Abstract
Mitochondrial RNA (mtRNA) polymerases are related to bacteriophage RNA polymerases, but contain a unique amino-terminal extension of unknown origin and function. In addition to harboring mitochondrial targeting information, we show here that the amino-terminal extension of yeast mtRNA polymerase is required for a mtDNA maintenance function that is separable from the known RNA polymerization activity of the enzyme. Deletion of 185 N-terminal amino acids from the enzyme results in a temperature-sensitive mitochondrial petite phenotype, characterized by increased instability and eventual loss of the mitochondrial genome. Mitochondrial transcription initiation in vivo is largely unaffected by this mutation and expression of just the amino-terminal portion of the protein in trans partially suppresses the mitochondrial defect, indicating that the amino-terminal extension of the enzyme harbors an independent functional domain that is required for mtDNA replication and/or stability. These results suggest that amino-terminal extensions present in mtRNA polymerases comprise functional domains that couple additional activities to the transcription process in mitochondria.
Expression of genes encoded by the mitochondrial genome is initiated by a distinct mitochondrial RNA (mtRNA) polymerase enzyme that is encoded by the nuclear genome and imported into the organelle. Consistent with a prokaryotic origin of mitochondria, mtRNA polymerases are homologous to the single-subunit RNA polymerases encoded by the bacteriophage genomes of T7, T3, and SP6 (1). In the yeast, Saccharomyces cerevisiae, similarity of mtRNA polymerase to these prokaryotic enzymes does not extend throughout the entire primary amino acid sequence, but rather is localized to eight, highly conserved regions that are dispersed within the C-terminal two-thirds of the protein. Based on structural and mutational analyses (2–5), these regions are postulated to be critical for promoter selectivity and catalytic activity of this class of RNA polymerases. However, yeast mtRNA polymerase has an amino-terminal extension of ≈400 aa that is not present in the bacteriophage enzymes (Fig. 1). A relatively small portion of this extension (the 29 N-terminal aa) is needed to target the enzyme to mitochondria, and an intact amino terminus has been proposed to be necessary, but not sufficient, for interaction with the mitochondrial transcription factor sc-mtTFB (Mtf1p)(6). Currently, it remains unclear whether the remainder of the amino-terminal extension has additional functions, other than localization, that allow a bacteriophage-like enzyme to function optimally in the context of the mitochondrial genetic system. For example, gene expression in yeast and vertebrate mitochondria requires a large number of RNA processing events that generate mature RNA species from polycistronic precursor transcripts (7–9). In addition, RNA transcript processing events have been implicated in formation of RNA primers for mtDNA replication (for review, see ref. 10). Thus, proper coordination of transcription and RNA processing activities is critical for mitochondrial gene expression and mtDNA replication. Reasoning that the amino-terminal extension of mtRNA polymerase may be involved in additional functions of the enzyme other than localization, we analyzed a series of deletion mutations in the yeast mtRNA polymerase gene (RPO41). These mutations were engineered to retain the mitochondrial localization signal, thus allowing the consequence of removing amino acids in the amino-terminal extension to be analyzed without affecting mitochondrial localization and import.
Figure 1.
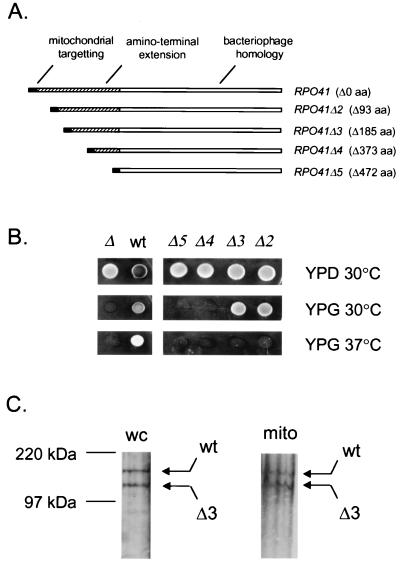
Characterization of amino-terminal deletions in mtRNA polymerase. (A) A schematic representation of yeast mtRNA polymerase and deletion mutations used in this study is shown with important features labeled as follows: mitochondrial targeting sequence, black box; region containing bacteriophage homology, open box; amino-terminal extension, hatched box. The number of amino acids deleted (Δ) in each protein is given in parentheses. (B) Phenotypic analysis of RPO41 deletion mutants by plasmid shuffle. Mitochondrial function was assessed in each strain by comparing growth on YPD and YPG at 30°C and 37°C. The RPO41 genotype of each strain after plasmid shuffle is given as follows: Δ, rpo41; wt, RPO41; Δ2-Δ5, rpo41Δ2-Δ5. (C) Western analysis of rpo41Δ3-encoded mtRNA polymerase from GS129. After growth at 37°C for ≈20 generations, protein from whole cells (wc) and from a purified mitochondrial fraction (mito) were analyzed. Signals for the wild-type (wt) and rpo41Δ3-encoded (Δ3) protein are indicated with arrows. Protein molecular mass standards are indicated on the left.
MATERIALS AND METHODS
Plasmid Construction and Plasmid Shuffle of RPO41 Alleles.
