Abstract
In a subset of infertile men, a spectrum of spermatogenic defects ranging from a complete absence of germ cells (sertoli cell only) to oligozoospermia is associated with microdeletions of the DAZ (deleted in azoospermia) gene cluster on human distal Yq. DAZ encodes a testis-specific protein with RNA-binding potential recently derived from a single-copy gene DAZL1 (DAZ-like) on chromosome 3. Y chromosomal DAZ homologues are confined to humans and higher primates. It remains unclear which function unique to higher primate spermatogenesis DAZ may serve, and the functional status of the gene recently has been questioned. To assess the extent of functional conservation we have tested the capacity of a human DAZ gene contained in a 225-kb yeast artificial chromosome to complement the sterile phenotype of the Dazl null mouse (Dazl−/−), which is characterized by severe germ-cell depletion and meiotic failure. Although Dazl−/− mice remained infertile when the DAZ transgene was introduced, histological examination revealed a partial and variable rescue of the mutant phenotype, manifest as a pronounced increase in the germ cell population of the seminiferous tubules and survival to the pachytene stage of meiosis. As well as constituting definitive proof of the spermatogenic role of the DAZ gene product, these findings confirm the high degree of functional conservation between the DAZ and DAZL1 genes, suggesting they may constitute a single target for contraceptive intervention and raising the possibility of therapeutic up-regulation of the DAZL1 gene in infertile men.
The existence of a Y borne fertility gene or “azoospermia factor” (AZF) was suggested first by the observation of microscopically visible deletions of distal Yq in a proportion of infertile or subfertile men. In the four examples tested, the deletions were shown to have arisen de novo, in the paternal germ line, supporting the likelihood of a causal link (1). Molecular techniques employing Y chromosome-specific markers subsequently revealed that 5–10% of severely oligozoospermic (abnormally low sperm count) or azoospermic (zero sperm count) men have cytogenetically invisible Yq microdeletions (2–7). These fall into three classes mapping to discrete subintervals of the Yq region, termed AZFa, b, and c (8), and positional cloning strategies have led to the isolation of candidate genes from each subinterval. The predicted product of the AZFa candidate resembles a ubiquitin C-terminal hydrolase (9) whereas both the AZFb and AZFc candidates encode distinct putative RNA-binding proteins, designated RBM (RNA-binding motif) and DAZ (deleted in azoospermia), respectively, which are expressed exclusively in germ cells (3, 10). Both gene products contain a single RNA-recognition motif (RRM), are present in multiple copies on the Y chromosome (3), and were derived from ancestral single-copy autosomal genes. Phylogenetic studies have revealed that in the case of RBM the transposition and amplification events preceded the radiation of mammals (10–13) whereas DAZ was copied to the Y chromosome much more recently, in the old-world primate lineage (14, 15). This difference in evolutionary history of the two genes is reflected in their patterns of functional conservation. The proposed role of RBM in spermatogenesis must reflect a degree of functional specialization subsequent to transposition because the progenitor, hnRNPG, is a ubiquitously expressed gene with a function in pre-mRNA metabolism and is not implicated in spermatogenesis (16). Because no single-copy RBM homolog is known in an organism amenable to genetic manipulation, such as Drosophila or mouse, experimental proof of its role in spermatogenesis is difficult to obtain.
In contrast to RBM, the proposed role of DAZ in gametogenesis is shared with its autosomal ancestor and is very ancient. The ancestral form of DAZ is identifiable by a high degree of sequence homology in phylogenetically diverse species and is implicated in the execution of both oogenesis and spermatogenesis (14, 15, 17–24). Targeted mutagenesis of the mouse DAZ homolog, Dazl, on chromosome 17 produced a null phenotype in the male of sterility, characterized by failure of germ–cell maintenance and maturation, which resembles defects seen in AZFc patients (25). Dazl is expressed in the germ cells of both sexes, and the null female exhibits complete failure of folliculogenesis and sterility. The human autosomal ancestor, DAZL1 (DAZ-like), which is 90% identical to DAZ, also is expressed in the germ cells of males and females (18), and the possibility of its implication in some cases of male and/or female infertility merits investigation. Intriguingly, Drosophila melanogaster and Caenorhabditis elegans exhibit reciprocal patterns of sex specificity in their requirements for the DAZ progenitor. Thus, whereas C. elegans daz-1 is required in the hermaphrodite for oogenesis and dispensable in the male (23), the Drosophila homolog, boule is required for spermatogenesis but not oogenesis (17). Although these differences between species in the effects of loss of DAZL function could signify some fundamental divergence in the gametogenesis mechanism, it seems more likely that another gene(s) can substitute for DAZL function in some contexts.
