Abstract
Methylation of cytosine residues in CpG dinucleotides is generally associated with silencing of gene expression. DNA methylation, as a somatic event, has the potential of diversifying gene expression in individual cells of the same lineage. There is little quantitative data available concerning the extent of methylation heterogeneity in individual cells across the genome. T cells from the peripheral blood can be grown as single-cell-derived clones and can be analyzed with respect to their DNA methylation patterns by restriction landmark genomic scanning. The use of the methylation-sensitive enzyme NotI to cut and end-label DNA fragments before their separation in two dimensions provides a quantitative assessment of methylation at NotI sites that characteristically occur in CpG islands. We have undertaken quantitative analysis of two-dimensional DNA patterns to determine the extent of methylation heterogeneity at NotI sites between peripheral blood single-cell-derived T cell clones from the same individual. A total of 1,068 NotI-tagged fragments were analyzed. A subset of 156 fragments exhibited marked methylation heterogeneity at NotI sites between clones. Their average intensity among clones correlated with their intensity in uncultured, whole-blood-derived T cells, indicating that the methylation heterogeneity observed in clones was largely attributable to methylation heterogeneity between the individual cells from which the clones were derived. We have cloned one fragment that exhibited variable NotI-site methylation between clones. This fragment contained a novel CpG island for a gene that we mapped to chromosome 4. The methylation status of the NotI site of this fragment correlated with expression of the corresponding gene. Our data suggest extensive diversity in vivo in the methylation and expression profiles of individual T cells at multiple unrelated loci across the genome.
Programmed changes in DNA methylation occur during development and result in establishment of tissue-specific DNA methylation patterns (1). There is increasing evidence that ectopic changes in these methylation patterns may alter expression of affected genes (2). There is also evidence that altered genomic methylation may be mutagenic and may affect genomic stability (3–6). This is best exemplified in neoplastic cells, which exhibit striking ectopic changes, including hypomethylation of some protooncogenes and/or hypermethylation of tumor suppressor genes and which also frequently exhibit genomic instability and an excess of mutations (7). It has been proposed that heterogeneity within a cell population with respect to methylation and other genetic changes may provide some cells with a growth advantage, resulting in their clonal expansion (7, 8). Such clonal expansion is a key element of neoplasia. An important requirement for our understanding of the basis and significance of DNA methylation changes observed in clonal proliferative disorders is to define the extent and functional significance of methylation heterogeneity among normal cells that belong to the same lineage. Additionally, an assessment of the extent of methylation heterogeneity between individual cells is relevant to our understanding of the effect of endogenous and exogenous factors on diversity in gene expression programs of lineage-related cells.
We have implemented a computer-based approach for the analysis of two-dimensional (2-D) separations of human genomic restriction fragments (9). This approach, which is designated restriction landmark genomic scanning (10), relies on radioisotope labeling of genomic fragments at cleavage sites for a rare cutting restriction enzyme. The labeled genomic digests are separated in a first dimension followed by in situ digestion before second-dimension separation. The reliance on the methylation-sensitive enzyme NotI, which recognizes the sequence GCGGCCGC, to digest genomic DNA before labeling allows visualization of DNA fragments that occur preferentially in CpG islands in the genome. The intensity of these fragments in 2-D patterns depends on the methylation status of NotI cut sites. This approach has led to the identification of imprinted gene(s) in the mouse (11), the identification of loci on the X chromosome that escape methylation inactivation (12, 13), and the identification of novel CG-rich DNA repeats that undergo demethylation in cancer (14, 15). A high degree of quantitative reproducibility in 2-D DNA patterns has been demonstrated (9, 13), thus allowing an assessment of methylation differences among different cell populations derived from an individual.
Circulating peripheral blood lymphoid cells can be subgrouped based on their surface marker expression and their functional characteristics. T cells from the peripheral blood can be grown as single-cell-derived clones and can be analyzed with respect to their DNA methylation patterns. In this study, we have undertaken a quantitative analysis of the intensity of NotI derived fragments in 2-D patterns of single-cell-derived T cell clones from the same individual. These clones were functionally and immunophenotypically identical. 2-D patterns were analyzed to determine the extent of NotI sites methylation heterogeneity between clones and the relationship between methylation heterogeneity of individual NotI fragments and their intensity in 2-D patterns of polyclonal peripheral blood T cells.