Site-directed mutagenesis was used to introduce a NheI restriction site in the RPO41 coding region, just beyond the mitochondrial targeting sequence (at nucleotides 78–83, according to ref. 1). The plasmid used for mutagenesis was a yeast shuttle plasmid pRS314 (11) harboring a 7.2-kb SalI–SpeI restriction fragment encompassing the RPO41 locus (pGS348). NheI sites also were engineered at four other positions within the RPO41 gene (at nucleotides 351–356, 627–632, 1191–1196, and 1497–1502). In-frame deletions then were made that extend from the NheI site beyond the mitochondrial targeting sequence to each NheI site within the coding region of mtRNA polymerase to create plasmids that contained the rpo41Δ2–5 alleles (pGS348Δ2-Δ5). The plasmid-shuffle protocol was performed as described (12, 13), except the strain GS112 (α his3-Δ200 leu2–3,-112 ura3–52 trp1-Δ1 ade2 rpo41Δ1∷HIS3 +pGS347[URA3,RPO41]) was used. The following strains were generated by plasmid shuffle and characterized further: GS122 (α his3-Δ200 leu2–3,-112 ura3–52 trp1-Δ1 ade2 rpo41Δ1∷HIS3 +[pGS348]); GS124-GS127 (α his3-Δ200 leu2–3,-112 ura3–52 trp1-Δ1 ade2 rpo41Δ1∷HIS3 +[pGS348Δ2-Δ5]); and GS128 (α his3-Δ200 leu2–3,-112 ura3–52 trp1-Δ1 ade2 rpo41Δ1∷HIS3 +[pRS314]).
The yeast high-copy plasmid YEp352 was used to promote overexpression of the sc-mtTFA and sc-mtTFB proteins. A 1.7-kb EcoRI fragment containing the ABF2 gene was inserted into the EcoRI site of YEp352 to create the plasmid YEp-mtTFA. A 2.8-kb XhoI fragment containing the MTF1 gene was inserted into the SalI site of YEp352 to create the plasmid YEp-mtTFB. As judged by Western analysis, yeast strains carrying YEp-mtTFA or YEp-mtTFB exhibit ≈10-fold overproduction of the respective protein (data not shown). The plasmid YEp-Δ3EXP was constructed by using a PCR cloning strategy. The 5′ PCR primer (5′-taatgtcgacgaaaatggcg) hybridized at a site encompassing a SalI site located ≈1,100 bp upstream of the RPO41 gene. The 3′ PCR primer (5′-attaaagcttacttcaaaagcatcgagcttg) hybridized to nucleotides 613–636 (according to ref. 1) and contains a HindIII restriction site. By using pGS348 as a template, a ≈1.8-kb PCR product was generated and ligated into YEp352 digested with SalI and HindIII.
Western Analysis.
Whole-cell yeast protein was extracted as described (14). Yeast mitochondria were prepared by a standard differential centrifugation protocol (15). Proteins from whole cells or the mitochondria-enriched pellet were separated on SDS/PAGE gels and transferred to nitrocellulose membranes by standard methods (16), and Western analysis was performed as described in the ECL Western blotting detection kit (Amersham Pharmacia), by using a yeast mtRNA polymerase polyclonal antibody.
4′,6-Diamidino-2-phenylindole (DAPI) Staining of Yeast Cells.
Before staining, ≈1.0 × 107 yeast cells from the indicated strains were fixed in 70% ethanol for 10 min on ice. The cells then were washed twice with 1 ml of deionized water and resuspended in 100 μl of 50 ng/ml DAPI (Sigma) in PBS for 10 min. DAPI fluorescence was observed by using an Olympus BX60 epi-fluorescence microscope.
Northern Analysis and S1 Nuclease Mapping.
Total yeast RNA was isolated as described (17) and treated with 3.5 units of RQ1 DNase I (Promega) for 30 min at 37°C. For Northern analysis, 15 μg of RNA was loaded into each lane of a 1.2% formaldehyde/agarose gel, separated by electrophoresis, and transferred to a nylon membrane essentially as described (16). The immobilized RNA then was probed by using a 32P-labeled, random-primed DNA probe, generated by using Ready-To-Go DNA labeling beads (Amersham Pharmacia) as described by the manufacturer and a 966-bp NdeI restriction fragment containing yeast ori5 as a template. Hybridization and washing conditions were exactly as described for use with Rapid-hyb buffer (Amersham Pharmacia). Ethidium bromide staining of mature rRNA species confirmed that nearly identical amounts of RNA were loaded in each lane (data not shown).
The S1 nuclease protection assay was performed essentially as described (16). RNA samples (10–20 μg) were coprecipitated with 0.7 μg of an end-labeled probe that was generated by digesting the plasmid pHS3324 with EcoRV, 5′ end-labeled with [γ-32P]ATP, and then digesting with BglII. This generated a 545-bp DNA probe that is complementary to the ori5 transcript and extends 331 nt beyond the predicted transcription initiation site. After the nucleic acid hybridization step, the S1 nuclease (Promega) reaction was carried out in the buffer provided by the manufacturer. Where indicated, samples were treated with 500 ng of RNase A for 10 min at 37°C before the hybridization step.
High-Copy Suppression of the rpo41Δ3 Temperature-Sensitive Phenotype.