Because a single-copy gene suffices to support spermatogenesis in all other species, the requirement for an additional Y chromosomal DAZ gene cluster in man and old-world primates requires explanation. In comparison with their autosomal counterpart, the DAZ genes have undergone a number of structural rearrangements (18). These include a tandem amplification of exon 7, giving rise to a 24-aa repeat with a copy number of between 7 and 14 (“DAZ repeat”). However, the functional significance of this feature is unknown, and throughout most of the coding region including the RRM, the sequence has been highly conserved, suggesting the retention of similar RNA targets. Although it is possible that the Y chromosomal genes have acquired some novel function, not shared by the ancestral form and required only in higher primates, the similar phenotypic consequences of loss of mouse Dazl and human DAZ also argue against such a functional distinction. Alternatively, the significance of the DAZ gene cluster may be only quantitative, serving to increase overall gene dosage and product levels, thereby enhancing spermatogenic efficiency. Gene amplification after transposition to the nonrecombining portion of the Y chromosome may have been favored further by the need to maintain gene function in the face of an accumulation of deleterious mutations through the operation of Müllers ratchet (26). In man, the variable penetrance of AZFc deletions that remove the entire DAZ gene cluster is consistent with a degree of genetic redundancy. The phenotypic spectrum ranges from a total lack of germ cells (sertoli cell only) to oligozoospermia, and, in one case, a male lacking the DAZ gene cluster is known to have fathered a son who was infertile (3, 8). An extreme view is that the Y chromosomal DAZ cluster is a nonfunctional relic. The infertility associated with DAZ deletions could be accounted for by the deletion of other, as yet uncharacterized genes within the interval with testis-specific expression patterns (27, 28). In support of this possibility there are no reported instances of DAZ point mutations in infertile men, and a recent study of nucleotide substitution rates within the DAZ locus revealed a pattern consistent with an absence of selective pressure (29).
Our approach to assessing the degree of functional overlap between DAZ and its autosomal counterparts was to test the capacity of a DAZ transgene to rescue the sterility of the Dazl knockout mouse. This report describes the introduction of a 225-kb yeast artificial chromosome (YAC) clone from the AZFc region containing a DAZ gene into mice lacking a functional copy of Dazl. The DAZ transgene displayed an appropriate male germ cell-specific pattern of expression in the mouse and conferred a partial rescue of the mutant phenotype manifest as a pronounced increase both in the survival of spermatogonia and the frequency of maturation to the pachytene stage of meiotic prophase. The implications of the present findings for the functional relationship between DAZ/Dazl are discussed.
MATERIALS AND METHODS
Introduction of YAC S12 into the DazlTm1Hgu/DazlTm1Hgu Genetic Background.
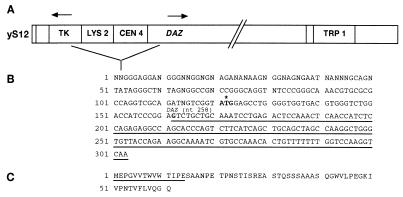
YAC S12 (yS12) contains a 225-kb human Y chromosome insert. The S12 YAC was transferred to the yeast host CGY2570 (30) and retrofitted with modified vector arms based on pCS990 (centromeric arm) (30) and pJS89 (acentric arm) (31). pCGS990 contains a conditional yeast centromere whose activity is abolished under selective growth conditions (30). Under these conditions, S12 YAC copy number increased 10- to 20-fold, allowing sufficient DNA to be prepared for injection (not shown). YAC DNA was prepared as described (32). S12 YAC DNA was injected into pronuclei at a concentration of 1 ng/μl. The G0 mice were F2 hybrids from a C57BL × CBA/Ca cross. The single G0 male transgenic mouse was backcrossed to a CB57BL × CBA/Ca F1 female to establish the S12 transgenic line, TgN (hDAZy1)1Hgu, which will be referred to as TgS12. TgS12 positive males were crossed to DazlTm1Hgu/+ females (25). DazlTm1Hgu/+;TgS12 individuals were mated to test for phenotypic rescue in DazlTm1Hgu/DazlTm1Hgu;TgS12 progeny. Dazl genotype was determined by PCR as described (25), and the presence of yS12 was detected by PCR with TRP1 primers f488 (5′-gctctcttgccttccaacc-3′) and f492 (5′-caacctaaggaggatgttttgg-3′).
Nucleic Acid Preparation.
Genomic DNA was prepared from mouse tail tips or kidney by using standard techniques. Total RNA was prepared by using RNAZol B reagent (AMS Biotechnology, Oxford, U.K.) according to the manufacturer’s instructions. Poly(A)+ RNA for 5′ rapid amplification of cDNA ends PCR was prepared by using oligo(dT)-coupled magnetic beads (Dynal, Great Neck, NY).
YAC PCR Analysis.
Y chromosome-specific sequence tagged site (STSs) used to map the extent of yS12 have been described (33). Primers dazint1 (5′-gtcgacaacaaagcagga-3′) and dazint2 (5′-gtcgacatcataattacg-3′) confirmed the retention of sequence immediately upstream of DAZ exon 2 in the S12 YAC. The absence of exon 1 in TgS12 transgenic mice was confirmed with primers daz1 (5′-ggttttccttacaccttagc-3′) and daz2 (5′-aggctcaaggaggaacagag-3′).
Reverse Transcription—PCR (RT-PCR).
First-strand cDNA synthesis reactions and RT-PCR were performed by using a preamplification system kit (GIBCO) with 1 μg of total RNA and oligo(dT) primer. RT-PCRs contained 1 μl cDNA and DAZ forward and reverse primers at 1 μM (forward primer, daz3 (5′-ccgaagcatacaaatggtgg-3′; reverse primer, daz4 (5′-tgttcacaggtacttcttgg-3′). Control reactions were performed by using S16 ribosomal protein primers (14).
5′ Rapid Amplification of cDNA Ends PCR.
