Abstract
Studies of biotic remains of polar ice caps have been limited to morphological identification of plant pollen and spores. By using sensitive molecular techniques, we now demonstrate a much greater range of detectable organisms; from 2000- and 4000-year-old ice-core samples, we obtained and characterized 120 clones that represent at least 57 distinct taxa and reveal a diversity of fungi, plants, algae, and protists. The organisms derive from distant sources as well as from the local arctic environment. Our results suggest that additional taxa may soon be readily identified, providing a plank for future studies of deep ice cores and yielding valuable information about ancient communities and their change over time.
Polar ice caps continually accumulate wind-blown organic material. Although fossil plant pollen and spores from several deep ice cores have been characterized and have provided information about ancient flora composition and dispersal (for examples, see refs. 1–6), most organic material found in fossil glacial ice does not lend itself to morphological identification (7, 8). We have therefore used a molecular approach to explore the biological context of two samples, dated approximately 2000 and 4000 years B.P., that were collected from an ice core drilled at the Hans Tausen ice cap in North Greenland, one of the northernmost glaciers in the world (82.5°N, 37.5°W, 1270 m above sea level). We report a hitherto unknown diversity of eukaryotic fungi, plants, algae, and protists of both local and distant origin, thereby expanding the range of detectable organic material in glacier ice. The study of deep ice cores at the molecular level has wide implications because of the possibility of increasing our knowledge of ancient communities and their changes through time.
MATERIALS AND METHODS
Sampling and Extraction.
Two samples of approximately 5 liters, each representing about 20 years of snow accumulation, were collected from the ice core sections with a band saw (treated with 5% sodium hypochlorite) and stored in sterile plastic bags (Stomacher, London) at −80°C. The samples had been dated to times, approximately 2000 or 4000 years B.P., by using a scale based on annual snow accumulation, layers of volcanic fallout of historically known eruptions, and ice-flow modeling. One to two centimeters was sliced from the surfaces of the samples with a microtome knife (Leica) that had been treated with 5% sodium hypochlorite for 24 h, leaving approximately 3 liters of ice per sample. The samples were then melted in UV-treated (45 W for 72 h) plastic buckets (USON, Herlev, Denmark), and each sample was filtered through a sterile 0.22-μm filter unit (Nalgene). The filter membrane was transferred to a 10-ml sterile plastic tube (Nunc), and DNA was extracted directly from the filter by a silica-based purification method (9, 10) with minor modifications (1 ml of extraction buffer was added to the filter, which was incubated at 65°C for 24 h with sporadic agitation). The extraction buffer was transferred to a 1.5-ml safe-lock tube (Eppendorf), and 15 μl of silica suspension was added. The DNA was eluted at 65°C in two aliquots of 15 μl of 1× TE (10 mM Tris/1 mM EDTA, pH 7.6) and stored at −20°C.
Primers, PCR Amplifications, and Sequencing.
The primers NS8 (11) and the reverse of nu-SSU-1627-31 (12) were used to amplify approximately 160 bp of the eukaryotic 18S rRNA gene. Four new primers were designed to amplify exclusively the rDNA sequences; designing was performed by using the program amplify 1.2 after manually aligning 69 eukaryotic and prokaryotic rDNA sequences from the small subunit (SSU) rRNA database (13) and the National Center for Biotechnology Information database: the pair nu-SSU-1288-(5′-TGGTGGTGCATGGCCGTTCTTAGTTGG-3′/nu-SSU-1420-(5′-ACATCTAAGGGCATCACAGACCTGTTA-3′) amplifies approximately 180 bp; and the primer pair nu-SSU-598-(5′-GCCGCGGTAATTCCAGCTCCAATAGCGT-3′)/nu-SSU-898-(5′-TCCAAGAATTTCACCTC-3′), amplifies approximately 340 bp. The primer pair nu-SSU-1288–5′ and NS8 potentially amplifies approximately 550 bp. PCR amplification was performed in 50-μl volumes with 10 mM Tris⋅HCl, 1.5 mM MgCl2, 50 mM KCl (pH 8.3), 0.8 mM dNTPs, 1 mM of each primer, and 1 unit of Pfu DNA polymerase (Stratagene). Thirty-six cycles (1 min at 94°C, 1 min at 45°C, 2 min at 72°C, and a final cycle of 10 min at 72°C) of PCR were carried out (GeneAmp PCR system 9600, Perkin–Elmer). A second amplification was performed on the amplification products and on the controls under identical conditions. The PCR products were cleaned (QIAquick PCR purification kit, Qiagen, Chatsworth, CA) and cloned [pCR-Script Amp SK(+) cloning kit, Stratagene]. Positive clones were amplified by using T3 and T7 primers for 31 cycles. The PCR products were purified with a kit (QIAquick), cycle-sequenced, and electrophoresed on a DNA sequencer (Prism 377, Applied Biosystems).