The rpo41Δ3 yeast strain (GS125) was transformed with a library of yeast genomic DNA contained in the plasmid YEp352. A pool of uracil+ yeast cells (representing ≈20,000 transformants) then was plated onto yeast extract/peptone/glycerol (YPG) medium and grown at 37°C to select for strains that were able to form colonies at the nonpermissive temperature. The library plasmid was isolated from each putative suppressor strain and used to transform a fresh strain of GS125, which ensured that the ability to grow on YPG at 37°C was linked to the plasmid. Those plasmids that conferred growth at 37°C were sequenced by using M13 reverse and M13 −40 primers that allowed the nucleotide sequence of each end of the yeast genomic DNA insert to be determined. This information was used to determine the precise genomic fragment that was present in each suppressor plasmid by searching the Saccharomyces Genome Database using the provided fasta algorithm.
RESULTS AND DISCUSSION
The Amino-Terminal Extension of Yeast mtRNA Polymerase Is Required for Mitochondrial Function.
A series of four deletions (rpo41Δ2–Δ5) was generated that removed increasing amounts of the amino terminus of mtRNA polymerase, while leaving the putative mitochondrial localization signal intact (Fig. 1A). The phenotype of each RPO41-deletion mutation was assessed by using a plasmid-shuffle protocol, after which, the ability of the mutant yeast strains to grow on yeast extract/peptone/dextrose (YPD) (glucose medium, on which mitochondrial function is not required for growth) and YPG (glycerol medium, on which mitochondrial function is required for growth) at 30°C and 37°C was determined (Fig. 1B). A positive control strain (GS122), which retained a wild-type RPO41 allele after plasmid shuffle, was able to grow on YPD and YPG at both temperatures; whereas a negative control strain (GS128), which is null for RPO41 after plasmid shuffle, was unable to grow on glycerol at any temperature. Inability to grow on glycerol as the sole carbon source indicates the loss of mitochondrial respiration (i.e., a petite phenotype) and is the documented phenotype of an RPO41 null mutation (18). Deletion of amino acids 28–119 (rpo41Δ2) or 28–211 (rpo41Δ3) resulted in a temperature-sensitive petite phenotype, whereas deletions that extended beyond amino acid 211 (rpo41Δ4 and rpo41Δ5) resulted in a RPO41 null phenotype. Thus, in addition to targeting the protein to mitochondria, the amino-terminal extension of yeast mtRNA polymerase provides at least one additional function that is essential for mitochondrial activity.
The rpo41Δ3 Mutation Causes an Irreversible Petite Phenotype Caused by Mitochondrial Genome Instability.
The fact that the relatively large deletion in the rpo41Δ3 allele (185 aa) did not completely inactivate the enzyme at normal growth temperatures, coupled with the knowledge that the deleted region did not remove any residues with homology to bacteriophage RNA polymerases, suggested that this region of the enzyme might be critical for a function other than transcription. To test this hypothesis, we characterized rpo41Δ3 mutant yeast strains in more detail. First, by using a yeast strain that expresses both the wild-type RPO41 and rpo41Δ3 alleles (GS129), we determined that the rpo41Δ3 mutation is recessive (data not shown). We used this same strain to demonstrate that, even after extended growth at 37°C, the rpo41Δ3 allele expressed a truncated mtRNA polymerase protein of predicted molecular mass (≈129 kDa) that associated with mitochondria at levels comparable to the wild-type protein (≈150 kDa) (Fig. 1C). This finding suggested that neither stability nor mitochondrial localization was dramatically affected by the mutation. We next determined whether the petite phenotype of the rpo41Δ3 mutation was reversible. Yeast strains derived from plasmid shuffle that contained a plasmid-borne copy of either the wild-type RPO41 (GS122) or rpo41Δ3 allele (GS125) were grown at the nonpermissive temperature for multiple generations. At specific time points, the cultures were diluted and plated onto YPG and YPD medium and allowed to grow at the permissive temperature (30°C) until colonies formed. Growth on YPG indicated maintenance or re-establishment of mitochondrial function, whereas growth on YPD served as a measure of cell viability. The percentage of viable GS125 cells that were unable to grow on YPG medium increased significantly after the third generation of growth at 37°C (data not shown). By the 12th generation more than than 97% of the cells were petite. As expected, the wild-type GS122 strain generated spontaneous petites, as do most laboratory strains of S. cerevisiae (19). However, in the wild-type case, nearly 70% of the cells were still competent for respiration after 12 generations at 37°C (compared with ≈3% of the rpo41Δ3 cells). In addition to the YPG growth defect, GS125 cells plated from 37°C cultures exhibited a red and white sectoring colony phenotype on YPD medium (data not shown). Formation of a red pigment in this strain (because of the ade2 genetic background) occurs only in the presence of a functional mitochondrial respiratory chain. Colonies ranged from pure white (petite) to varying mixtures of red (respiration competent) and white cells. The total number of pure white colonies, as well as the number of mixed colonies that contained predominantly white cells, increased with each generation of growth at the nonpermissive temperature. Because the vast majority of GS125 cells remain petite even after an extended growth at the permissive temperature, we concluded that an irreversible mitochondrial defect eventually is manifested at 37°C as a result of the rpo41Δ3 mutation.