MATERIALS AND METHODS
T Cell Cloning and Phenotyping.
Tetanus toxoid reactive T cell clones were derived from a healthy male donor, referred to as the primary subject. The clones were established by limiting dilution, expanded by weekly rechallenge with autologous irradiated peripheral blood mononuclear cells and antigen as described (16). The clones included in this study were all CD4+α+β+ and exhibited similar growth rates. Clones were harvested after a maximum of 3–4 months of culture, the minimum amount of time necessary for their expansion in sufficient number to permit their characterization. Where indicated, freshly isolated peripheral blood mononuclear cells from the primary subject were isolated by density gradient centrifugation, stimulated with phytohemagglutinin and expanded by culture in IL-2-containing media for 6–10 days as described (17, 18). T cells from the primary subject and from additional healthy male donors also were isolated by e-rosetting (19). IL-4 and IFN-γ secretion was measured by culturing clones in flat bottom microtiter wells coated with anti-CD3 (Coulter, Hialeah FL) for 18 hours, then quantitating cytokines by using commercially available kits. For methylation inhibition, cells were cultured with 5-aza-2′deoxycytidine (Sigma) for 6 days.
2-D Gel Electrophoresis.
High-molecular-weight DNA was extracted from unfractionated peripheral blood T cells and from T cell clones. DNA was subjected to 2-D electrophoresis, as described (20). In brief, 3 μg of DNA was used for digestion with NotI and EcoRV. The NotI-derived 5′ protruding ends were labeled with 32P-marked nucleotides (EcoRV yields blunt ends). The labeled fragments were directly applied onto first-dimension agarose gels before electrophoresis. Separated fragments were treated in situ with HinfI for further cleavage before second-dimension separation in acrylamide slab gels. The gels subsequently were dried, were exposed to PhosphorImager plates (Molecular Dynamics) for 3 days, and were scanned with a PhosphorImager at a resolution of 176 microns per pixel. For most samples, the DNA extraction and 2-D DNA electrophoresis were undertaken at least twice to ensure consistency.
Construction of NotI-EcoRV Genomic Fragment Library.
The genomic DNA used to construct the library was extracted from peripheral blood lymphocytes from the primary subject, was first digested with NotI, and was ligated to the NotI- and EcoRV-digested plasmid vector pBluescript SK(+) (Strategene) with T4 ligase (Boehringer Mannheim). After the first ligation and phenol/chloroform extraction, the DNA was digested with EcoRV and was purified with phenol/chloroform extraction-ethanol precipitation. The EcoRV ends were ligated. The circular DNA was introduced into Epicurian Coli, XL1-blue MRF′ cells (Stratagene) by electroporation according to the conditions recommended by the manufacturer.
Cloning of NotI Genomic DNA Fragments.
Fragments were cloned from a preparative genomic DNA library 2-D gel in which 1.1 μg of library DNA that was labeled at the NotI site was mixed with unlabeled library DNA and was subjected to 2-D electrophoresis. The resulting gel was exposed against x-ray film with an intensify screen for 3 days at −80°C. The location of some fragments that exhibited variable intensity between clones was determined in the library gel pattern. These fragments, designated M0442, M0584, M0967, and M0053, were extracted from the library 2-D gel and were ligated into modified pBluescript SK(+) vector sets, followed by electroporation into Epicurian Coli, XL1-blue MRF cells and transformed bacteria colony selection. Plasmids containing these DNA fragments were obtained by mini-preparation and were subjected to sequencing with an automated DNA sequencer at the University of Michigan Sequencing Core (Ann Arbor, MI).
RNA Samples and Reverse Transcription (RT)–PCR.
Total RNA was extracted from T cell clones by using Trizol Reagent (GIBCO/BRL). A portion (1.25 μg) of each mRNA was reverse transcribed into single-stranded DNA with Superscript II reverse transcriptase (GIBCO/BRL) and random primers according to the conditions recommended by the manufacturer. The product (2 μl) was added to 50 μl of reaction mixture that contained 5 units of Taq DNA polymerase (GIBCO/BRL), 50 pM of each (primer 1× buffer with 1.5 mM MgSO4), 12.5 nM of each dNTP, with the cDNA added last to the mixed ingredients. The amplification process consisted of 30 cycles of 61°C for 1 min, 72°C for 2 min, and 94°C for 1 min after starting with a denaturation step at 94°C for 4 min and ended at 72°C for 7 min. The PCR reactions were performed in the presence or absence of primers for β actin. M0442 primers were designed according to the matched expressed sequence tag sequence: forward primer, 5′AGAATCACTACAGCCCACGG; reverse primer, 5′CTGAGATAGGCCCTTCCCTC. β actin primers were forward, 5′GTGGGGCGCCCCAGGCACCA; reverse, 5′CTCCTTAATGTCACGCACGATTTC.