Reactions were performed by using the Marathon cDNA Amplification kit (CLONTECH). First-round PCR used DAZ gene-specific primer 1 (5′-gaaccgtatctaccaaagcagcttcc-3′). Nested PCR used DAZ gene-specific primer 2 (5′-gccaagccgtggaatggtagcaatg-3′). PCR products were gel-purified (Qiagen gel-extraction kit), cloned into a plasmid vector by using the pGEM-T kit (Promega), and sequenced by using a BigDye Terminator sequencing kit and automated sequencing apparatus (Applied Biosystems).
Filter-Hybridization Analysis.
Northern and Southern hybridization was carried out according to standard methods by using nylon filters and hybridizing in 7% SDS/0.5 M sodium phosphate, pH 7, at 65°C. Southern hybridization probes were either restriction fragments isolated from cloned genomic EcoRI fragments in plasmid pUC19 (p7C46A/pG5D provided by P. Yen, Harbor–UCLA Medical Center, Los Angeles, CA) or gel-purified PCR products (p8A/DAZ3TER). The 1.5-kb probe p8A (DAZ 5′ flanking) was generated by using forward primer l411P4 (5′-gaccggaggctatgaacagg-3′) and reverse primer l411P5 (5′-caattggcccttgaggagcc-3′) on a human genomic DNA template. p8A is located 10 kb upstream of DAZ exon 1 in cosmid l411 (D. Page, available at http://www.genome.wi.mit.edu). DAZ3TER (DAZ 3′ untranslated region) was generated by using forward primer daz3 and the T3 vector primer (5′-aattaaccctcactaaaggg-3′) on a DAZ cDNA template cloned in phagemid vector pBK-CMV (provided by P. Yen). The Northern hybridization probes were DAZ3TER or mouse β-actin (mouse actin PCR product cloned into pGEM1) (34).
Immunohistochemistry and in Situ Hybridization.
Mouse testes were fixed in Bouins for 6–8 h and stored in 70% ethanol processed into paraffin wax, and 5-μm sections were cut. Immunohistochemistry for proliferating cell nuclear antigen was performed by using a mAb at 1:100 (Dako); positive nuclei were visualized by using standard methods (35). Radiolabeled sense and antisense riboprobes were synthesized from DAZ3TER subcloned in pCRIITOPO (Invitrogen) and used to perform in situ hybridization according to standard methods (35); sections were exposed to emulsion for 33 days.
Western Blotting.
Methods for protein extraction, Western blotting, and antibody detection are described in ref. 36. DAZ antibody 133 is specific for human DAZ (R. Reijo, personal communication).
RESULTS
Introduction of a YAC from the Human DAZ Genomic Region Gene into Mice.
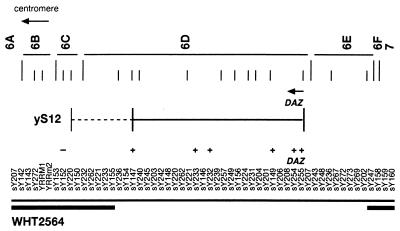
YAC clone S12 (yS12) contains a 225-kb insert originally reported to span the AZFc deletion in an azoospermic patient KLARD (10). Further analysis of the STS content of yS12 revealed that the YAC extends over only a portion of the minimal AZFc region and tests positive for sY254 and sY255 located within the DAZ gene (3) plus additional 3′ flanking STS markers (Fig. 1) and is negative for STSs representing CDY, PRY, TTY2, and BPY2 (27).
Figure 1.
STS map of the AZFc region, including the positions of the DAZ gene cluster and RBM (YRRM) genes adapted from Reijo et al. (3). At the top are 22 intervals defined by patient deletions and YAC endpoints. Below that are minimum and maximum possible extents of yS12 revealed by testing for the presence (+) or absence (−) of indicated STS markers. The broken bar indicates the minimal AZFc region as defined by the deletion in azoospermic patient WHT2564 (3). All STSs have been described (3, 41).
yS12 was introduced into a C57BL × CBA/Ca F1 mouse background by pronuclear injection. A single transgenic G0 male (M1) was obtained, and Southern hybridization to M1 genomic DNA with probes specific to each YAC vector arm indicated the presence of a complete YAC (not shown). M1 was backcrossed to C57BL × CBA/Ca F1 females to generate the transgenic mouse line TgN(hDAZy1)1Hgu, referred to hereafter as TgS12. Fluorescence in situ hybridization (FISH) with a YAC vector arm probe revealed an integration site close to the centromere of chromosome 7 (not shown). A crude estimate of YAC copy number was obtained from the FISH signal in interphase nuclei. A maximum of two hybridization signals were seen per nucleus indicating a probable copy number of 1 or 2 (not shown).
yS12 Contains a Single Copy of DAZ.
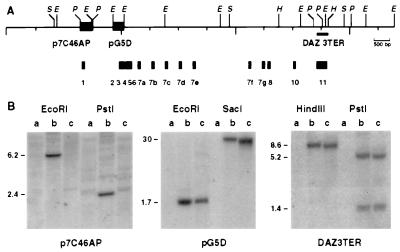
To assess the integrity of the DAZ sequence within the TgS12 mouse, Southern blots of TgS12 mouse and human genomic DNA were probed with sequences derived from the indicated locations within the DAZ genomic region (Fig. 2A). Probe pG5D, a genomic EcoRI fragment representing exons 2–6, and DAZ3TER, a DAZ-specific cDNA-derived PCR product representing the 3′ untranslated region, gave similar patterns of hybridization in human and mouse, but p7C46AP, the genomic EcoRI/PstI fragment including DAZ exon 1, failed to produce a hybridization signal in the TgS12 mouse (Fig. 2B). The presence of a breakpoint in yS12 between exons 1 and 2 was confirmed by PCR (Materials and Methods). The absence of sequences further upstream of exon 1 was confirmed by hybridization with an additional probe p8A (not shown). Coupled with the STS content, this structural information is consistent with the interpretation that TgS12 contains a single, incomplete copy of the DAZ gene, excluding exon 1 and promoter sequences, and probably derived from the centromere proximal end of the DAZ gene cluster (Fig. 1).