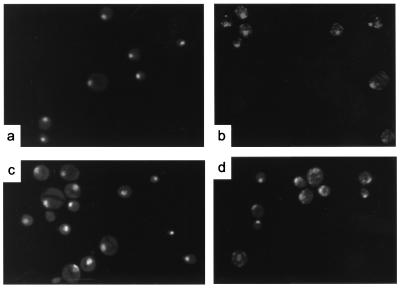
The irreversible nature of the rpo41Δ3 temperature-sensitive defect, as well as the mixed colony color phenotype, could be explained by loss or mutation of mitochondrial genomes. To investigate this possibility, we examined the petite colonies, resulting from growth of GS125 at 37°C, for the presence of mtDNA by using the nucleic acid stain DAPI (4′,6-diamidino-2-phenylindole) and fluorescence microscopy (Fig. 2). We stained cells from cultures of GS125 grown at the permissive temperature and the isogenic strain (GS125 rhoo), that was depleted of mtDNA ([rhoo]) by treatment with ethidium bromide, to serve as positive (Fig. 2b) and negative (Fig. 2a) controls for mtDNA staining, respectively. Approximately half of the GS125 petite colonies were homogeneous populations of cells that exhibited no mtDNA signal [rhoo] (Fig. 2c), while the other half were heterogeneous populations, consisting of [rhoo] cells and cells that exhibited some mtDNA staining (Fig. 2d). Given that deletion of the RPO41 gene previously has been shown to result in increased formation of deleted mitochondrial genomes ([rho−]), and eventual loss of mtDNA (18, 20), we propose that the petite cells that stain positive for mtDNA in our experiments are likewise [rho−]. In addition, segregation of a mixture of [rho+] and [rho−] genomes during cell division after plating and growth at the permissive temperature would produce red and white daughter cells in the same colony, providing a likely explanation for the observed sectoring phenotype of GS125 after growth at 37°C.
Figure 2.
The rpo41Δ3 mutation results in mtDNA instability. (a) GS125 rhoo was used as a control for cells completely lacking mtDNA. (b) GS125 was grown at 30°C in YPG medium and used as a positive control for mtDNA staining. Punctate cytoplasmic mtDNA staining was observed in these cells in addition to nuclear DNA staining. Representative petite colonies generated by growth of GS125 for 12 generations at 37°C are shown in c and d. In ≈50% of the colonies tested, none of the cells exhibited an mtDNA signal, [rhoo] (c), while the remainder consisted of a mixture of [rhoo] cells and cells that exhibited some mtDNA signal (d).
The rpo41Δ3 Mutation Does Not Result in Major Transcriptional Defects in Mitochondria.
Loss of mtRNA polymerase function could lead to loss of mtDNA by either direct or indirect mechanisms. It is documented that disruption of mitochondrial protein synthesis can lead to mitochondrial genome instability (21), thus loss of mitochondrial transcription could lead to mtDNA instability by this mechanism. To address this possibility, we determined whether the rpo41Δ3 mutation results in defective transcription in vivo. In yeast, two mitochondrial proteins are known to affect transcription by mtRNA polymerase (22). The sc-mtTFB protein originally was identified as a high-copy suppressor of a temperature-sensitive RPO41 allele (23) and is required for mitochondrial transcription initiation (13, 24, 25). The sc-mtTFA protein (Abf2p) also influences mitochondrial transcription in vitro (26) and is required for mtDNA maintenance under certain growth conditions (27). We introduced multicopy plasmids that overproduced the sc-mtTFA and sc-mtTFB proteins into GS125 and determined that, while each strain was competent for growth on YPG at 30°C, neither was able to grow on YPG at 37°C (Fig. 3A). Thus, overexpression of the mitochondrial transcription factor (sc-mtTFB) or the abundant mitochondrial high mobility group-box protein (sc-mtTFA) cannot compensate for the rpo41Δ3 defect.
Figure 3.
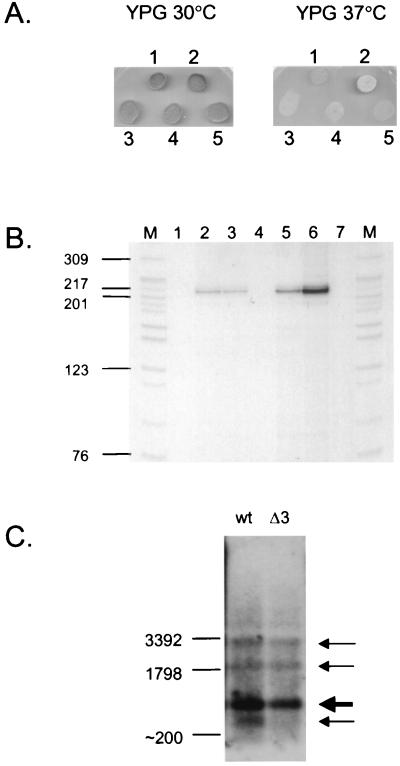
The mtDNA instability phenotype of the rpo41Δ3 mutation does not result from loss of mitochondrial transcription. (A) The sc-mtTFA and sc-mtTFB overexpression plasmids, YEp-mtTFA and YEp-mtTFB, and a control plasmid YEp352 each were introduced into GS125, and the ability of the resulting strains to grow on YPG at 30°C and 37°C is shown labeled as follows: 1, GS125(rpo41Δ3); 2, GS122(RPO41); 3, GS125(rpo41Δ3)+YEp-mtTFA; 4, GS125 (rpo41Δ3)+YEp-mtTFB; 5, GS125(rpo41Δ3)+YEp352. (B) S1 nuclease protection analysis of mitochondrial transcripts from the yeast strain (BS127 HS3324 ΔRPO41) containing either no mtRNA polymerase (lanes 1 and 4), the RPO41-containing plasmid, pGS348 (lanes 2 and 5) or an rpo41Δ3-containing plasmid, pGS348Δ3 (lanes 3 and 6), after growth at 30°C (lanes 1–3) or 37°C (lanes 4–7) for ≈20 generations. Lane 7, same as lane 6, except the sample was pretreated with RNase A. M, HpaII-digested pBR322 markers (length in nucleotides is indicated on the left). (C) Northern analysis of ori5 transcripts in BS127 HS3324 ΔRPO41 strains grown at 37°C: wt, wild-type mtRNA polymerase; Δ3, rpo41Δ3 mtRNA polymerase. The major ori5 RNA transcript is indicated by the bold arrow. Two additional minor transcripts of ≈2 kb and ≈3 kb, corresponding to two and three ori5 repeat lengths, are indicated by thin arrows, as is a subrepeat length transcript that was observed only in the wild-type strain. Indicated on the left is the position where known RNA species migrated.