Quantitative Analysis.
2-D images were translated into 1,024 × 1,024 pixel formats, suitable for visage software (BioImage, Ann Arbor, MI), which was used to perform spot (fragment) detection and quantitation. The best gel for each clone was selected for careful quantitation and matching. For each image, spot boundaries were reviewed and edited, resulting in finalized spot-lists for each image. Spot-lists were matched to a master image, which initially was a copy of the image for one of the clones, as described (9, 13). Spots detected in study gels that were not detected in the master image had spot-list entries added to the master.
After matching, 1,068 fragments that were not overlapped and that did not appear to represent fragments present in more than two copies in the genome (such as ribosomal DNA) were selected on the master for analysis. In a preliminary analysis of the data, 382 reference fragments were chosen as being likely to represent DNA fragments present in two copies per genome whose NotI sites were unmethylated in all of the clones. The integrated intensities of individual fragments were adjusted for each fragment in the study by using it’s 12 nearest neighboring reference fragments. The integrated intensities of the 12 neighbors were trimmed by discarding the two highest and two lowest values, and the remaining 8 values were averaged by computing the antilogarithm of the mean of the logarithms of the integrated intensities. The study fragment’s integrated intensity was adjusted by dividing by this value.
RESULTS
Heterogeneity in the Methylation Patterns of Single-Cell-Derived T Cell Clones.
A 2-D pattern of peripheral blood T cells of a tetanus toxoid reactive T cell clone derived from a healthy adult man is shown in Fig. 1. A subset of five single-cell derived clones were selected among clones grown from peripheral blood T cells of this subject. These clones were selected on the basis of exhibiting similar growth characteristics. All were CD4+α+β+ and tetanus toxoid reactive. Because the extent of variation in methylation at a given site in the genome between cell populations may be marked or subtle, we wished to limit our analysis to the assessment of marked variation in intensity between clones for the selected group of 1,068 fragments. The criteria we defined for marked variation were met if the smallest integrated intensity of a fragment in a clone was <0.25 and the largest intensity among the clones was at least 0.30 larger than the smallest measure. A total of 156 of 1,068 analyzed exhibited marked variation based on the above criteria. Four examples of fragments that met the criteria are shown in Fig. 2. There were additional fragments that exhibited less striking differences in intensity between clones, for which no clone had a fragment with integrated intensity >0.30 and, similarly, for which no clone had fragments with integrated intensity <0.25. Thus, the set of 156 variable intensity fragments is only a subset of the fragments with clonal heterogeneity.
Figure 1.
2-D pattern of a tetanus toxoid reactive T cell clone from a healthy adult male. Three boxed areas that contained fragments that exhibited marked variation between clones are shown in Fig. 2 to illustrate clonal heterogeneity.
Figure 2.
Close-up sections (A, B, and C) of 2-D patterns of three T cell clones. Control polyclonal T cells from the same individual and of 5-aza-2′deoxycytidine treated polyclonal T cells are also from the same individual. Arrows point to some of the fragments that exhibited marked variation between clones. The arrows in C point to fragment 0442, which was cloned.
For 95 of the 156 fragments, the difference between the smallest and largest adjusted integrated intensity for the fragment was >0.50, and such differences are striking, as can be seen in Fig. 2. A total of 79 fragments were each found to be missing in at least one clone (Table 1). Of these, 47 never had intensities >0.7 whereas, for the remaining 32, there were clones with fragment intensities >0.7. Fragment intensities >0.7 would be expected in cases in which both copies of a fragment are unmethylated (9). To exclude the possibility that the heterogeneity could be attributable to Th1 and Th2 subsets, the two clones demonstrating the greatest number of differences for the set of 156 spots were tested for secretion of IFN-γ and IL-4. Both secreted IFN-γ but not IL-4, indicating that both were phenotypically Th1 cells.
Table 1.
