Abstract
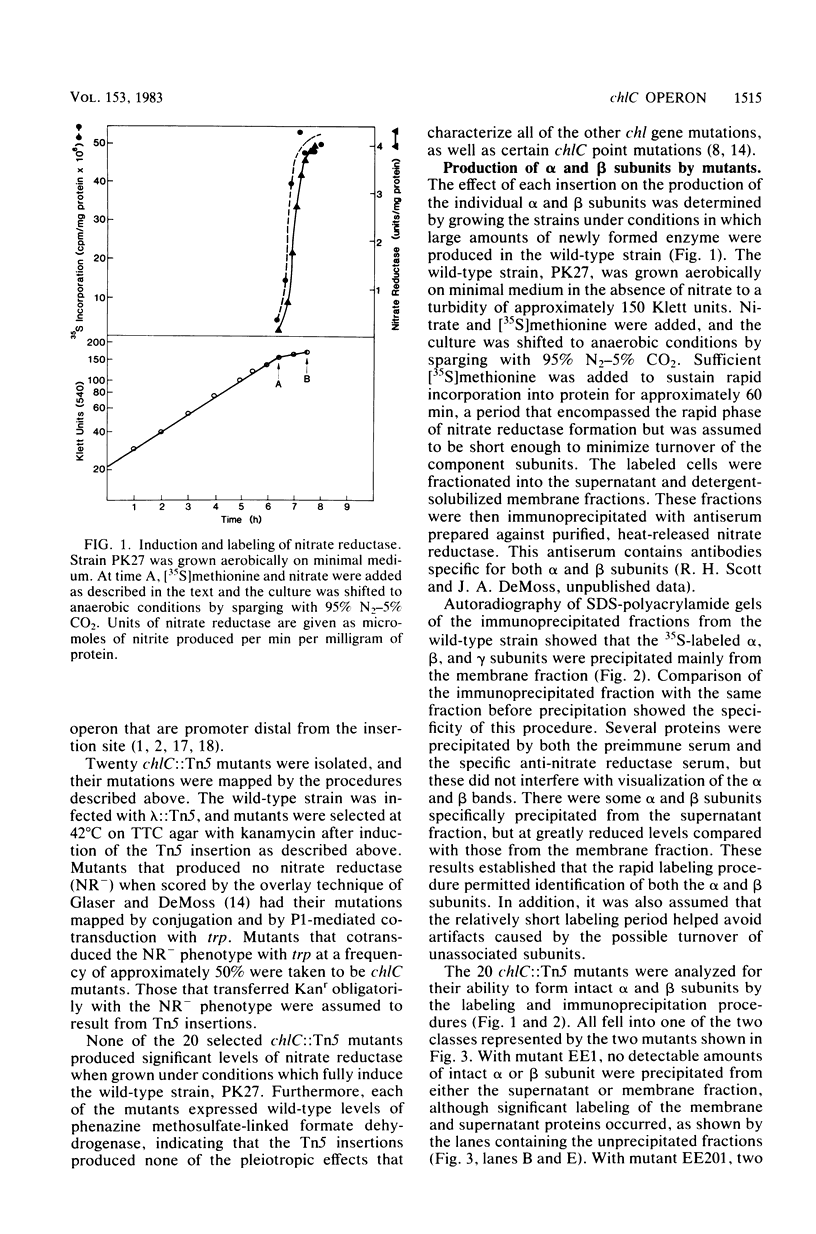
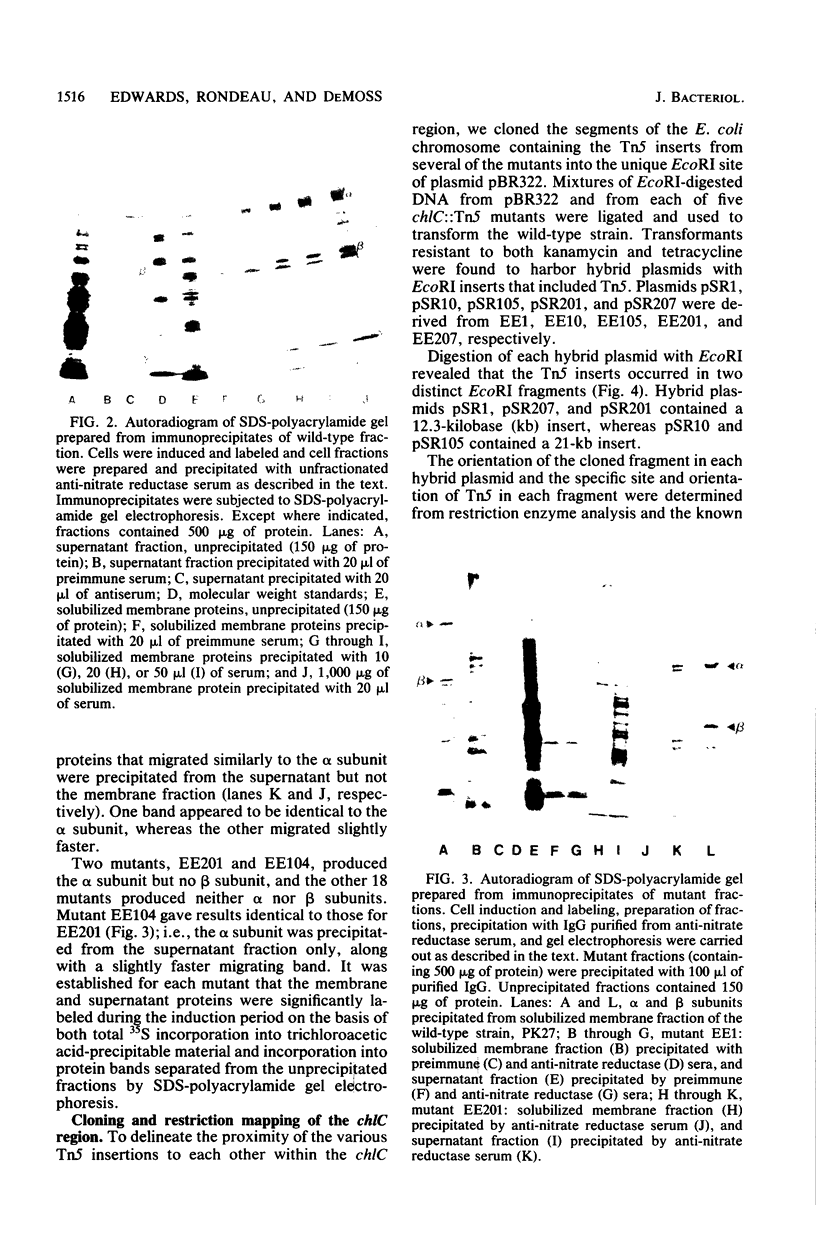
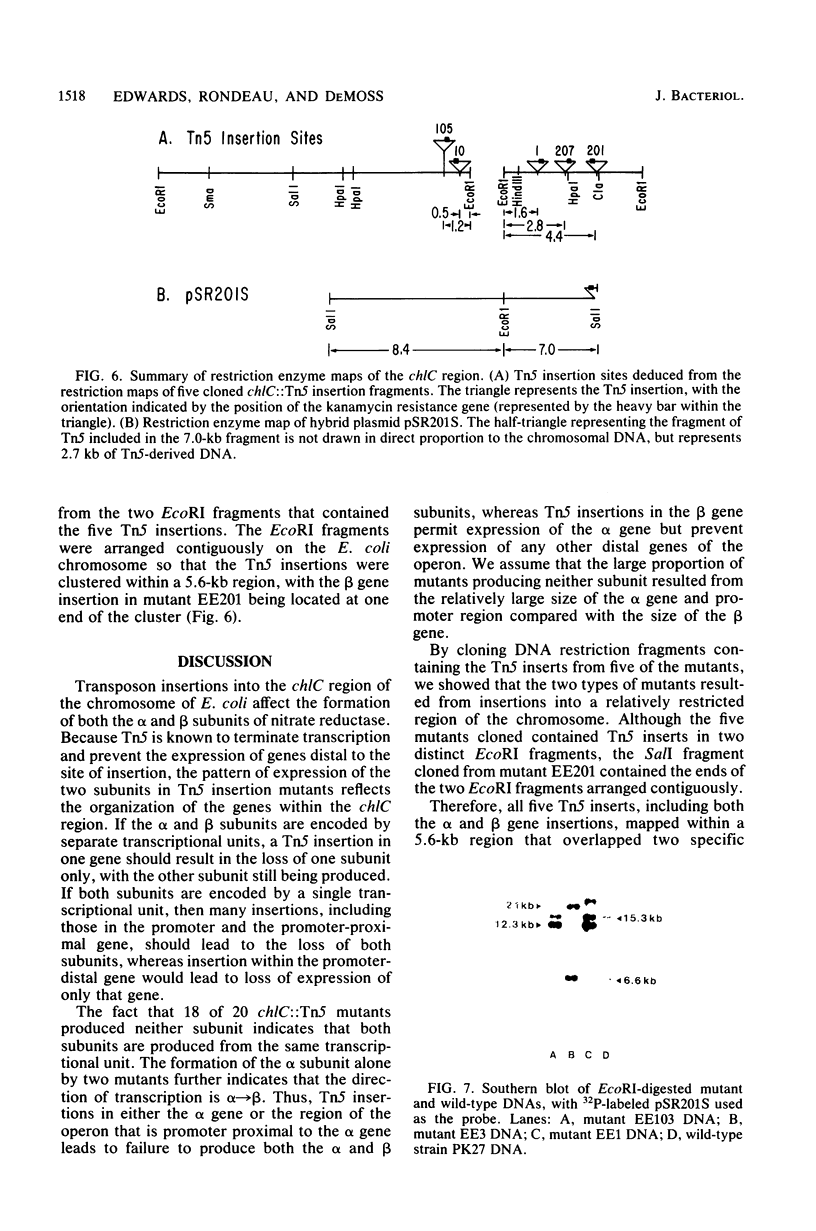
The synthesis of the alpha and beta subunits of nitrate reductase by 20 chlC::Tn5 insertion mutants of Escherichia coli was determined by immune precipitation of the subunits from fractions of cell extracts. Only two of the mutants produced either subunit in detectable amounts; these two accumulated the alpha subunit, but no beta subunit. In both cases the alpha subunit was present in the cytosolic fraction, in contrast to wild-type cells, in which both subunits are present mainly in the membrane fraction. EcoRI restriction fragments containing the Tn5 inserts from five of the mutants were cloned into pBR322. The insertions were localized on two contiguous EcoRI fragments spanning a 5.6-kilobase region that overlapped the contiguous ends of the two fragments. An insertion that permitted alpha subunit formation defined one end of the 5.6-kilobase region. The results indicated that the genes encoding the alpha and beta subunits of nitrate reductase were part of a chlC (nar) operon that is transcribed in the direction alpha leads to beta.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Weiss A., Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980 May;142(2):439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bonnefoy-Orth V., Lepelletier M., Pascal M. C., Chippaux M. Nitrate reductase and cytochrome bnitrate reductase structural genes as parts of the nitrate reductase operon. Mol Gen Genet. 1981;181(4):535–540. doi: 10.1007/BF00428749. [DOI] [PubMed] [Google Scholar]