Next, we investigated whether the rpo41Δ3-encoded mtRNA polymerase is capable of supporting transcription at the nonpermissive temperature in vivo. Because [rho+] mitochondrial genomes are unstable in RPO41 mutant or null genetic backgrounds (refs. 18 and 20, this study), we used a yeast strain (BS127 HS3324 ΔRPO41) that harbors an ori5 [rho−] mitochondrial genome that is stable in the RPO41 null background (28). The 966-bp, head-to-tail ori5 repeat in this strain harbors a mitochondrial promoter that allowed us to examine the production of a specific RNA transcript in vivo. First, we analyzed the 5′ ends of in vivo transcripts generated at either the permissive or nonpermissive temperature by using an S1 nuclease protection assay. As expected, under both conditions the parental strain, which contains no mtRNA polymerase, did not produce mtRNA transcripts that could hybridize to and protect the end-labeled antisense ori5 DNA probe (Fig. 3B, lanes 1 and 4). In contrast, when either the wild-type RPO41 or rpo41Δ3 allele was provided in this strain on a plasmid, a single major radiolabeled fragment was protected from S1 nuclease digestion that was independent of the growth temperature of the strain (Fig. 3B, lanes 2, 3, 5 and 6). Even with twice as much input RNA analyzed in the rpo41Δ3, 37°C experiment (Fig. 3B, lane 6), to allow visualization of minor transcription initiation differences, the observed S1 digestion pattern was virtually identical to that observed in the wild-type control strains (Fig. 3B, lanes 2 and 5). The major S1-protected DNA fragment migrated with a mobility nearly identical to a single-stranded DNA marker band of 217 nt, consistent with the 5′ end of the transcripts mapping to the ori5 promoter (the expected S1-protected fragment is 218 nt in this case). This mapping assignment was confirmed by running the samples next to a DNA sequencing ladder generated by using a primer that matched the labeled 5′ end of the S1 probe (data not shown). Finally, pretreatment of the rpo41Δ3, 37°C sample with RNase A resulted in loss of the protected fragment (Fig. 3B, lane 7), demonstrating that the observed S1 protection pattern depends on RNA/DNA hybrids. We next compared the quantity and size distribution of ori5 transcripts generated in these strains by Northern analysis. When either a wild-type RPO41 or rpo41Δ3 allele was provided on a plasmid, one major RNA transcript (corresponding to the 966-bp ori5 repeat size) and two minor transcripts (corresponding to two and three ori5 repeat units) were observed in the ori5 hypersuppressive strain, even after extended growth at 37°C (Fig. 3C). These data, together with the results of our S1 nuclease protection experiments, clearly demonstrate that the rpo41Δ3-encoded mtRNA polymerase is capable of entering mitochondria and sustaining normal levels of accurate transcription in vivo at both the permissive and nonpermissive temperature. Our Northern analysis did reveal a potentially relevant difference between the wild-type and rpo41Δ3 strains. A minor, subrepeat-sized RNA species was observed consistently only in the wild-type control experiment (Fig. 3C), suggesting that there may be an RNA-related defect in the rpo41Δ3 strain. Given that our S1 nuclease protection analysis revealed primarily a single 5′ end distribution pattern in both the wild-type and rpo41Δ3 strains under these same conditions (Fig. 3B), we conclude that the absence of this RNA species in the mutant strain most likely results from an RNA stability and/or RNA processing difference between the two strains, and not altered transcription initiation capacity. Thus, the rpo41Δ3 mutation appears to uncouple the mtDNA maintenance function from the known transcription initiation and RNA polymerization activities of mtRNA polymerase.
The Amino-Terminal Extension of Yeast mtRNA Polymerase Harbors an Independent Functional Domain.