Precautions and Controls.
Complete physical separation of filtration, extraction, and PCR set-up from running, cloning, and sequencing was achieved by using two different laboratories, one pre- and one post-PCR, each completely separately equipped with lab clothes, pipettes, chemicals, and reagents. No DNA work had previously been carried out in the pre-PCR laboratory. All pre- and post-PCR work was done in laminar positive-flow hoods (model 1.2, Holten, Brøndby, Denmark) with frequently changed sterilized gloves (Gammex, Ansell Medical, Munich) and facemasks. Tools and containers for chemicals and reagents in the pre-PCR room were cleaned with 5% sodium hypochlorite. All of the pipettes, including the pistons, were cleaned for at least 48 h with 2.5 M HCl or 5% sodium hypochlorite, followed by extensive rinsing in double-distilled, UV-treated, and autoclaved water. Only pipette filter tips (Alpha Laboratory, Hants, U.K.) were used.
Aliquots of chemicals and reagents were frozen at −20°C immediately after purchase. The water (double-distilled, UV-treated, and autoclaved), dNTP-mix, Taq buffer, ethanol, and acetone were centrifuged through 30 K filter units (NMWL Ultrafree-MC, Millipore) and the primer solutions were centrifuged through 50 K filter units (Millipore) immediately before use. The silica suspension was autoclaved immediately before being used for extraction; the extraction and washing buffers and the EDTA and Tris for the 10 mM Tris/1 mM EDTA (pH 7.6) were incubated with silica suspension for at least 48 h before extraction. All of the tubes (including the centrifugation tubes) used in the pre-PCR processes were washed with 2.5 M HCl/soap in 128-ml sterile specimen cups (Elkay, Shrewsbury, MA) for a minimum of 48 h with constant stirring. Thereafter, the tubes were washed twice with water (double-distilled, UV-treated, and autoclaved) in 128-ml sterile specimen cups under constant stirring for a minimum of 48 h; they were then dried in sterile specimen cups at 65°C, also for a minimum of 48 h.
Five types of controls were used to check for possible contamination by extraneous DNA: (i) An extraction from an empty filter, (ii) an extraction from an empty tube, (iii) an extraction from clean water from a sampling bucket, (iv) a no-template control, and (v) a sterile control (0.22-μm filter unit, Nalgene), which monitored the air inside the laminar-flow hood used for pre-PCR work for possible contamination. No amplification products were obtained in the blank controls. The air control (v) produced an approximately 180-bp-long amplification product, which was cloned and sequenced. The 10 clones that were investigated contained the same sequence, one belonging to a loculoascomycetous fungus that was not found in the ice core samples.
Data Analysis.
The sequences of the clones that we obtained were classified through GenBank comparisons by using the gapped blast program (14), and they were aligned manually with sequences from the SSU rRNA database (13). The clones were compared for the percentage of sequence similarity by using the genedoc program (version 2.2.000, B. Nicholas and H. B. Nicholas, information available at website www.cris.com/∼ketchup/genedoc.shtml). Possible recombination among the clones was investigated by means of the phylpro program (15). The overall frequency of amplification errors in the ancient DNA sequences was estimated at approximately 1% to 2% on the basis of substitution differences in cloned PCR products of mitochondrial DNA from the “Iceman,” a mummified human body approximately 5,000 years old that is found in a glacier in the Tyrolean Alps (16), and a Neanderthal bone (17).
RESULTS AND DISCUSSION
Amplification products were obtained from the sample extracts by using three sets of versatile eukaryotic primers designed to amplify a 160-, 180-, or 340-bp fragment of the eukaryotic 18S rRNA gene. No products were obtained with the primer set expected to amplify a fragment of 550 bp. The PCR products showed an inverse relationship between amplification efficiency and fragment length that is typical of ancient DNA (16, 18). Twenty clones per primer set per ice sample, 120 in total, were sequenced.