- Clegg R. A. Purification and some properties of nitrate reductase (EC 1.7.99.4) from Escherichia coli K12. Biochem J. 1976 Mar 1;153(3):533–541. doi: 10.1042/bj1530533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMoss J. A. Role of the chlC gene in formation of the formate-nitrate reductase pathway in Escherichia coli. J Bacteriol. 1978 Feb;133(2):626–630. doi: 10.1128/jb.133.2.626-630.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoss J. A., Fan T. Y., Scott R. H. Characterization of subunit structural alterations which occur during purification of nitrate reductase from Escherichia coli. Arch Biochem Biophys. 1981 Jan;206(1):54–64. doi: 10.1016/0003-9861(81)90065-5. [DOI] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The role of a novel cytochrome b-containing nitrate reductase and quinone in the in vitro reconstruction of formate-nitrate reductase activity of E. coli. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1234–1241. doi: 10.1016/s0006-291x(74)80416-x. [DOI] [PubMed] [Google Scholar]
- Fimmel A. L., Haddock B. A. Use of chlC-lac fusions to determine regulation of gene chlC in Escherichia coli K-12. J Bacteriol. 1979 Jun;138(3):726–730. doi: 10.1128/jb.138.3.726-730.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestone G. L., Heath E. C. The cyclic AMP-mediated induction of alkaline phosphatase in mouse L-cells. J Biol Chem. 1981 Feb 10;256(3):1396–1403. [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Keesey J. K., Jr, Bigelis R., Fink G. R. The product of the his4 gene cluster in Saccharomyces cerevisiae. A trifunctional polypeptide. J Biol Chem. 1979 Aug 10;254(15):7427–7433. [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lund K., DeMoss J. A. Association-dissociation behavior and subunit structure of heat-released nitrate reductase from Escherichia coli. J Biol Chem. 1976 Apr 25;251(8):2207–2216. [PubMed] [Google Scholar]