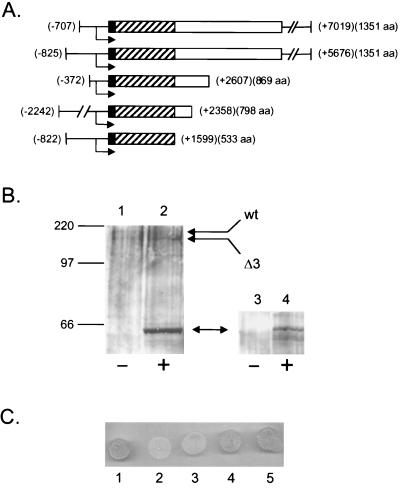
To gain additional insight into the nature of the defect caused by the rpo41Δ3 mutation, genes that were able to suppress the mitochondrial phenotype of the rpo41Δ3 mutation were sought. We transformed GS125 with a multicopy plasmid library of yeast genomic DNA and selected for growth on YPG at 37°C. We repeatedly recovered suppressors that contained at least part of the RPO41 gene (Fig. 4A). Two independent isolates contained the entire RPO41 reading frame as well as intact upstream and downstream sequences, consistent with the recessive nature of the rpo41Δ3 allele. However, four independent suppressors were isolated that represented three different truncated versions of the RPO41 locus (Fig. 4A). Each isolate harbored the RPO41 gene promoter, but contained only a portion of the RPO41 ORF. In fact, the smallest suppressing fragment isolated is predicted to encode only the first 533 aa of the RPO41 ORF. Remarkably, this peptide corresponds almost exactly to the amino-terminal extension of mtRNA polymerase that is missing in the bacteriophage RNA polymerase homologs, suggesting that this region of mtRNA polymerase can function as a separate domain in trans. However, the fact that all of the RPO41-containing suppressors also contained regions of homology both upstream and downstream of the deleted sequences in the rpo41Δ3 allele, it is possible that suppression is mediated indirectly by a recombination event between the suppressor plasmid and the rpo41Δ3 allele, resulting in the production of a wild-type RPO41 allele. To address this possibility, we tested the strain harboring the smallest RPO41-containing suppressor plasmid for the production of full-length mtRNA polymerase as well as the predicted amino-terminal peptide. We did not observe any full-length RPO41 gene product by Western analysis; however, a truncated mtRNA polymerase polypeptide of predicted molecular mass (≈57 kDa) that copurified with mitochondria was produced consistently in the suppressed strain (Fig. 4B). Finally, we constructed a plasmid (YEp-Δ3EXP) that expresses precisely the amino-terminal portion of mtRNA polymerase that is missing in the rpo41Δ3 allele, but does not contain downstream sequences in the RPO41 gene. This plasmid is not competent for generating a wild-type RPO41 allele by homologous recombination, but nonetheless conferred significant suppression of the temperature-sensitive phenotype of the rpo41Δ3 mutation (Fig. 4C). Altogether, the results of this genetic analysis indicate that the amino-terminal extension of mtRNA polymerase harbors an independent functional domain of the enzyme that is required for mtDNA stability and is capable of functioning to a significant degree in trans.
Figure 4.
Expression of an amino-terminal domain of mtRNA polymerase suppresses the rpo41Δ3 mutant phenotype. Portions of the RPO41 locus that conferred growth of GS125 on YPG at 37°C are diagrammed in A the same as in Fig. 1. All of the isolates contained the RPO41 promoter (bent arrow). The upstream endpoint is indicated by negative numbers in parentheses, indicating the distance upstream of the A of the AUG start codon for RPO41. The amount of genomic DNA downstream of the AUG start codon is given by the positive numbers in parentheses. The number in parentheses next to the positive values indicates the number of amino acids (aa) of mtRNA polymerase that is predicted to be expressed from each isolate. (B) A truncated mtRNA polymerase peptide is expressed in suppressor plasmid-containing strains. Whole-cell protein (lanes 1 and 2) and protein from a purified mitochondrial fraction (lanes 3 and 4) was isolated from yeast strains that either did (+) or did not (−) contain the suppressor plasmid harboring the smallest complementing fragment (the first 533 RPO41 codons) and subjected to Western analysis. GS129, which expresses both the RPO41 and rpo41Δ3 alleles, is shown in lanes 1 and 3. The rpo41Δ3 strain (GS125) harboring the suppressor plasmid is shown in lanes 2 and 4. The truncated mtRNA polymerase protein (≈57 kDa) expressed in the suppressor-containing strain is indicated by the double-headed arrow. The wild-type (wt) and rpo41Δ3-encoded (Δ3) mtRNA polymerase are indicated with arrows (note the absence of wild-type mtRNA polymerase in lane 2). Protein molecular mass standards are indicated on the left. (C) Expression of the precise mtRNA polymerase peptide missing in the rpo41Δ3 allele suppresses the temperature-sensitive petite phenotype in trans. Shown are the following strains grown on YPG medium at the nonpermissive temperature (36°C): 1, GS122(RPO41); 2, GS128(rpo41); 3, GS125(rpo41Δ3)+YEp352; 4, GS125(rpo41Δ3)+YEp-Δ3EXP; 5, GS125(rpo41Δ3)+ original suppressor plasmid (encoding amino acids 1–533 of mtRNA polymerase).
Potential Roles for the Amino-Terminal Domain of Yeast mtRNA Polymerase.