Each clone was compared with sequences deposited in GenBank as of August 3, 1998. For the 107 clones displaying 90% to 100% percent similarity to GenBank sequences, identification to the level of class was attempted by noting the consensus taxon of the database sequences displaying the highest similarity to the clones, and noting the consensus taxon of those GenBank sequences displaying 1% to 3% dissimilarity to the sequences with the highest score. By using this criterion, 66 clones could be assigned to different classes of fungi (ascomycetes and basidiomycetes), to various classes of protists (including green algae, alveolates, and stramenopiles), and finally, to one class of conifers (Table 1). By using the same criterion, 24 of the 30 clones that displayed 99% to 100% similarity to GenBank sequences were identified to the level of order or family, revealing the presence of several different orders and families of fungi, one family of green algae, and one family of conifers (Table 2).
Table 1.
Clones identified to the level of class
| Age, yr | No. of clones | Kingdom | Phylum | Class | Similarity to database sequences, % |
|---|---|---|---|---|---|
| 2000 | 2 | Fungi | Ascomycota | Euascomycetes (2–22) | 96–99 |
| 8 | Hemiascomycetes (2–6) | 98–100 | |||
| 2 | Basidiomycota | Urediniomycetes (2–8) | 98–99 | ||
| 3 | Hymenomycetes (1–3) | 96–100 | |||
| 18 | Viridiplantae | Cholorophyta | Chlorophyceae (2–18) | 93–99 | |
| 3 | Coniferophyta | Coniferopsida (3–4) | 99 | ||
| 1 | “Alveolata” | Ciliophyta | Oligohymenophora (1) | 94 | |
| 4000 | 14 | Fungi | Ascomycota | Euascomycetes (1–90) | 92–100 |
| 2 | Hemiascomycetes (4–5) | 97–100 | |||
| 6 | Basidiomycota | Urediniomycetes (1–7) | 94–100 | ||
| 2 | Hymenomycetes (1–3) | 93–97 | |||
| 5 | “Stramenopiles” | — | Chrysophyceae (4–5) | 91–92 |
Numbers in parentheses after class names indicate the number of genera in that class matching the assigned clones in the GenBank database with less than 4% dissimilarity. Final column indicates the highest percent similarities to GenBank sequences.
Table 2.
Clones of ice core sequences displaying 0% to 4% dissimilarity to the GenBank taxa that are closest (within 1% of identity) to the ice core sequences
| Age, yr | Primer, bp | No. of clones | Dissimilarity
|
Family or order | ||||
|---|---|---|---|---|---|---|---|---|
| 0% | 1% | 2% | 3% | 4% | ||||
| 2000 | 160 | 4 | Saccharomycetaceae (1) | Saccharomycetaceae (2) | Saccharomycetaceae (5) | Saccharomycetaceae (3) | — | Saccharomycetaceae |
| 160 | 2 | NSD | Saccharomycetaceae (1) | Saccharomycetaceae (2) | Saccharomycetaceae (5) | Saccharomycetaceae (4) | Saccharomycetaceae | |
| 180 | 1 | Saccharomycetaceae (1) | NSD | NSD | Saccharomycetaceae (1) | — | Saccharomycetaceae | |
| 180 | 1 | NSD | Sporidiales (3) | Sporidiales (1) | NSD | Unclassified* (4) | Sporidiales | |
| Unclassified* (1) | Unclassified* (1) | Sporidiales (1) | ||||||
| 180 | 3 | NSD | Chlamydomonadaceae (2) | Chlamydomonadaceae (1) | NSD | Chlamydomonadaceae (2) | Chlamydomonadaceae | |
| 320 | 2 | NSD | Pinaceae (1) | Pinaceae (1) | Pinaceae (2) | Pinaceae (1) | Pinaceae | |
| 320 | 1 | NSD | Pinaceae (1) | Pinaceae (1) | Pinaceae (3) | NSD | Pinaceae | |
| 320 | 1 | NSD | Hymenochaetaceae (1) | NSD | NSD | NSD | Hymenochateraceae | |
| 4000 | 160 | 1 | Saccharomycetaceae (1) | Saccharomycetaceae (2) | Saccharomycetaceae (5) | Saccharomycetaceae (4) | — | Saccharomycetaceae |
| 160 | 2 | Chaetothyriales (2) | NSD | NSD | NSD | — | Chaetothyriales | |
| 180 | 1 | Ophiostomataceae (1) | NSD | Ophiostomataceae (1) | NSD | — | Ophiostomataceae | |
| 180 | 1 | Trichocomaceae (10) | Trichocomaceae (8) | Trichocomaceae (3) | Trichocomaceae (3) | — | Eurotiales | |
| Monacaceae (1) | ||||||||
| 180 | 1 | Sporidiales (3) | Sporidiales (1) | NSD | Unclassified* (1) | — | Sporidiales | |
| Unclassified* (1) | Unclassified* (1) | Sporobolomycetaceae (2) | ||||||
