALK collection. Homozygous 35S-AtPrxII E and KD-AtPrxII E T3 plants subjected to RT-PCR and protein gel blot analysis indicated that the level of PrxII E was ∼150% in the 35S-AtPrxII E line and ∼20% in the KD-AtPrxII E line compared with the wild type (see Supplemental Figure 1 online). Four-week-old KD-AtPrxII E and 35S-AtPrxII E plants showed no significant differences in growth, chlorophyll content, and fresh weight compared with wild-type plants (see Supplemental Figure 2 online).
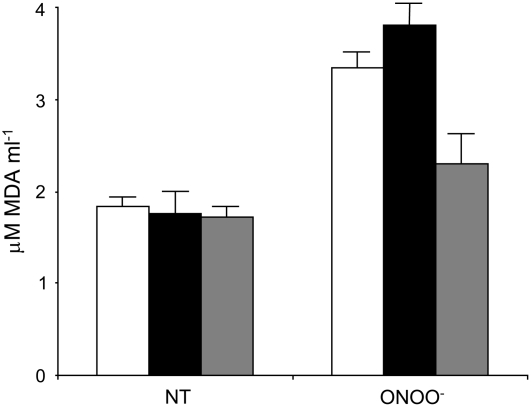
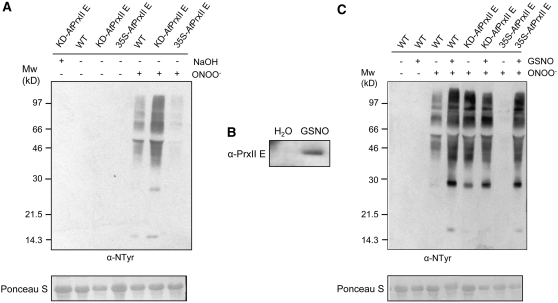
Leaves from wild-type, KD-AtPrxII E, and 35S-AtPrxII E plants were challenged with Pst AvrB to compare the progression of the defense response. The altered expression of PrxII E in KD-AtPrxII E and 35S-AtPrxII E plants did not affect bacterial growth, the development of hypersensitive cell death, the accumulation of H2O2, and the abundance of nitrated proteins (data not shown). This lack of differences among plants altered in the expression of PrxII E strongly supports the proposed loss of functionality of PrxII E during the HR. This effect was unequivocally demonstrated by analyzing the ONOO− detoxification activity of PrxII E in naïve Arabidopsis plants in which PrxII E was not S-nitrosylated. We first analyzed lipid peroxidation by measuring the concentration of thiobarbituric acid–reactive substances (TBARS) in wild-type, KD-AtPrxII E, and 35S-AtPrxII E leaves infiltrated with ONOO−. In wild-type plants, ONOO− caused an increase in TBARS that was more pronounced in KD-AtPrxII E plants but significantly reduced in 35S-AtPrxII E plants (Figure 6). Then, total proteins extracted from the same leaves were subjected to protein gel blot analysis using an anti-nitrotyrosine antibody. KD-AtPrxII E showed a significant increase in nitrated proteins compared with the wild type, while protein nitration was hardly detectable in 35S-AtPrxII E (Figure 7A). These results confirm that PrxII E detoxifies ONOO− in planta.
Figure 6.
PrxII E Functions in Lipid Peroxidation in Arabidopsis Plants.
Lipid peroxidation was determined in wild-type (white bars), KD-AtPrxII E (black bars), and 35S-AtPrxII E (gray bars) Arabidopsis leaves untreated (NT) or 24 h after infiltration with 3 mM ONOO−. This experiment was repeated three times with similar results. The asterisk indicates a value statistically different from that of ONOO−-treated wild-type leaves (Student's t test, P < 0.05). MDA, malondialdehyde.
Figure 7.
PrxII E Functions in Protein Nitration in Arabidopsis Plants.
(A) Protein extracts from wild-type, KD-AtPrxII E, and 35S-AtPrxII E Arabidopsis leaves at 5 min after infiltration with 3 mM ONOO− were subjected to protein gel blot analysis with an anti-nitrotyrosine antibody. Extracts from plants infiltrated with vehicle (NaOH) were used as a control.
(B) Protein extracts from Arabidopsis leaves at 1 h after infiltration with 2 mM GSNO or H2O as a control were subjected to the biotin-switch assay, purified on neutravidin-agarose, and then revealed by protein gel blot analysis with an anti-PrxII E antibody.