Based on these results, we conclude that the unique amino-terminal extension of yeast mtRNA polymerase constitutes a functional domain of the enzyme that has provided a simple bacteriophage-related RNA polymerase with additional capabilities. In addition to targeting the enzyme to mitochondria, we have shown that this domain is required for maintenance of a wild-type [rho+] mitochondrial genome and that this function appears to be separable from the documented transcription initiation and RNA polymerization activities of mtRNA polymerase. While the precise mtDNA maintenance function of mtRNA polymerase remains to be determined, several possibilities can be proposed based on the current state of knowledge. First, it is possible that the amino-terminal domain of mtRNA polymerase is necessary for the proper interaction with the transcription initiation factor sc-mtTFB. Deletions that remove either the amino or carboxyl terminus of mtRNA polymerase have been reported to disrupt the interaction with sc-mtTFB, yet neither of these regions is sufficient for the interaction (6). However, the fact that transcription initiation is not dramatically affected by the rpo41Δ3 mutation (which removes 185 amino-terminal residues)(Fig. 3B) and that higher than normal levels of sc-mtTFB do not rescue the rpo41Δ3 defect (Fig. 3A) is not consistent with this proposal. Nonetheless, it is possible that partial disruption of the sc-mtTFB/mtRNA polymerase interaction could inhibit a process other than transcription initiation, leading to the observed mtDNA instability. A second possibility is that disruption of the amino-terminal domain of mtRNA polymerase results in genome instability, because of increased aberrant recombination within and/or between mtDNA molecules. For example, the Mgt1p and sc-mtTFA proteins have been shown to affect mtDNA stability, presumably by altering or stabilizing the level of recombination intermediates (29, 30). It is possible that disruption of the mtDNA maintenance function of mtRNA polymerase promotes mtDNA instability by a similar mechanism. Although our results showing that overproduction of sc-mtTFA does not rescue the rpo41Δ3 defect could be taken as evidence against this possibility, this result must be interpreted with caution, because overproduction of sc-mtTFA alone causes temperature-sensitive mtDNA instability (31). Lastly, it is possible that [rho+] yeast mtDNA, like its vertebrate counterpart, is replicated by a transcription-primed mechanism and inactivation of the amino-terminal domain of mtRNA polymerase disrupts this process. In vertebrates, mtDNA replication is primed by transcription because RNA transcripts initiated at the light-strand promoter are processed to form primers for DNA synthesis at the heavy-strand origin (OH) of replication (10). Although the precise RNA processing events that lead to RNA primer formation have not been elucidated fully, evidence has accumulated that suggests a stable RNA-DNA hybrid formed at OH is a requisite step in this process (32, 33). This RNA-DNA hybrid is thought to serve as a substrate for RNA processing activities, such as RNase mitochondrial RNA processing (34, 35), that cleave the hybridized transcript to generate RNA primers (36). In yeast, putative origins of mtDNA replication (so-called ori or rep elements) (37, 38) are similar in nucleotide sequence and structure to vertebrate OH, including an upstream transcription promoter. These similarities, coupled with additional biochemical and genetic data, also have implicated transcription-derived RNA primers in initiation of yeast mtDNA replication (18, 23, 39–43). However, because certain deleted forms of mtDNA can be propagated in the absence of RPO41-dependent transcription (20, 28, 44), models for a transcription-dependent mtDNA replication mechanism in yeast have remained controversial. The identification of a functional domain of mtRNA polymerase that is required to propagate [rho+] mtDNA in yeast is the strongest evidence to date supporting a general model for transcription-dependent mtDNA replication for wild-type mtDNA in eukaryotes (10). If the amino-terminal domain of yeast mtRNA polymerase is required for transcription-primed mtDNA replication, our data would suggest that it is most likely needed at a step downstream of transcription initiation. Events required for mtDNA replication that might be coupled to transcription via the amino-terminal domain of mtRNA polymerase include the formation of a properly configured RNA/DNA hybrid or RNA transcript processing at origins of replication.
Whether the mtDNA maintenance activity identified in these studies resides in the amino-terminal domain itself or is provided by other factors that interact with the amino-terminal domain remains a critical question. In either case it appears that additional activities, other than those required for transcription initiation, are coupled to the transcription process in mitochondria via the mtRNA polymerase amino-terminal domain (e.g., mtRNA processing activities). Identification of the amino-terminal extension of mtRNA polymerase as an independent functional domain provides a focal point for future experiments aimed at understanding the precise function of this key regulatory enzyme in mitochondrial gene expression and mtDNA replication. Of particular importance will be to determine whether a similar functional domain exists within the divergent amino-terminal extension of human mtRNA polymerase (45) and its potential relevance to human diseases that result from defective maintenance and expression of the mitochondrial genome (46, 47).
Acknowledgments
We thank Dr. Heather E. Lorimer for providing the BS127 HS3324 ΔRPO41 yeast strain and pHS3324 plasmid harboring the cloned yeast ori5 repeat and Dr. Richard A. Kahn for providing the yeast high-copy genomic library. We also thank Drs. A. H. Corbett, P. W. Doetsch, J. D. Garman, S. M. Kaech, and S. T. Warren for critical reading of the manuscript. This research was supported by a National Institutes of Health grant from the National Heart, Lung and Blood Institute (HL-59655) awarded to G.S.S.