Figure 2.
(A) Restriction map of DAZ gene indicating the positions of probe fragments and DAZ exons 1–11. Scale in bp is shown. (B) Southern analysis of genomic DNA from wild-type mouse (a), human male (b), and TgS12 transgenic mouse (c) with probes derived from the DAZ genomic region. Probes (p7C46AP, pG5D, and DAZ3TER) used are indicated at the bottom.
Tissue Specificity of Transgene Expression.
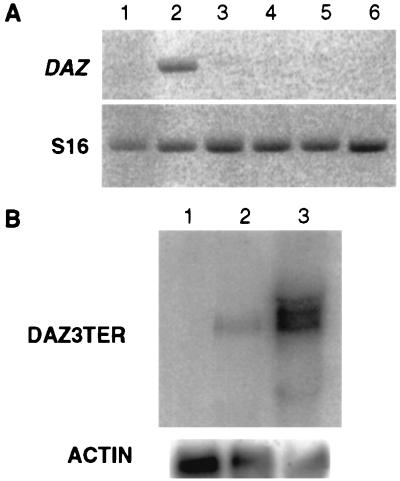
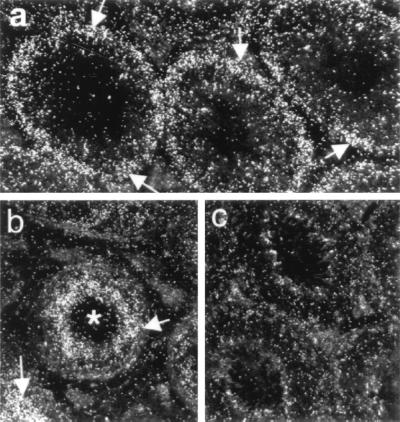
In the TgS12 transgenic mouse DAZ transcripts were detectable by RT-PCR in testis RNA but not in ovary RNA or in the range of nongonadal tissues that were examined, indicating an appropriate tissue distribution of DAZ transgene expression (Fig. 3A). Northern hybridization with DAZ 3TER confirmed the presence of DAZ transcript in the transgenic testis, although comparison with the signal in human testis RNA indicated a much reduced level of transcript in the TgS12 mouse (Fig. 3B). At least three bands are seen in the human testis RNA, presumably representing transcripts containing different numbers of DAZ repeats (37). A single size class of transcript, within the range seen in human, is apparent in the TgS12 mouse testis. RNA in situ hybridization to sectioned adult testes of TgS12 transgenic mice with a DAZ-specific probe revealed a pattern of cell type specificity that mirrored DAZ expression in human testis. Transcripts were confined to the germ-cell component of the testis lumen and appeared most abundant in spermatocyte stages (Fig. 4). Thus, despite its structural rearrangement, the DAZ transgene displayed a highly conserved pattern of spatial regulation.
Figure 3.
(A) Transcription profile of DAZ transgene in a range of tissues (lane 1, ovary; lane 2, testes; lane 3, female brain; lane 4, male brain; lane 5, female kidney; and lane 6, male kidney) from TgS12 transgenic mice assayed by RT-PCR. DAZ and S16 ribosomal protein primers described in Materials and Methods. (B) Northern hybridization to adult testis RNA of wild-type mouse (lane 1), TgS12 transgenic mouse (lane 2), and human (lane 3) with probe DAZ3TER (Upper). The diffuse signal in human RNA represents a range of DAZ transcripts containing different numbers of the 24-aa DAZ repeat. There is no apparent cross-hybridization to mouse Dazl. Signal obtained with a mouse actin probe is shown (Lower).
Figure 4.
Detection of DAZ mRNA transcripts in human testis and in a Dazl−/+;TgS12 mouse by in situ hybridization. Abundant silver grains were present within the seminiferous epithelium (∗) of human (b, positive control) and the Dazl−/+;TgS12 transgenic mouse (a). Most mRNA appeared to be localized in pachytene spermatocytes (arrows). As expected, no DAZ mRNA was detected in wild-type mice (c).
Structure and Expression of the DAZ Transgene.
Because Southern blotting indicated that exon 1 and the promoter region are absent from the DAZ transgene, 5′ rapid amplification of cDNA ends (RACE) analysis was performed to determine the structure of the DAZ transcript in the TgS12 mouse. Sequence analysis of RACE products confirmed the absence of DAZ exon 1 from the cDNA sequence and revealed the attachment of at least 150 bp of herpes simplex virus thymidine kinase (HSV-TK) sequence to the second exon of the DAZ transcript at nucleotide position 258 (Fig. 5B). This arrangement most probably is the result of an interstitial deletion of the yS12 construct fusing DAZ exon 2 with the HSV-TK gene located upstream in inverted orientation within the YAC left arm (Fig. 5A). DAZ exon 1 encodes only the initial methionine. The predicted product of the fusion transcript therefore would consist of the remaining amino acids of DAZ plus an N-terminal sequence of 14 foreign aa (Fig. 5C). Western analysis with a DAZ-specific antibody confirmed the presence of an immunoreactive transgene product and revealed variegation of transgene expression between individual males (Fig. 6). The retarded migration of the transgene product relative to DAZ protein from the testis of an individual human male may be attributable to a difference in DAZ repeat number in addition to the N-terminal extension of the fusion protein. The lower abundance of the transgene protein product may reflect the observed difference in transcript level seen by Northern hybridization (Fig. 3B) and/or reduced translational efficiency.