(C) Protein extracts from wild-type, KD-AtPrxII E, and 35S-AtPrxII E Arabidopsis leaves at 5 min after infiltration with 3 mM ONOO− were subjected to protein gel blot analysis with an anti-nitrotyrosine antibody. Where indicated, leaves were preinfiltrated with 2 mM GSNO at 1 h before infiltration with ONOO−.
A portion of Ponceau-stained membrane corresponding to ribulose-1,5-bis-phosphate carboxylase/oxygenase is shown as a loading control. These findings are representative of at least two independent experiments.
Since GSNO was able to S-nitrosylate PrxII E in vivo (Figure 7B), we mimicked the S-nitrosylation of PrxII E during the HR by pretreating plants with GSNO. Protein extracts from wild-type leaves treated with ONOO− showed a significant increase in nitrated proteins when pretreated with GSNO, as revealed by protein gel blot analysis with the anti-nitrotyrosine antibody. No signal could be observed in wild-type leaves treated only with GSNO (Figure 7C). Finally, the level of protein nitration observed in KD-AtPrxII E leaves treated with ONOO− was very high and comparable to that observed in wild-type and 35S-AtPrxII E leaves pretreated with GSNO and then treated with ONOO− (Figure 7C). This observation provides the experimental evidence that S-nitrosylation inhibits the ONOO− detoxification activity of PrxII E in vivo. The lack of correspondence with the profile of nitrated proteins observed during Pst AvrB infection (Figure 4) is likely due to the high level of nitrated proteins, caused by the high amount of ONOO− added to show clear and convincing differences between plants expressing different amounts of PrxII E, which did not allow us to reveal weak or moderately intense bands because of the very short exposure to film.
The availability of genetic material that boosts ONOO− bioactivity prompted us to examine the toxicity of this potent oxidant. Whereas exposure of animal cells to ONOO− in the range 1 to 1000 μM causes concentration-dependent cell death (Lin et al., 1995; Foresti et al., 1999), trypan blue staining and ion leakage measurements did not reveal any differences in dead cells between wild-type and KD-AtPrxII E untreated leaves or at 24 h after infiltration with up to 3 mM ONOO− (data not shown). These data confirm our previous observations that ONOO− is not an essential component of NO/ROS-mediated cell death (Delledonne et al., 2001).
DISCUSSION
S-Nitrosylation, the covalent attachment of an NO group to the thiol side chain of Cys, is considered the most widespread and functionally important form of physiological NO-dependent posttranslational modification (Hess et al., 2005). In plants, the experimental evidence of regulation through S-nitrosylation is currently available for only four proteins: nonsymbiotic hemoglobin (Perazzolli et al., 2004), glyceraldehyde 3-phosphate dehydrogenase (Lindermayr et al., 2005), Met adenosyltransferase (Lindermayr et al., 2006), and metacaspase 9 (Belenghi et al., 2007). Here, we demonstrated by several methods that NO mediates the S-nitrosylation of PrxII E, a type II peroxiredoxin that reduces H2O2 and alkyl hydroperoxides to H2O and the corresponding alcohol (Dietz, 2003). During H2O2 reduction, the catalytic Cys residues of peroxiredoxins undergo oxidation and must be reduced by electron donors such as thioredoxins, glutaredoxins, or cyclophilins before the next catalytic cycle (Horling et al., 2003). We found that S-nitrosylation severely inhibits the peroxidase activity of PrxII E, thus revealing a novel regulation mode for peroxiredoxins.
Recently, alkyl hydroperoxide reductase subunit C, a bacterial peroxiredoxin, was reported to possess peroxynitrite reductase activity (Bryk et al., 2000). Subsequently, ONOO−-detoxifying activity was also reported for the plant 2-Cys Prx overexpressed in E. coli (Sakamoto et al., 2003). However, peroxynitrite reductase activity cannot be generalized to all members of this family of proteins, as certain bacterial peroxiredoxins do not detoxify ONOO− (Comtois et al., 2003). Since systems for ONOO− scavenging have not hitherto been identified in plants, we investigated whether PrxII E could afford protection against ONOO−. We found that PrxII E possesses peroxynitrite reductase activity and that S-nitrosylation inhibits this activity.
By itself, ONOO− is considered the major toxic reactive nitrogen species in animal cells (Stamler et al., 1992) and is responsible for Tyr nitration and oxidation of lipids (Schopfer et al., 2003). Its function in plants is still unknown. Nevertheless, as the formation of ROS and NO is a normal event in plant cellular metabolism, it may be hypothesized that ONOO− is continuously formed in healthy cells. It should also be noted that during the onset of a pathogen-induced hypersensitive reaction, the formation of ONOO− is promoted by the rate of NO reaction with O2−, which is approximately three times faster than the reaction of O2− with superoxide dismutase (Ischiropoulos and al-Mehdi, 1995), the latter being responsible for the catalysis of O2− disproportionation to H2O2 and O2 during the oxidative burst (Delledonne et al., 2001).