| 180 | 1 | NSD | Sporidiales (3) | Sporidiales (1) | NSD | Unclassified* (1) | Sporidiales | |
| Unclassified* (1) | Unclassified* (1) | Sporobolomycetaceae (2) | ||||||
| 320 | 1 | NSD | Sordariaceae (1) | Sordariaceae (2) | NSD | Chaetomiaceae (1) | Sordariales | |
| Lasiosphaeriaceae (1) | Unclassified* (1) | |||||||
| 320 | 1 | NSD | Pleosporaceae (4) | Pleosporaceae (4) | Pleosporaceae (1) | NSD | Dothideales | |
| Unclassified* (1) | Dorthideacae (1) | Unclassified* (1) | ||||||
Numbers in parentheses after order and family names indicate the number of genera in that family or order matching the assigned clones in the GenBank database with less then 4% dissimilarity. Final column gives the assigned order or family name. NSD, no sequences in the database match the clones.
Sequences not classified to a particular order or family in GenBank database.
The clones assigned to the conifer family Pinaceae show the presence in the ice of material transported from long distances; this finding agrees with previous detections of both conifer pollen and wood fragments in Greenland ice cores (5, 6). Several of the clones assigned to groups of fungi and algae show high similarity to GenBank taxa previously recorded in polar and alpine environments. These include some of the clones allocated to the fungus order Sporidiales and display strong resemblance to genera such as Leucosporidium (100% similarity) and Rhodotorula (99% similarity), recorded from Antarctica (19, 20). One clone assigned to the fungus class Hymenomycetes could be grouped with the genera Mrarkia (97% similarity), also previously found in Antarctica (19). Another clone allocated to the class Euascomycetes shows high similarity (99%) to a viable fungus recovered from the clothing of the Tyrolean “Iceman” (21). All clones allocated to the green algae family Chlamydomonadaceae were grouped exclusively with the genera Chlamydomonas and Chloromonas (99% similarity), which comprise the majority of snow-inhabiting algae and are responsible for the red coloration of snow fields (22). Other clones display resemblance to green algal genera such as Raphidonema (98% similarity) or Stichococcus (96% similarity), both common in European snowfields and also recorded in Alaska (23, 24). A few clones were grouped entirely with a group of unclassified heterotrophic flagellates, Heteromita (95% similarity), including cold-tolerant species recorded from Antarctic fell-fields. (25). Our results therefore suggest that material from long-distance dispersal and material from the local arctic environment are both present in the ice. Some of this material is probably part of an ancient snow-living microbial community that inhabited the surface layers of the ice cap.
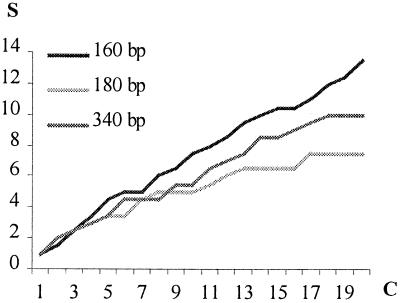
The 120 clones included 78 different sequences. Infrequently intraspecific variation has been reported within the hypervariable domains of the 18S rRNA gene (26), and although experimental errors can arise during PCR [in this case primarily because of age-introduced template damage (17)], clones displaying 96% to 100% similarity are here considered to belong to the same taxa. We used this criterion to assemble the 120 clones into 57 distinct clusters (Fig. 1). However, this number is likely to be a minimal estimate of the diversity of organisms present in the samples, because (i) an applied threshold value of 4% will result in physiologically distinct taxa being grouped together because of nearly identical 18S rDNAs, (ii) the blast results revealed several instances in which different taxa that have identical sequences in the regions amplified or clones that are 100% identical can belong to different taxonomic groups, and (iii) the “diversity curve” for at least one of the primer sets was found to be approximately linear (Fig. 2). The curves reach a plateau when the same sequences are repeatedly recorded (27); therefore, further sampling, even from the same PCR, is likely to reveal several new taxa.