Characteristics for variable intensity fragments that are absent in at least one clone
| Intensity of largest spot on clone | Number of spots | Average mean spot size for clones | Average size for spots on uncloned T cells |
|---|---|---|---|
| <0.3 | 0 | — | — |
| 0.3–0.4 | 11 | 0.14 | 0.19 |
| 0.4–0.5 | 19 | 0.20 | 0.16 |
| 0.5–0.6 | 7 | 0.22 | 0.17 |
| 0.6–0.7 | 10 | 0.28 | 0.32 |
| 0.7–0.8 | 9 | 0.28 | 0.32 |
| 0.8–0.9 | 6 | 0.30 | 0.20 |
| 0.9–1.0 | 4 | 0.34 | 0.44 |
| 1.0–1.1 | 7 | 0.41 | 0.40 |
| 1.1–1.2 | 3 | 0.50 | 0.49 |
| >1.2 | 3 | 0.54 | 0.71 |
| Total | 79 | 0.27 | 0.28 |
*The size of the largest adjusted integrated intensity measurement for a spot among the 5 clones.
The Mean Fragment Intensity for Clones Is Correlated with Fragment Intensity in Peripheral Blood T Cells.
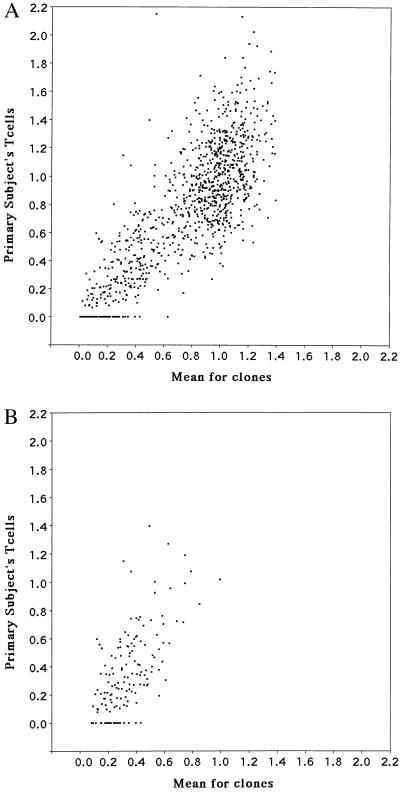
Plots of the mean fragment integrated intensity for the clones vs. fragment integrated intensity for polyclonal peripheral blood T cells obtained from the same individual are shown in Fig. 3 for all fragments as well as for the subset of 156 clearly variable fragments. The plots show the mean for the clones to be generally highly predictive of the fragment intensity in peripheral blood T cells (r = 0.81, P < 0.0001) and, in particular, that the reduced intensity of some of the fragments in peripheral blood T cells is predicted by the average fragment intensity for the clones (r = 0.64, P < 0.0001). The methylation intensity variability for the subset of 156 variable fragments was not particularly attributable to any single clone. For these fragments, each clone had the most reduced intensity for a fragment, between 27 and 53 times, and had the highest fragment intensity, between 23 and 39 times. It can be inferred from the data that the heterogeneity in methylation at a given site(s) among individual cells results in a spectrum of methylation levels at different sites for the whole population. Also, for the subset of 156 variable fragments, the methylation status of their NotI sites in the progeny of individual cells in a population does not undergo a rapid change to reach a methylation level equivalent to the level observed in uncloned peripheral blood T cells.
Figure 3.
Plot of mean intensity distribution of fragments in clones vs. peripheral blood T cells from the same individual for all fragments (A) and for the subset of 156 variable fragments (B).