The dangerous potential of ONOO− contrasts with several beneficial properties ascribed to NO (Delledonne, 2005), and the ability of plants to cope with reactive nitrogen species like ONOO− is central in determining the physiological consequences of NO accumulation. Therefore, the discovery of the ONOO− detoxification activity of PrxII E and of a novel regulatory mechanism for this class of proteins based on S-nitrosylation prompted us to investigate the NO/ROS cooperation in transducing and executing cellular functions.
In animal models, ONOO− causes apoptotic or necrotic cell death depending on its concentration (Bonfoco et al., 1995). Conversely, in plants, ONOO− does not appear to be an essential mediator of NO/ROS-induced cell death. Although 1 mM ONOO− was found to produce necrotic lesions in Arabidopsis leaves (Alamillo and Garcia-Olmedo, 2001), we previously reported that cellular viability remains unaltered in soybean (Glycine max) cell suspensions exposed up to 1 mM ONOO− or up to 5 mM SIN-1, an NO donor that provides a continuous source of ONOO− because it gradually decomposes to yield equimolar amounts of NO and O2− (Delledonne et al., 2001). Here, we provide further evidence that despite causing a dramatic increase in the level of nitrated proteins, exposure to 3 mM ONOO− does not cause cell death even in the PrxII E mutant line, thus confirming that, at least in our hands, ONOO− is not an essential intermediate of NO-induced cell death. Nonetheless, it is expected to have important physiological and signaling functions in plants, as SIN-1 was found to induce the accumulation of the transcript encoding PR-1 in tobacco (Nicotiana tabacum) leaves (Durner et al., 1998) and ONOO− was found to induce protein nitration in soybean and tobacco (Delledonne et al., 2001; Saito et al., 2006).
We observed an increase in nitrotyrosine immunoreactivity during the progression of the HR, and this observation is consistent with the high number of bands showing nitrotyrosine immunoreactivity in antisense nitrite reductase tobacco plants, which are characterized by a strong increase of NO emission (Morot-Gaudry-Talarmain et al., 2002). Transgenic plants altered in the expression of PrxII E did not show any difference in terms of the abundance of nitrated proteins when challenged with an avirulent pathogen, confirming that S-nitrosylation inhibits PrxII E activity in the wild type and in the overexpressing line causing the same phenotype observed in the knockdown line. As S-nitrosylation occurs on Cys residues and PrxII E possesses only two Cys residues, both of which are involved in the active site, a mutated form of PrxII E insensitive to S-nitrosylation would not allow us to analyze the effect of PrxII E overexpression under S-nitrosylating conditions. Future experiments with plants modulated in S-nitrosoglutathione reductase level (Feechan et al., 2005) may help to overcome this problem, although the broad changes in the S-nitrosylated proteome expected in those plants will make it difficult to unequivocally link the observed phenotype to the lack of S-nitrosylation of PrxII E.
To confirm that PrxII E–dependent detoxification of ONOO− and modulation of this activity by NO through S-nitrosylation are biologically relevant, we analyzed the ONOO− detoxification function of PrxII E in naïve plants. Both lipid peroxidation and nitrotyrosine residue formation were reduced in 35S-AtPrxII E and increased in KD-AtPrxII E plants compared with wild-type plants. Furthermore, ONOO−-dependent nitrotyrosine formation in wild-type and 35S-AtPrxII E plants previously treated with the S-nitrosylating agent GSNO was similar to that observed in untreated KD-AtPrxII E plants, confirming genetically that S-nitrosylation causes the complete inhibition of PrxII E activity and the consequent release of ONOO− bioactivity.
Protein nitration attracts considerable interest in biomedical research as a biomarker of NO-dependent oxidative stress (Radi, 2004) and because it can alter catalytic activity and interfere with cellular signaling processes (Greenacre and Ischiropoulos, 2001; Schopfer et al., 2003). In fact, it has been demonstrated that protein nitration is a reversible and selective process (Koeck et al., 2004) that can also interfere with signaling processes associated with protein Tyr phosphorylation, either keeping the protein from performing the normal task of the phosphorylated form or mimicking the structural changes imposed by protein phosphorylation (Klotz et al., 2002). Therefore, Tyr phosphorylation is a good candidate for mediating signaling events induced by nitrating agents like ONOO−, and depending on ONOO− local concentrations, the nitration and phosphorylation of critical Tyr residues may be competing processes (Brito et al., 1999). Despite its overwhelming importance in animals, Tyr phosphorylation in plants has been largely neglected, because a typical Tyr kinase has not been found (Luan, 2002). However, recent studies have characterized several protein Tyr phosphatases from Arabidopsis and other species, implying that Tyr phosphorylation and dephosphorylation also serve important functions in plant biology (Luan, 2002).