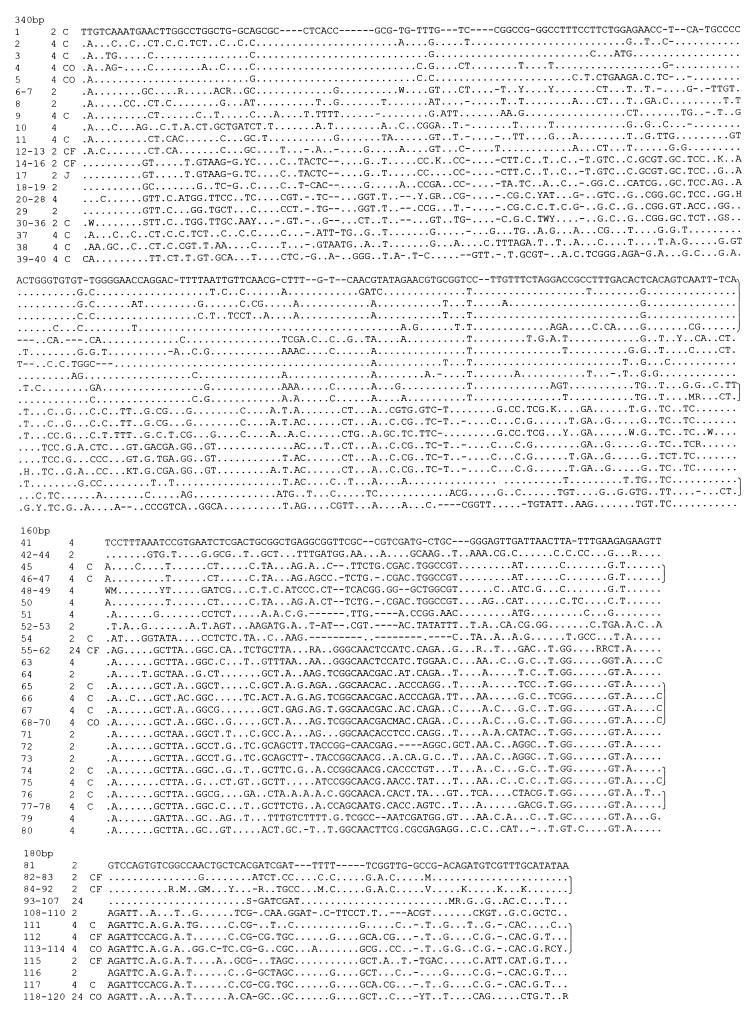
Figure 1.
DNA sequences of clones from 3 distinct fragments of the 18S rRNA gene amplified from ice core samples. Only variable sites are shown. Dots indicate identity with sequences on top of each alignment and dashes indicate insertions/deletions. Clones that are less than 4% different from each other are grouped together by using standard abbreviations at nonhomogeneous sites. C, O, and F indicate clones identified to the level of class, order, and family, respectively; J indicates a clone created by possible jumping PCR events. Numbers in first column represent clone number, and nos. 2 and 4 in second column refer to the approximate age (×1000 years) of the clone samples. Brackets at right indicate clones identified to the same class.
Figure 2.
Diversity curves. The number of clone clusters obtained by a single primer set S plotted against the number of clones with different sequences (4% level) for that primer set C. The curves are an average of the values obtained for the 2000- and 4000-year-old samples. Approximate lengths (160, 180, and 340 bp) of amplified fragments are specified for each primer set.
Because the primer sets span different regions, species overlap is possible among some of the distinct clones. However, the GenBank comparisons reveal that the primer sets preferentially amplify different kingdoms (by χ2 analysis, P < 0.05), making a possible overlap unlikely. One clone that appeared to represent a recombination product between other sequences in the sample was probably an artifact created during PCR, a phenomenon previously observed in investigations of ancient DNA (16, 18) (Fig. 1). Because no other chimeras were detected, we believe that only a small part of the observed diversity could have been caused by recombination events.
Biological studies of deep ice cores have previously been restricted to morphological investigations of fossil plant pollen and spores (e.g., see refs. 1–6). The method applied in this study makes possible the identification of a much wider range of organisms. The results show that polar ice caps can contain a diverse assemblage of eukaryotic organisms of both local and distant origin. Although one can only speculate about the diversity of prokaryotes present in fossil glacier ice, the eukaryotic diversity seen here raises expectations about these widely spread groups of organisms.