Figure 5.
(A) Structural rearrangement of yS12 resulting in fusion of HSV-TK and DAZ sequences. Positions and orientations of the DAZ gene and the LYS2, TRP1, and HSV-TK genes within the modified YAC are indicated. (B) Interstitial deletion has resulted in a fusion of HSV-TK sequence to the DAZ gene with elimination of DAZ exon 1 (including initiation codon) and intron 1. The location of a potential initiation codon is indicated by an asterisk. (C) Predicted amino acid sequence of the fusion transcript. DAZ nucleotide and amino acid sequences (in B) and amino acids introduced by fusion (in C) are underlined.
Figure 6.
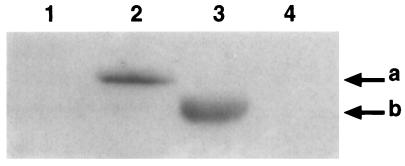
Western blot of adult human and mouse testis proteins probed with DAZ-specific antibody 133. Lanes: 1, TgS12 transgenic mouse number 1; 2, TgS12 transgenic mouse number 2; 3, human; 4, wild-type mouse. Signals corresponding to the 50-kDa immunoreactive protein in the TgS12 transgenic mouse number 2 (a) and the 45-kDa DAZ protein (b) are indicated. TgS12 mice are Dazl+/+.
yS12 Confers Partial Rescue of the Dazl Phenotype.
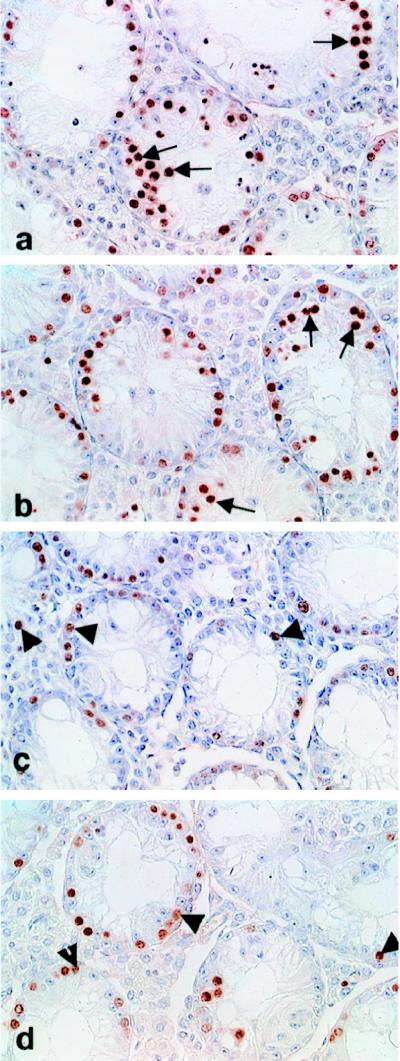
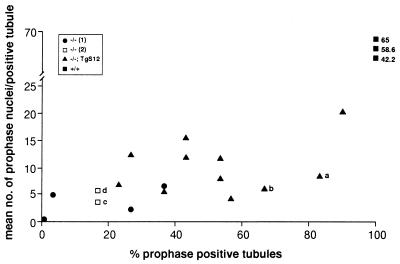
To test its ability to complement the mouse Dazl mutation, yS12 was introduced into a DazlTm1Hgu/DazlTm1Hgu knockout background (hereafter referred to as Dazl−/−) by genetic crossing (see Materials and Methods). Progeny homozygous null for Dazl and carrying the yS12 DAZ transgene (Dazl−/−;TgS12) were assessed immunohistochemically for phenotypic rescue by using sectioned adult testes stained with an anti-proliferating cell nuclear antigen antibody to detect dividing cells (Fig. 7 A and B). In comparison with testis sections from Dazl−/− controls lacking the DAZ transgene (Fig. 7 C and D), these mice exhibited clear evidence of phenotypic rescue. Although fertility was not restored and there was no evidence of complete spermatogenesis, a high proportion of seminiferous tubules in the transgenic males contained substantial germ-cell numbers and a lumen populated with numerous prophase spermatocytes. In contrast, tubules of control mice often were devoid of germ cells or contained only a few, peripherally located spermatogonial cells, which rarely progressed into meiosis. The scatter diagram in Fig. 8 illustrates data from 19 adult males: 11 genotype Dazl−/−;TgS12, five Dazl−/−, and 3 wild type. Individuals were scored for the dual criteria of percentage of prophase-positive tubules and number of prophase nuclei per positive tubule. Although individual males varied considerably in the degree of phenotypic rescue, as the distribution in Fig. 8 shows, Dazl−/−;TgS12 animals exhibited consistent and, in some cases, dramatic enhancement compared with Dazl−/− controls. In the five Dazl−/− individuals shown, the percentage of prophase-positive tubules ranges from 3% to 38% (mean, 18%). Corresponding values in Dazl−/−;TgS12 individuals range from 38% to 90% (mean, 60%). On the criterion of number of prophase nuclei per positive tubule, Dazl−/−;TgS12 mice scored from 6 to 20 compared with a range of 3 to 7 in Dazl−/− controls (means of 10 and 3, respectively). No correlation between the degree of rescue and the homozygous or hemizygous status of the YAC transgene (as determined by fluorescence in situ hybridization analysis on metaphase chromosome spreads; not shown) was detected in the subset of seven Dazl−/−;TgS12 animals tested.