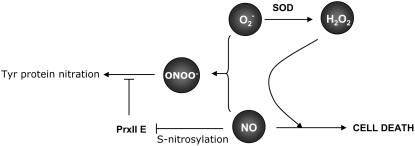
In conclusion, the observed link of S-nitrosylation to the transduction of NO- and ROS-mediated signaling events suggests that NO regulates and fine-tunes the effects (both damage and signaling) of its own radicals, such as ONOO−, through the S-nitrosylation of crucial components of the antioxidant defense machinery, such as peroxiredoxins (Figure 8). This reinforces the model of NO/ROS crosstalk and sheds new light on the combined action of reactive oxygen and nitrogen species, paving the way for studies of the modification of Tyr residues in proteins to yield nitrotyrosine.
Figure 8.
Proposed Mechanism of Action of Reactive Oxygen and Nitrogen Species.
While the NO/H2O2 cooperation triggers cell death, the NO/O2− reaction leads to the formation of ONOO−, the protein-nitrating activity of which is released by the inhibition of peroxiredoxin function through NO-mediated nitrosylation of a critical Cys. SOD, superoxide dismutase.
METHODS
Transgenic Plants
The full-length cDNA of prxII E (accession number At3g52960) was amplified by RT-PCR using the primers prxIIE-BamHI-F (5′-ATATAGGATCCATGGCGACTTCTCTCTC-3′) and prxIIE-SacI-R (5′-ATATAGAGCTCTCAGAGAGCTTTAAGCAT-3′) and cloned into the XhoI site of the 35S-cauliflower mosaic virus cassette of a modified pGreenII 0229 vector (Hellens et al., 2000). Agrobacterium tumefaciens–transformed Arabidopsis thaliana ecotype Columbia plants (GV3101; Ti helper plasmid pMP90RK; pGreen vector) were selected on Murashige and Skoog agar plates (7.5 mg phosphinotricin/L) and tested for the presence of the 35S-cauliflower mosaic virus cassette and for PrxII E transcript levels by PCR. For the initial mutant phenotype characterization, we examined several independent mutant lines. Further detailed analyses were performed with two representative overexpressing lines (A and B) that showed similar behavior. The results presented here correspond to data obtained with the 35S-AtPrxII E-B line. Insertion of the T-DNA was tested in the T3 mutant generation of KD-AtPrxII E (SALK line 064512) (Alonso et al., 2003) by PCR using gene-specific primers and a primer anchored in the T-DNA border. The following primers were used for prxII E: prxIIE-f1 (5′-GCGCATAAGTTGCGATGGATACCATAT-3′) and prxIIE-r (5′-AGTCCCACAGGCTTATCC-3′), as well as a primer for the SALK T-DNA left border (5′-ATGGTTCCACGTAGTGGGCCATC-3′).
Expression and Purification of Recombinant PrxII E
Arabidopsis PrxII E cDNA (without the 70–amino acid signal peptide) was amplified by RT-PCR with the two primers prxIIE-F (5′-GCCTCCATTTCCGTCGGAGA-3′) and prxIIE-R (5′-TCAGAGAGCTTTAAGCATATCC-3′). The identification of the processing site was based on a TargetP prediction according to the Munich Information Center for Protein Sequences database (http://mips.gsf.de/projects/plants/). Recombinant protein was generated as described (Horling et al., 2003).
Biotin-Switch Assay
To detect S-nitrosylated proteins in plants, we adopted the biotin-switch method, a three-step procedure that converts S-nitrosylated Cys residues into biotinylated Cys residues (Jaffrey et al., 2001). Subsequently, proteins were subjected to protein gel blot analysis using an anti-biotin antibody from Sigma-Aldrich (1:10,000 dilution). Otherwise, biotin-labeled proteins were purified by incubation with neutravidin-agarose beads, and bound proteins were eluted with the addition of β-mercaptoethanol to a final concentration of 150 mM.
Fluorometric Detection of S-Nitrosothiols
S-Nitrosothiols were detected using the NAT/DAN fluorometric assay, as described previously (Ghelardoni et al., 2003). This method measures the stoichiometric conversion by nitrous acid of DAN in its fluorescent derivative NAT. Nitrous acid production is due to NO release from S-nitrosothiols and S-nitrosylated proteins by treatment with mercuric chloride in an acidic environment (Gu et al., 2002). Purified recombinant PrxII E and BSA as a positive control were incubated with 1 mM GSNO (Sigma-Aldrich) for 30 min at room temperature to induce S-nitrosylation. The proteins were then subjected twice to cold acetone precipitation to remove GSNO before S-nitrosothiol measurement by NAT fluorescence. Serial NAT dilutions were used to construct a standard curve.