Because DNA is apparently well-preserved in polar ice caps, and several deep ice cores span more than 100,000 years, the results suggest that molecular studies of fossil glacier ice can provide new information about the diversity of both local and distant ancient biotic communities and their changes in composition through time.
Acknowledgments
We thank J. Bourgeois, N. Daugbjerg, A. Gargas, S. Nyakaana, S. Pääbo, and M. Sogin for help and discussions; and the Aage V. Jensen Fonde, the Japetus Steenstrup Foundation, and the Nordic Council of Ministers for financial support.
Footnotes
References
- 1.Lichti-Federovich S. Geological Survey of Canada. 1975;75-1Part A:441–444. [Google Scholar]
- 2.MacAndrews J H. Quaternary Res. 1984;22:68–73. [Google Scholar]
- 3.Bourgeois J C. Boreas. 1986;15:345–354. [Google Scholar]
- 4.Koerner R M, Bourgeois J C, Fisher D A. Ann Glaciology. 1988;10:85–91. [Google Scholar]
- 5.Fredskild B, Wagner O. Boreas. 1974;3:105–108. [Google Scholar]
- 6.Bourgeois J C. J Glaciology. 1990;36:340–342. [Google Scholar]
- 7.Hammer C U, Clausen H B, Dansgaard W, Neftel A, Kristinsdottir P, Johnson E. Geophys Monogr. 1985;33:90–94. [Google Scholar]
- 8.Kumai M, Langway C C., Jr Ann Glaciology. 1988;10:208. (abstr.). [Google Scholar]
- 9.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Höss M, Pääbo S. Nucleic Acids Res. 1993;21:3913–3914. doi: 10.1093/nar/21.16.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White T J, Bruns T, Lee S, Taylor J W. In: PCR Protocols: A Guide to Methods and Application. Innis M A, Gelfans D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 315–322. [Google Scholar]
- 12.Gorgas A, DePriest P T. Mycologia. 1996;88:745–748. [Google Scholar]
- 13.Van de Peer Y, Jansen J, De Rijk P, De Wachter R. Nucleic Acids Res. 1997;25:111–116. doi: 10.1093/nar/25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiller G F. Mol Biol Evol. 1998;15:326–335. doi: 10.1093/oxfordjournals.molbev.a025929. [DOI] [PubMed] [Google Scholar]
- 16.Handt O, Richards M, Trommsdorff M, Kilger C, Simanainen J, Georgiev O, Bauer K, Stone A, Hedges R, Schaffner W, et al. Science. 1994;264:1775–1778. doi: 10.1126/science.8209259. [DOI] [PubMed] [Google Scholar]
- 17.Krings M, Stone A, Schmitz R W, Krainitzki H, Stoneking M, Pääbo S. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 18.Handt O, Höss M, Krings M, Pääbo S. Experientia. 1994;50:524–529. doi: 10.1007/BF01921720. [DOI] [PubMed] [Google Scholar]
- 19.Hawkswoth D L, Kirk P M, Sutton B C, Pegler D N. Ainsworth and Bisby’s Dictionary of the Fungi. Wallingford, Oxon, U.K.: CAB International; 1995. [Google Scholar]
- 20.Paterson R A, Knox J S. In: Conservation Problems in Antarctica. Parker B C, editor. Lawrence, KS: Allan; 1972. pp. 185–192. [Google Scholar]
- 21.Ubaldi M, Sassaroli S, Rollo F. Ancient Biomolecules. 1996;1:35–42. [Google Scholar]
- 22.Hoham R W. In: Phytoflagellates. Cox E R, editor. New York: Elsevier/North–Holland; 1980. pp. 61–84. [Google Scholar]
- 23.Kol E. Smithsonian Miscellaneous Collections. 1942;101:1–36. [Google Scholar]
- 24.Kol E. Annales Historico-Naturales Musei Nationalis Hungarici. 1972;64:63–70. [Google Scholar]
- 25.Hughes J, Smith H G. In: University Research in Antarctica, Proceedings of British Antarctic Survey, Antarctic Special Topic Award, Scheme Symposium, 9–10 November 1988. Heywood R B, editor. Natural Environment Research Council, Cambridge, U.K.: British Antarctic Survey; 1989. pp. 117–122. [Google Scholar]
- 26.Hillis D M, Dixon M T. Q Rev Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 27.Magurran A E. Ecological Diversity and Its Measurement. London: Chapman and Hall; 1996. [Google Scholar]