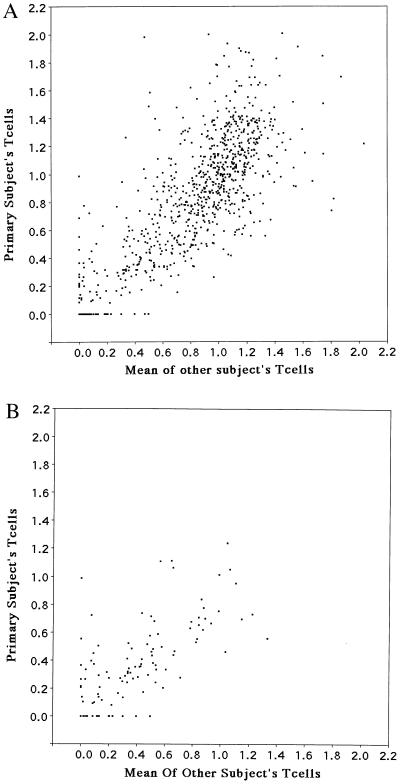
We wished to determine whether the intensity of fragments in the peripheral blood T cell 2-D patterns of the individual from whom the T cell clones were derived correlated with their intensity in other individuals’ peripheral blood T cell 2-D patterns. A total of 121 fragments were excluded from this analysis because they were previously found to be genetically polymorphic in family studies of 2-D restriction fragment patterns (9, 13). For the remainder of the fragments included in this analysis, there was a high degree of correlation of fragment intensity between individuals (r = 0.814, P < 0.0001, Fig. 4). A plot also is shown for the subset of variable spots that also showed a strong correlation (Fig. 4).
Figure 4.
Plot of intensity distribution of fragments in peripheral blood T cells of the individual from whom T cell clones were derived vs. mean intensity distribution of fragments from peripheral blood T cells of four adult males for all fragments (A) and for the subset of variable fragments (B).
Effect of 5-aza-2′deoxycytidine Treatment on NotI Fragment Intensity.
To further rule out that fragments that exhibited variable intensity between clones represent a subset of fragments that undergo labile methylation at their NotI sites, we examined the effect of treatment with the methylation inhibitor 5-aza-2′deoxycytidine on fragment intensity. In particular, we examined whether mitogenic stimulation of peripheral blood T cells in the presence or absence of 5-aza-2′deoxycytidine changed the methylation status of variable fragments. Peripheral blood T cells from the individual from whom the T cell clones were established were used for 5-aza-2′deoxycytidine treatment. Mitogenic stimulation and culture of peripheral blood T cells for 8 days in the absence of 5-aza-2′deoxycytidine had no effect on fragment intensity compared with freshly isolated peripheral blood T cells. Treatment with 5-aza-2′deoxycytidine of phytohemagglutinin-stimulated cells for 6 days at concentrations of 0.5 and 1.0 μM had a minimal effect on the set of 156 variable fragments. All 25 fragments in this set that were detected on some of the clones but that were missing in the polyclonal T cell pattern remained undetectable after 5-aza-2′deoxycytidine treatment of the peripheral blood T cell population. Only 12 of the 131 fragments in this set that were detectable in the peripheral blood T cell patterns in the absence of 5-aza-2′deoxycytidine exhibited an increase (up to a 2-fold) in their intensity after 5-aza-2′deoxycytidine treatment. Similar results were obtained for one of the clones (clone 44G) after treatment with 5-aza-2′deoxycitidine (0.5 and 1.0 μM). It should be pointed out that 5-aza-2′deoxycytidine treatment of both the clone and the polyclonal T cell population resulted in the occurrence, in 2-D patterns, of several repetitive DNA units which had previously been observed in 2-D patterns of a variety of tumors whose occurrence in 2-D patterns is attributable to demethylation at their NotI sites (Fig. 5) (14, 15).
Figure 5.
Close-up sections of 2-D patterns of polyclonal T cells and a T cell clone. Patterns are shown for untreated (control) and 5-aza-2′deoxycytidine treated cells. Arrows point to the location of demethylated repetitive DNA.
Cloning of Fragments that Varied in Intensity Between T Cell Clones.
Four NotI/HinfI DNA fragments that varied in intensity between T cell clones were directly cloned from a preparative 2-D gel of a NotI-EcoRV genomic library that we constructed from peripheral blood lymphocytes of a healthy man. The cloned inserts corresponded in size to the size of the fragments observed in 2-D gels. All four cloned genomic DNA fragments were part of CpG islands (Table 2). Three cloned fragments yielded novel sequences that had no match in sequence databases searched with blast-nr. The clone M0442 is a 794-bp NotI-HinfI genomic DNA fragment, of which the 20-bp sequence from the NotI end is CG rich and contains 27 CpG pairs. There is an ≈242-bp Alu repetitive sequence near the HinfI end. Clone M0442, beginning 520 bp from the NotI end, had a 220-bp sequence identity with a Homo sapiens DNA sequence contained in a NotI linking clone (accession no. Z22241), and 414 bp from the NotI end matched the entire sequence of two overlapping human expressed sequence tags (accession nos. M78232 and AA845651). We inferred that the genomic fragment of M0442 is part of a gene and contained at least one exon. We have undertaken radiation hybrid mapping of this fragment, which resulted in its mapping in the vicinity of marker SHGC-51332 on chromosome 4.