Mass Spectrometric Analysis of S-Nitrosylated Peptides
Purified PrxII E was treated with 1 mM GSNO or 1 mM DEA-NONOate (Cayman Chemicals) for 20 min at room temperature in the dark. The protein was then precipitated with acetone (25 min, −20°C), dissolved in 100 mM ammonium bicarbonate, and digested with trypsin (trypsin spin columns; Sigma-Aldrich). To reduce interference from a near-isobaric peptide (m/z 904.89), some PrxII E was digested with both trypsin and AspN (protease profiler kit; Sigma-Aldrich) before treatment with either DEA-NONOate or GSNO, and the peptide reaction mixture was simply acidified with 5% formic acid before MS analysis.
Nanoflow LC-MS/MS analysis was performed using a LTQ mass spectrometer (Thermo Electron) employing automated data-dependent acquisition. A nanoflow HPLC system (Surveyor; Thermo Electron) was used to deliver a flow rate of ∼250 nL/min to the mass spectrometer. Peptides were desalted using a precolumn (C18 pepmap100; LC Packings) that was switched in-line to the analytical self-packed C18, 8-cm analytical column (Picotip 75 μm i.d., 15 μm tip; New Objective). Peptides were eluted by a gradient of 2 to 40% acetonitrile over 20 min. The mass spectrometer was operated in positive ion mode with a nanospray source and a capillary temperature of 200°C; no sheath gas was employed, and the source voltage and focusing voltages were optimized for the transmission of angiotensin. Data-dependent analysis consisted of the six most abundant ions in each cycle: MS m/z, 300 to 2000; minimum signal, 1000; collision energy, 25; five repeat hits; 300-s exclusion. Isolation width for MS2 analysis was m/z 2. For selected ion programs, the inclusion list was m/z 890.9, 905.5, 906.8, and 907.9.
Raw data were processed using BioWorks 3.2 and TurboSEQUEST (Thermo Electron) and searched against the Arabidopsis genome supplemented with common contaminants (trypsin and keratins; sequences collated by Thermo Electron) with variable Cys modifications (sulfide or sulfinic acid [+31.9988] or nitrosylation [+28.9982]). Peptide hits were filtered by Xcorr and charge state (xc [+1, 2, 3] 2.0, 2.2, 3.5). The spectra were also examined manually, and individual data files were submitted to Mascot (Matrix Science) and searched against the Arabidopsis genome with the above modifications.
Peroxidase Activity
PrxII E peroxidase activity was detected by the ferrithiocyanate assay as described previously (Konig et al., 2003). Briefly, PrxII E (2.5 μM) was incubated with 2 mM DTT in 40 mM KPi buffer for 5 min at room temperature. The reaction was started by adding 50 μM H2O2 as peroxide substrate. Between 0 and 10 min, 50-μL aliquots were taken and transferred into 800 μL of trichloroacetic acid (12.5%) to stop the reaction. Then, 200 μL of Fe(II)(NH4)2(SO4)2 (10 mM) and 100 μL of KSCN (2.5 M) were added to visualize the remaining H2O2 as a red-colored complex, which was quantified at 480 nm. To detect the effect of S-nitrosylation on PrxII E activity, the protein was preincubated with different concentrations of GSNO (0 to 1 mM) for 30 min at room temperature, and GSNO was then removed by passage across a MicroBioSpin 6 gel filtration device (Bio-Rad). After GSNO incubation, the protein was also subjected to the biotin-switch method followed by protein gel blot analysis with an anti-biotin antibody. To test the reversibility of S-nitrosylation after GSNO treatment, PrxII E was incubated for 1 h with an excess of DTT (8 mM), which was then removed as described above for GSNO.
Peroxynitrite Reductase Activity
ONOO−-mediated oxidation of dihydrorhodamine 123 to rhodamine was followed by adding 1 μM ONOO− to a 1-mL reaction containing 50 mM KPi buffer, pH 7.0, 20 μM dihydrorhodamine 123 (Sigma-Aldrich), 20 μM diethylenetriaminepentaacetic acid (Sigma-Aldrich), and different concentrations of PrxII E. Rhodamine was then measured at 500 nm as reported previously (Bryk et al., 2000). BSA, which does not possess any peroxyitrite reductase activity, was used as a negative control. To test further the ability of PrxII E to protect dihydrorhodamine 123 against ONOO−-dependent oxidation, the protein (10 μM) was incubated on ice with 10 mM DTT, 10 mM H2O2, 1 mM GSNO, or a stoichiometric amount of ONOO− with or without pretreatment with 10 mM DTT for 30 min followed by purification through PD10 columns (Amersham Bioscience).