- MacGregor C. H. Anaerobic cytochrome b1 in Escherichia coli: association with and regulation of nitrate reductase. J Bacteriol. 1975 Mar;121(3):1111–1116. doi: 10.1128/jb.121.3.1111-1116.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H. Biosynthesis of membrane-bound nitrate reductase in Escherichia coli: evidence for a soluble precursor. J Bacteriol. 1976 Apr;126(1):122–131. doi: 10.1128/jb.126.1.122-131.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A., Normansell D. E. Purification and properties of nitrate reductase from Escherichia coli K12. J Biol Chem. 1974 Aug 25;249(16):5321–5327. [PubMed] [Google Scholar]
- MacGregor C. H. Solubilization of Escherichia coli nitrate reductase by a membrane-bound protease. J Bacteriol. 1975 Mar;121(3):1102–1110. doi: 10.1128/jb.121.3.1102-1110.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H. Synthesis of nitrate reductase components in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1975 Mar;121(3):1117–1121. doi: 10.1128/jb.121.3.1117-1121.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Showe M. K., DeMoss J. A. Nitrate reductase complex of Escherichia coli K-12: isolation and characterization of mutants unable to reduce nitrate. J Bacteriol. 1969 Mar;97(3):1291–1297. doi: 10.1128/jb.97.3.1291-1297.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showe M. K., DeMoss J. A. Localization and regulation of synthesis of nitrate reductase in Escherichia coli. J Bacteriol. 1968 Apr;95(4):1305–1313. doi: 10.1128/jb.95.4.1305-1313.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]