ABBREVIATIONS
- mtRNA
mitochondrial RNA
- YPG
yeast extract/peptone/glycerol
- YPD
yeast extract/peptone/dextrose
References
- 1.Masters B S, Stohl L L, Clayton D A. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 2.Bonner G, Patra D, Lafer E M, Sousa R. EMBO J. 1992;11:3767–3775. doi: 10.1002/j.1460-2075.1992.tb05462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner L P, Mookhtiar K A, Coleman J E. Biochemistry. 1997;36:2908–2918. doi: 10.1021/bi962397i. [DOI] [PubMed] [Google Scholar]
- 4.Gross L, Chen W, McAllister W T. J Mol Biol. 1992;228:488–505. doi: 10.1016/0022-2836(92)90837-a. [DOI] [PubMed] [Google Scholar]
- 5.Patra D, Lafer E M, Sousa R. J Mol Biol. 1992;224:307–318. doi: 10.1016/0022-2836(92)90996-w. [DOI] [PubMed] [Google Scholar]
- 6.Cliften P F, Park J, Davis B P, Jang S, Jaehning J A. Genes Dev. 1997;11:2897–2909. doi: 10.1101/gad.11.21.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Constanzo M C, Fox T D. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- 8.Ojala D, Montoya J, Attardi G. Nature (London) 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 9.Tzagoloff A, Myers A M. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- 10.Shadel G S, Clayton D A. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 11.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sikorski R S, Boeke J D. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 13.Shadel G S, Clayton D A. Mol Cell Biol. 1995;15:2101–2108. doi: 10.1128/mcb.15.4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yaffe M P, Schatz G. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaffe M P. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Schmitt M E, Brown T A, Trumpower B L. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenleaf A L, Kelly J L, Lehman I R. Proc Natl Acad Sci USA. 1986;83:3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dujon B. In: The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritence. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 505–637. [Google Scholar]
- 20.Fangman W L, Henly J W, Brewer B J. Mol Cell Biol. 1990;10:10–15. doi: 10.1128/mcb.10.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers A M, Papa L K, Tzagoloff A. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shadel G S, Clayton D A. J Biol Chem. 1993;268:16083–16086. [PubMed] [Google Scholar]
- 23.Lisowsky T, Michaelis G. Mol Gen Genet. 1988;214:218–223. doi: 10.1007/BF00337714. [DOI] [PubMed] [Google Scholar]
- 24.Jang S, Jaehning J A. J Biol Chem. 1991;266:22671–22677. [PubMed] [Google Scholar]
- 25.Xu B, Clayton D A. Nucleic Acids Res. 1992;20:1053–1059. doi: 10.1093/nar/20.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parisi M A, Xu B, Clayton D A. Mol Cell Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diffley J F X, Stillman B. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorimer H E, Brewer B J, Fangman W L. Mol Cell Biol. 1995;15:4803–4809. doi: 10.1128/mcb.15.9.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockshon D, Zweifel S G, Freeman-Cook L L, Lorimer H E, Brewer B J, Fangman W L. Cell. 1995;81:947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 30.MacAlpine D M, Perlman P S, Butow R A. Proc Natl Acad Sci USA. 1998;95:6739–6743. doi: 10.1073/pnas.95.12.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zelenaya-Troitskaya O, Newman S M, Okamoto K, Perlman P S, Butow R A. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee D Y, Clayton D A. J Biol Chem. 1996;271:24262–24269. doi: 10.1074/jbc.271.39.24262. [DOI] [PubMed] [Google Scholar]
- 33.Xu B, Clayton D A. EMBO J. 1996;15:3135–3143. [PMC free article] [PubMed] [Google Scholar]
- 34.Chang D D, Clayton D A. EMBO J. 1987;6:409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stohl L L, Clayton D A. Mol Cell Biol. 1992;12:2561–2569. doi: 10.1128/mcb.12.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee D Y, Clayton D A. Genes Dev. 1997;11:582–592. doi: 10.1101/gad.11.5.582. [DOI] [PubMed] [Google Scholar]
- 37.Blanc H, Dujon B. Proc Natl Acad Sci USA. 1980;77:3942–3946. doi: 10.1073/pnas.77.7.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Zamaroczy M, Marotta R, Faugeron-Fonty G, Goursot R, Mangin M, Baldacci G, Bernardi G. Nature (London) 1981;292:75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]
- 39.Baldacci G, Chérif-Zahar B, Bernardi G. EMBO J. 1984;3:2115–2120. doi: 10.1002/j.1460-2075.1984.tb02099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt M E, Clayton D A. Curr Opin Genet Dev. 1993;3:769–774. doi: 10.1016/s0959-437x(05)80097-8. [DOI] [PubMed] [Google Scholar]
- 41.Xu B, Clayton D A. Mol Cell Biol. 1995;15:580–589. doi: 10.1128/mcb.15.1.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graves T, Dante M, Eisenhour L, Christianson T W. Nucleic Acids Res. 1998;26:1309–1316. doi: 10.1093/nar/26.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Dyck E, Clayton D A. Mol Cell Biol. 1998;18:2976–2985. doi: 10.1128/mcb.18.5.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fangman W L, Henly J W, Churchill G, Brewer B J. Mol Cell Biol. 1989;9:1917–1921. doi: 10.1128/mcb.9.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiranti V, Savoia A, Forti F, D’Apolito M, Centra M, Rocchi M, Zeviani M. Hum Mol Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 46.Larsson N-G, Clayton D A. Annu Rev Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 47.Wallace D C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]