Lipid Peroxidation
Lipid peroxidation was determined by measuring the concentration of TBARS as described previously (Buege and Aust, 1978). The concentration of TBARS formed from 200 μL of extract was obtained by measuring the A532. The value of TBARS was expressed in moles of malondialdehyde per milliliter by a calibration curve constructed from malondialdehyde. Wild-type, KD-AtPrxII E, and 35S-AtPrxII E Arabidopsis leaves were infiltrated with 3 mM ONOO−. Leaves were homogenized at 24 h after ONOO− infiltration in MAE buffer (25 mM HEPES, 1 mM EDTA, and 0.2% Triton X-100, pH 7.7) containing complete protease inhibitor cocktail (Sigma-Aldrich). The extract was centrifuged for 30 min at 4°C, and 200 μL of the supernatant was used for the lipid peroxidation assay.
Immunodetection of Nitrated Proteins
Wild-type, KD-AtPrxII E, and 35S-AtPrxII E Arabidopsis leaves were infiltrated with 3 mM ONOO−, and control leaves were infiltrated with 15 mM NaOH, corresponding to the concentration at which ONOO− was prepared. When required, leaves were infiltrated with 2 mM GSNO for 1 h before ONOO− infiltration. Five minutes after ONOO− infiltration, leaves were harvested and total proteins were extracted prior to protein gel blot analysis. Nitration was detected using an anti-nitrotyrosine antibody (Sigma-Aldrich; 1:2000 dilution).
HR Analysis
Arabidopsis ecotype Columbia leaves were infiltrated with 6 μL of a bacterial suspension containing 1 × 108 colony-forming units/mL avirulent Pseudomonas syringae pv tomato carrying the AvrB avirulence gene in 10 mM MgCl2. One day after infiltration, dead cells were visualized by trypan blue staining (Koch and Slusarenko, 1990). The production of H2O2 was visualized in situ by 3,3′-diaminobenzidine staining as described previously (Thordal-Christensen et al., 1997). Nitrated proteins were immunodetected as described above.
Protein Gel Blot Analysis
Immunoblotting was performed using standard protocols (Sambrook and Russell, 2001). The polyvinylidene difluoride membranes were stained with Ponceau red for loading and transfer control. Probing and detection of immunocomplexes were performed as described for the SuperSignal West Pico detection system (Pierce).
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At3g52960.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of PrxII E in Arabidopsis Mutant Lines.
Supplemental Figure 2. Phenotypes of Arabidopsis Wild-Type Plants and Mutants Altered in PrxII E Level.
Supplementary Material
Acknowledgments
M.C.R.-P. acknowledges the Ministerio de Educación, Ciencia, y Deportes and the Junta de Andalucía for a postdoctoral fellowship and contract, respectively. K.-J.D. acknowledges support by the Deutsche Forschungsgemeinschaft (Grants Di 346/6 and FOR 387 TP3). M.D. acknowledges support by the European Molecular Biology Organization Young Investigators Program. This work was supported by a grant to M.D. from the Ministero dell'Università e della Ricerca in the framework of the program Components of the Nitric Oxide Signaling Pathways in Plants.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Karl-Josef Dietz (karl-josef.dietz@uni-bielefeld.de) and Massimo Delledonne (massimo.delledonne@univr.it).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alamillo, J.M., and Garcia-Olmedo, F. (2001). Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J. 25 529–540. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Belenghi, B., Romero-Puertas, M.C., Vercammen, D., Brackenier, A., Inze, D., Delledonne, M., and Van Breusegem, F. (2007). Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J. Biol. Chem. 282 1352–1358. [DOI] [PubMed] [Google Scholar]
- Bonfoco, E., Krainc, D., Ankarcrona, M., Nicotera, P., and Lipton, S.A. (1995). Apoptosis and necrosis: Two distinct events induced, respectively, by mild and intense insults with N-methyl-D-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc. Natl. Acad. Sci. USA 92 7162–7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito, C., Naviliat, M., Tiscornia, A.C., Vuillier, F., Gualco, G., Dighiero, G., Radi, R., and Cayota, A.M. (1999). Peroxynitrite inhibits T lymphocyte activation and proliferation by promoting impairment of tyrosine phosphorylation and peroxynitrite-driven apoptotic death. J. Immunol. 162 3356–3366. [PubMed] [Google Scholar]