Figure 7.
Immunohistochemical staining of sectioned adult mouse testes with anti-proliferating cell nuclear antigen antibody to detect proliferating cells. In the Dazl−/− mouse a proportion of tubules appears devoid of proliferating germ cells, and the remainder contain only peripherally located, premeiotic spermatogonial stages (c and d, arrowheads). The Dazl−/−;TgS12 transgenic mouse exhibits a pronounced increase in the spermatogonial population and the presence of numerous prophase spermatocytes (arrows) in the tubular lumen, indicating partial restoration of meiosis (a and b). The Dazl−/−;TgS12 and control Dazl−/− mice were derived from the same cross of DazlTm1Hgu/+;TgS12 × DazlTm1Hgu/+;TgS12 animals. The individual animals are identified as a and b (Dazl−/−;TgS12) and c and d (Dazl−/−) in Fig. 8.
Figure 8.
Scatter diagram indicating the range of phenotypic rescue conferred by yS12. Testis sections from adult mice of genotype +/+, Dazl−/−;TgS12 (−/−;S12), and Dazl−/− (−/−) were probed with anti-PCNA antibody and scored for the percentage of prophase-positive tubules and mean number of prophase nuclei per positive tubule. Each point represents data from at least 10 (+/+) or 30 tubules (Dazl−/−;TgS12 and Dazl−/−) of an individual male. Dazl−/− individuals were generated in one of two genetic crosses: DazlTm1Hgu/+ × DazlTm1Hgu/+ (1) or DazlTm1Hgu/+;TgS12 × DazlTm1Hgu/+;TgS12 (2). Points corresponding to the Dazl−/−;TgS12 individuals (a and b) and Dazl−/− individuals (c and d) represented in Fig. 7 are indicated.
DISCUSSION
We have shown that a YAC clone containing a truncated copy of a human DAZ gene is capable of partially rescuing the infertility of the male Dazl knockout mouse. Consistent with the data from deletion analysis of infertile men, the present findings constitute persuasive evidence that the product of human DAZ is active in spermatogenesis and has retained functional homology with its murine autosomal ancestor. The probability of a causal link between the elimination of DAZ sequences and spermatogenic impairment in men with AZFc deletions thereby is strengthened.
Although the failure of the S12 YAC to restore fertility could reflect incomplete functional homology between the DAZ and Dazl gene products, it equally could result from a quantitative insufficiency or structural impairment of the DAZ transgene. Both Northern and Western analyses indicated a low level of DAZ expression in the transgenic mouse testis relative to human testis. This reduced expression level simply may reflect a difference in DAZ gene copy number because the Y chromosomal DAZ cluster is thought to consist of up to seven gene copies (38). Additional possibilities are incomplete conservation of cis- or trans-acting regulatory elements, a negative chromosomal position effect on transgene expression, or both. From our structural analysis of the DAZ transgene, which revealed the absence of exon 1 and 5′ flanking sequences, it seems likely that an enhancer element could be lacking, although the observed testis-specific expression pattern does suggest that at least partial regulatory information resides internally or 3′ of the transcribed region. The variability of both transgene expression as revealed by Western blotting and phenotypic rescue would be consistent with a degree of transgene positional silencing, possibly in consequence of the centromeric-insertion site. Furthermore, as a result of structural rearrangement, DAZ exons 2–11 are fused with the upstream HSV-TK YAC gene, leading to the addition of at least 14 foreign, N-terminal amino acids and the elimination of the native initial methionine. It is possible that these alterations have compromised the function of the transgene and the substitution for the native ATG of a relatively inefficient translation initiation site may have contributed to the reduction in transgene protein product.
Several lines of evidence point to there being a quantitative requirement in gametogenesis for DAZ gene family products. In humans, the penetrance of the phenotype in men with Y chromosomal DAZ deletions is highly variable, ranging from a complete lack of germ cells (Sertoli Cell Only syndrome) to meiotic arrest at various stages. Even within an individual testis, variegation is apparent between seminiferous tubules (3, 8). In both mouse and Drosophila, heterozygosity for the DAZ autosomal homologue reveals a degree of haploinsufficiency, and only partial rescue of the null phenotype was provided by a boule transgene (17, 25). Mouse Dazl is known to be expressed throughout much of the spermatogenic program, from the primordial germ-cell stage up to sperm maturation (25, 39, 40). Therefore, it is likely that the gene operates on different RNA targets and exercises different functions at different stages. The Dazl phenotype encompasses at least three classes of defect. The earliest apparent aberration is a reduction in germ-cell numbers, which is followed at puberty by a meiotic failure manifest as an absence of spermatogenic stages beyond pachytene. The heterozygous phenotype reveals a later function in spermiogenesis. These distinct functions of Dazl protein may be differentially dosage-sensitive. Specifically, our results suggest that the level of product provided by the DAZ transgene is sufficient to increase germ-cell survival and permit the initiation of meiosis in the absence of Dazl but below the threshold necessary to support completion of the spermatogenic program. To test the possibility that a higher dosage of DAZ protein could provide full phenotypic rescue, we currently are generating further transgenic lines carrying a range of DAZ constructs to obtain increased levels of gene expression.