- Bryk, R., Griffin, P., and Nathan, C. (2000). Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407 211–215. [DOI] [PubMed] [Google Scholar]
- Buege, J.A., and Aust, S.D. (1978). Microsomal lipid peroxidation. Methods Enzymol. 52 302–310. [DOI] [PubMed] [Google Scholar]
- Comtois, S.L., Gidley, M.D., and Kelly, D.J. (2003). Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology. 149 121–129. [DOI] [PubMed] [Google Scholar]
- Delledonne, M. (2005). NO news is good news for plants. Curr. Opin. Plant Biol. 8 390–396. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Xia, Y., Dixon, R.A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394 585–588. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 98 13454–13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denninger, J.W., and Marletta, M.A. (1999). Guanylate cyclase and the (NO)-N-./cGMP signaling pathway. Biochim. Biophys. Acta. 1411 334–350. [DOI] [PubMed] [Google Scholar]
- Dietz, K.J. (2003). Plant peroxiredoxins. Annu. Rev. Plant Biol. 54 93–107. [DOI] [PubMed] [Google Scholar]
- Dubuisson, M., Vander Stricht, D., Clippe, A., Etienne, F., Nauser, T., Kissner, R., Koppenol, W.H., Rees, J.F., and Knoops, B. (2004). Human peroxiredoxin 5 is a peroxynitrite reductase. FEBS Lett. 571 161–165. [DOI] [PubMed] [Google Scholar]
- Durner, J., Wendehenne, D., and Klessig, D.F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95 10328–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan, A., Kwon, E., Yun, B.-W., Wang, Y., Pallas, J.A., and Loake, G.J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 102 8054–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti, R., Sarathchandra, P., Clark, J.E., Green, C.J., and Motterlini, R. (1999). Peroxynitrite induces haem oxygenase-1 in vascular endothelial cells: A link to apoptosis. Biochem. J. 339 729–736. [PMC free article] [PubMed] [Google Scholar]
- Ghelardoni, S., Frascarelli, S., Ronca-Testoni, S., and Zucchi, R. (2003). S-Nitrosothiol detection in isolated perfused rat heart. Mol. Cell. Biochem. 252 347–351. [DOI] [PubMed] [Google Scholar]
- Greenacre, S.A., and Ischiropoulos, H. (2001). Tyrosine nitration: Localisation, quantification, consequences for protein function and signal transduction. Free Radic. Res. 34 541–581. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Kaul, M., Yan, B., Kridel, S.J., Cui, J., Strongin, A., Smith, J.W., Liddington, R.C., and Lipton, S.A. (2002). S-Nitrosylation of matrix metalloproteinases: Signaling pathway to neuronal cell death. Science 297 1186–1190. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hess, D.T., Matsumoto, A., Kim, S.O., Marshall, H.E., and Stamler, J.S. (2005). Protein S-nitrosylation: Purview and parameters. Nat. Rev. Mol. Cell Biol. 6 150–166. [DOI] [PubMed] [Google Scholar]
- Horling, F., Lamkemeyer, P., Konig, J., Finkemeier, I., Kandlbinder, A., Baier, M., and Dietz, K.J. (2003). Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol. 131 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischiropoulos, H., and al-Mehdi, A.B. (1995). Peroxynitrite-mediated oxidative protein modifications. FEBS Lett. 364 279–282. [DOI] [PubMed] [Google Scholar]
- Jaffrey, S.R., Erdjument-Bromage, H., Ferris, C.D., Tempst, P., and Snyder, S.H. (2001). Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3 193–197. [DOI] [PubMed] [Google Scholar]
- Jia, L., Bonaventura, C., Bonaventura, J., and Stamler, J.S. (1996). S-Nitrosohaemoglobin: A dynamic activity of blood involved in vascular control. Nature 380 221–226. [DOI] [PubMed] [Google Scholar]
- Klotz, L.O., Schroeder, P., and Sies, H. (2002). Peroxynitrite signaling: Receptor tyrosine kinases and activation of stress-responsive pathways. Free Radic. Biol. Med. 33 737–743. [DOI] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck, T., Fu, X., Hazen, S.L., Crabb, J.W., Stuehr, D.J., and Aulak, K.S. (2004). Rapid and selective oxygen-regulated protein tyrosine denitration and nitration in mitochondria. J. Biol. Chem. 279 27257–27262. [DOI] [PubMed] [Google Scholar]