The quantitative nature of the requirement for DAZ protein may explain the presence of the DAZ multigene family on the human Y chromosome, in addition to its autosomal progenitor DAZL1, if this results in a selective advantage of increased spermatogenic efficiency. Alternatively, the structural changes after transposition to the Y chromosome, including acquisition of the DAZ repeat, may have engendered subtle functional divergence such that DAZ provides a function complementary to DAZL1. The present study points to the feasibility of probing the structure/function relationships of the human DAZ and DAZL1 genes in a mouse model system. From the cross-species conservation demonstrated by the partial rescue of the Dazl phenotype it follows also that DAZ has at least partial identity of function with its human autosomal counterpart, DAZL1. In the clinical field, a therapeutic possibility arising from partial redundancy of DAZ and DAZL1 in the human male would be the mitigation of spermatogenic defects in DAZ-deleted men by up-regulation of the function of their autosomal DAZL1 genes.
Acknowledgments
We thank M. Dalrymple and colleagues at PPL Ltd. (Edinburgh) for YAC DNA injections, M. Jobling (Leicester University) for providing yS12, K. Ma for advice on yS12 content, P. Yen (Harbor–University of California at Los Angeles Medical Center) for providing DAZ probes and information on DAZ gene structure, R. Reijo (University of California at San Francisco) for the DAZ 133 antibody and advice on Western technology, M. Lee for technical assistance, J. Moss for the actin probe, T. Vogel for assistance with Fig. 2A, and D. Stuart for help with figure production. This work was supported by the U.K. Medical Research Council and European Union Biotechnology Grant E321/223 (B.G.).
ABBREVIATIONS
- DAZ
deleted in azoospermia
- Dazl
mouse autosomal DAZ-like
- DAZL1
human autosomal DAZ-like
- AZF
azoospermia factor
- RBM
RNA-binding motif
- STS
sequence-tagged site
- YAC
yeast artificial chromosome
- RT-PCR
reverse transcription–PCR
- HSV-TK
herpes simplex virus thymidine kinase
References
- 1.Tiepolo L, Zuffardi O. Hum Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi K, Mizuno K, Hida A, Komaki R, Tomita K, Matsushita I, Namiki M, Iwamoto T, Tamura S, Minowada S, et al. Hum Mol Genet. 1994;3:1965–1967. doi: 10.1093/hmg/3.11.1965. [DOI] [PubMed] [Google Scholar]
- 3.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 4.Najmabadi H, Huang V, Yen P, Subbarao M N, Bhasin D, Banaag L, Naseeruddin S, Dekretser D M, Baker H W G, McLachlan R I, et al. J Clin Endocrinol Metab. 1996;81:1347–1352. doi: 10.1210/jcem.81.4.8636331. [DOI] [PubMed] [Google Scholar]
- 5.Qureshi S J, Ross A R, Ma K, Cooke H J, Intyre M A, Chandley A C, Hargreave T B. Mol Human Reprod. 1996;2:775–779. doi: 10.1093/molehr/2.10.775. [DOI] [PubMed] [Google Scholar]
- 6.Stuppia L, Mastroprimiano G, Calabrese G, Peila R, Tenaglia R, Palka G. Cytogenet Cell Genet. 1996;72:155–158. doi: 10.1159/000134174. [DOI] [PubMed] [Google Scholar]
- 7.Pryor J L, Kent-First M, Muallem A, Van B A, Nolten W E, Meisner L, Roberts K P. N Engl J Med. 1997;336:534–539. doi: 10.1056/NEJM199702203360802. [DOI] [PubMed] [Google Scholar]
- 8.Vogt P H, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Kohn F M, Schill W B, Farah S, Ramos C, et al. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 9.Brown G M, Furlong R A, Sargent C A, Erickson R P, Longepied G, Mitchell M, Jones M H, Hargreave T B, Cooke H J, Attara N A, et al. Hum Mol Genet. 1998;7:97–107. doi: 10.1093/hmg/7.1.97. [DOI] [PubMed] [Google Scholar]
- 10.Ma K, Inglis J D, Sharkey A, Bickmore W A, Hill R E, Prosser E J, Speed R M, Thomson E J, Jobling M, Taylor K, et al. Cell. 1993;75:1287–1295. doi: 10.1016/0092-8674(93)90616-x. [DOI] [PubMed] [Google Scholar]
- 11.Laval S H, Glenister P H, Rasberry C, Thornton C E, Mahadevaiah S K, Cooke H J, Burgoyne P S, Cattanach B M. Proc Natl Acad Sci USA. 1995;92:10403–10407. doi: 10.1073/pnas.92.22.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott D J, Ma K, Kerr S M, Thakrar R, Speed R, Chandley A C, Cooke H. Hum Mol Genet. 1996;5:869–874. doi: 10.1093/hmg/5.7.869. [DOI] [PubMed] [Google Scholar]
- 13.Delbridge M L, Harry J L, Toder R, Oneill R J W, Ma K, Chandley A C, Graves J A M. Nat Genet. 1997;15:131–136. doi: 10.1038/ng0297-131. [DOI] [PubMed] [Google Scholar]
- 14.Cooke H J, Lee M, Kerr S, Ruggiu M. Hum Mol Genet. 1996;5:513–516. doi: 10.1093/hmg/5.4.513. [DOI] [PubMed] [Google Scholar]
- 15.Reijo R, Seligman J, Dinulos M B, Jaffe T, Brown L G, Disteche C M, Page D C. Genomics. 1996;35:346–352. doi: 10.1006/geno.1996.0366. [DOI] [PubMed] [Google Scholar]
- 16.Soulard M, Dellavalle V, Siomi M C, PinolRoma S, Codogno P, Bauvy C, Bellini M, Lacroix J C, Monod G, Dreyfuss C, et al. Nucleic Acids Res. 1993;21:4210–4217. doi: 10.1093/nar/21.18.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberhart C G, Maines J Z, Wasserman S A. Nature (London) 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 18.Saxena R, Brown L G, Hawkins T, Alagappan R K, Skaletsky H, Reeve M P, Rejio R, Rozen S, Dinulos M, Disteche C M, et al. Nat Genet. 1996;14:292–298. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 19.Shan Z, Hirschmann P, Seebacher T, Edelmann A, Jauch A, Morell J, Urbitsch P, Vogt P H. Hum Mol Genet. 1996;5:2005–2011. doi: 10.1093/hmg/5.12.2005. [DOI] [PubMed] [Google Scholar]
- 20.Yen P H, Chai N N, Salido E C. Hum Mol Genet. 1996;5:2013–2017. doi: 10.1093/hmg/5.12.2013. [DOI] [PubMed] [Google Scholar]
- 21.Seboun E, Barbaux S, Bourgeron T, Nishi S, Algonik A, Egashira M, Nikkawa N, Bishop C, Fellous M, McElreavey K, et al. Genomics. 1997;41:227–235. doi: 10.1006/geno.1997.4635. [DOI] [PubMed] [Google Scholar]
- 22.Carani C, Gromoll J, Brinkworth M H, Simoni M, Weinbauer G F, Nieschlag E. Mol Hum Reprod. 1997;3:479–483. doi: 10.1093/molehr/3.6.479. [DOI] [PubMed] [Google Scholar]
- 23.Karashima T, Sugimoto A, Yamamoto M A. Worm Breeders Gazette. 1997;15:65–66. [Google Scholar]
- 24.Houston D W, Zhang J, Maines J Z, Wasserman S A, King M L. Development (Cambridge, UK) 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- 25.Ruggiu M, Speed R, Taggart M, Mckay S J, Kilanowski F, Saunders P, Dorin J, Cooke H J. Nature (London) 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 26.Rice W R. Science. 1994;263:230–232. doi: 10.1126/science.8284674. [DOI] [PubMed] [Google Scholar]
- 27.Lahn B T, Page D C. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 28.Yen P H. Genomics. 1998;54:5–12. doi: 10.1006/geno.1998.5526. [DOI] [PubMed] [Google Scholar]
- 29.Agulnik A I, Zharkikh A, Boettger-Tong H, Bourgeron T, McElreavey K, Bishop C E. Hum Mol Genet. 1998;7:1371–1377. doi: 10.1093/hmg/7.9.1371. [DOI] [PubMed] [Google Scholar]
- 30.Smith D R, Smyth A P, Strauss W M, Moir D T. Mamm Genome. 1993;4:141–147. doi: 10.1007/BF00352229. [DOI] [PubMed] [Google Scholar]
- 31.Shero J H, McCormick M K, Antonarakis S E, Hieter P. Genomics. 1991;10:505–508. doi: 10.1016/0888-7543(91)90343-d. [DOI] [PubMed] [Google Scholar]
- 32.Schedl A, Grimes B, Montoliu L. Methods Mol Biol. 1996;54:293–306. doi: 10.1385/0-89603-313-9:293. [DOI] [PubMed] [Google Scholar]
- 33.Vollrath D, Foote S, Hilton A, Brown L G, Beerromero P, Bogan J S, Page D C. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- 34.Tokunaga K, Taniguchi H, Yoda K, Shimizu M, Sakiyama S. Nucleic Acids Res. 1986;14:2829. doi: 10.1093/nar/14.6.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majdic G, Sharpe R M, O’Shaughnessy P J, Saunders P T. Endocrinology. 1996;137:1063–1070. doi: 10.1210/endo.137.3.8603575. [DOI] [PubMed] [Google Scholar]
- 36.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 37.Yen P H, Chai N N, Salido E C. Mamm Gen. 1997;8:756–759. doi: 10.1007/s003359900560. [DOI] [PubMed] [Google Scholar]
- 38.Glaser B, Yen P H, Schempp W. Chromosome Res. 1998;6:481–486. doi: 10.1023/a:1009256613348. [DOI] [PubMed] [Google Scholar]
- 39.Seligman J, Page D C. Biochem Biophys Res Commun. 1998;245:878–882. doi: 10.1006/bbrc.1998.8530. [DOI] [PubMed] [Google Scholar]
- 40.Habermann B, Mi H F, Edelmann A, Bohring C, Backert I T, Kiesewetter F, Aumuller G, Vogt P H. Hum Reprod. 1998;13:363–369. doi: 10.1093/humrep/13.2.363. [DOI] [PubMed] [Google Scholar]
- 41.Foote S, Vollrath D, Hilton A, Page D C. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]