- Konig, J., Lotte, K., Plessow, R., Brockhinke, A., Baier, M., and Dietz, K.J. (2003). Reaction mechanism of plant 2-Cys peroxiredoxin. Role of the C terminus and the quaternary structure. J. Biol. Chem. 278 24409–24420. [DOI] [PubMed] [Google Scholar]
- Koppenol, W.H., Moreno, J.J., Pryor, W.A., Ischiropoulos, H., and Beckman, J.S. (1992). Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem. Res. Toxicol. 5 834–842. [DOI] [PubMed] [Google Scholar]
- Lin, K.T., Xue, J.Y., Nomen, M., Spur, B., and Wong, P.Y. (1995). Peroxynitrite-induced apoptosis in HL-60 cells. J. Biol. Chem. 270 16487–16490. [DOI] [PubMed] [Google Scholar]
- Lindermayr, C., Saalbach, G., Bahnweg, G., and Durner, J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 281 4285–4291. [DOI] [PubMed] [Google Scholar]
- Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis thaliana. Plant Physiol. 137 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Yan, Y., Zeng, M., Zhang, J., Hanes, M.A., Ahearn, G., McMahon, T.J., Dickfeld, T., Marshall, H.E., Que, L.G., and Stamler, J.S. (2004). Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116 617–628. [DOI] [PubMed] [Google Scholar]
- Luan, S. (2002). Tyrosine phosphorylation in plant cell signaling. Proc. Natl. Acad. Sci. USA 99 11567–11569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick, J.B., and Schonhoff, C.M. (2004). NO means no and yes: Regulation of cell signaling by protein nitrosylation. Free Radic. Res. 38 1–7. [DOI] [PubMed] [Google Scholar]
- Martinez-Ruiz, A., and Lamas, S. (2004). S-Nitrosylation: A potential new paradigm in signal transduction. Cardiovasc. Res. 62 43–52. [DOI] [PubMed] [Google Scholar]
- Morot-Gaudry-Talarmain, Y., Rockel, P., Moureaux, T., Quillere, I., Leydecker, M.T., Kaiser, W.M., and Morot-Gaudry, J.F. (2002). Nitrite accumulation and nitric oxide emission in relation to cellular signaling in nitrite reductase antisense tobacco. Planta 215 708–715. [DOI] [PubMed] [Google Scholar]
- Neill, S.J., Desikan, R., and Hancock, J.T. (2003). Nitric oxide signalling in plants. New Phytol. 159 11–35. [DOI] [PubMed] [Google Scholar]
- Perazzolli, M., Dominici, P., Romero-Puertas, M.C., Zago, E., Zeier, J., Sonoda, M., Lamb, C., and Delledonne, M. (2004). Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi, R. (2004). Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. USA 101 4003–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas, M.C., Campostrini, N., Mattè, A., Righetti, P.G., Perazzolli, M., Zolla, L., Roepstorff, P., and Delledonne, M. (2008). Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics, in press. [DOI] [PubMed]
- Saito, S., Yamamoto-Katou, A., Yoshioka, H., Doke, N., and Kawakita, K. (2006). Peroxynitrite generation and tyrosine nitration in defense responses in tobacco BY-2 cells. Plant Cell Physiol. 47 689–697. [DOI] [PubMed] [Google Scholar]
- Sakamoto, A., Tsukamoto, S., Yamamoto, H., Ueda-Hashimoto, M., Takahashi, M., Suzuki, H., and Morikawa, H. (2003). Functional complementation in yeast reveals a protective role of chloroplast 2-Cys peroxiredoxin against reactive nitrogen species. Plant J. 33 841–851. [DOI] [PubMed] [Google Scholar]
- Sakamoto, A., Ueda, M., and Morikawa, H. (2002). Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett. 515 20–24. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schopfer, F.J., Baker, P.R., and Freeman, B.A. (2003). NO-dependent protein nitration: A cell signaling event or an oxidative inflammatory response? Trends Biochem. Sci. 28 646–654. [DOI] [PubMed] [Google Scholar]
- Stamler, J.S., Lamas, S., and Fang, F.C. (2001). Nitrosylation: The prototypic redox-based signaling mechanism. Cell 106 675–683. [DOI] [PubMed] [Google Scholar]
- Stamler, J.S., Singel, D.J., and Loscalzo, J. (1992). Biochemistry of nitric oxide and its redox-activated forms. Science 258 1898–1902. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11 1187–1194. [Google Scholar]
- Wendehenne, D., Durner, J., and Klessig, D.F. (2004). Nitric oxide: A new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 7 449–455. [DOI] [PubMed] [Google Scholar]
- Zaninotto, F., La Camera, S., Polverari, A., and Delledonne, M. (2006). Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol. 141 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.