Table 2.
Characteristics of four cloned fragments
| Clone name | Clone length | C+G rich region | C+G content | CpG pairs |
|---|---|---|---|---|
| M0442 | 794 bp | 522 bp | 61.1% | 27 |
| M0584 | 826 bp | 826 bp | 64.3% | 94 |
| M0967 | 718 bp | 718 bp | 71.6% | 63 |
| M0053 | 1,300 bp | 553 bp | 67.3% | 51 |
Quantitative RT-PCR was undertaken to determine whether the variable intensity of the M0442 fragment in 2-D patterns of clones correlated with expression of the corresponding gene. Three T cell clones, 18I, 19F, and 48E, were analyzed for M0442 expression. The M0442 RT-PCR results of these clones were normalized with control RT-PCR for β-actin performed simultaneously. Fragment intensity and PCR results for these clones are shown in Table 3. M0442 expression differences between the clones were well correlated with M0442 fragment intensity differences between the clones. We infer that the NotI site methylation of M0442 correlated with down-regulation of M0442 gene expression. This finding was further substantiated by a demonstration of a 10-fold increase in M0442 expression in T cell clone 48e after treatment with 5-aza-2′deoxycytidine.
Table 3.
Fragment 0442 intensity and expression as measured by RT-PCR
| T cell clones | 18I | 19f | 48e |
|---|---|---|---|
| 2-D spot intensity | 0.39 | 1.00 | 0.06 |
| RT-PCR | 0.49*/0.47† | 1.00*/1.00† | 0.14*/0.09† |
Values for RT-PCR obtained with M0442 and β-actin amplified in separate reactions, but simultaneously.
Values for RT-PCR obtained with M0442 and β-actin amplified in same reactions.
DISCUSSION
The intensity of most of the 1,068 NotI fragments included in our analysis of 2-D patterns did not vary between clones, indicating similar levels of methylation at most NotI sites. However, we have uncovered 156 fragments in the set of 1,068 fragments we have analyzed that exhibited marked heterogeneity in their intensity between clones, reflecting their differential methylation. On average, these 156 fragments occurred at reduced intensity in the polyclonal T cell population from which the clones were derived as well as in polyclonal T cells from other individuals compared with other fragments in the study. We and others have previously identified the chromosomal origin of most fragments observed in genomic NotI-tagged 2-D patterns of polyclonal cells based on the analysis of 2-D patterns of individual chromosomes obtained by chromosome flow cytometry of a lymphoid cell line (21, 22). The set of 156 variable fragments we have uncovered included 92 that were detectable in the whole genome 2-D pattern of the lymphoid cell line used for chromosome flow cytometry. Overall, there was no evidence for a preponderance of variable fragments being derived from any particular chromosome. As an example, 8 of the subset of 92 variable fragments were attributable to chromosome 1 (21).
The methylation heterogeneity between clones is not simply attributable to stochastic processes during in vitro growth. The individual cells from which the clones were grown must have been cleavable at both, one, or neither of the two NotI sites, in the case of fragments that occur in two copies in the genome. During expansion of the clone, the methylation profile of the daughter cells may deviate from the initial status of 0, one or two copies methylated, by failure to copy methylation patterns on newly synthesized DNA strands or by de novo methylations or demethylations. In cases in which these stochastic processes quickly reach an equilibrium, we would expect to observe fragments of approximately equal intensity in all of the clones. Such fragments would not have been singled out as variable. However, if the cell population from which clones were established was heterogeneous with respect to methylation and if equilibrium is reached only slowly, there would still be detectable differences between the clones after they have been grown to sufficient population sizes to permit assay on our gels. The occurrence of fragments with marked differences in their integrated intensities between clones indicates that an equilibrium that reflects their intensity in the polyclonal population was not reached for these fragments in individual clones. The average intensity in the clones for the set of fragments we uncovered that exhibited NotI site methylation variability between clones correlated with their intensity in the polyclonal T cell population from which they were derived. Therefore, it seems most likely that intensity differences between clones for this subset of fragments largely reflect methylation differences between the initial single cells from which the clones were derived and that these methylation differences were maintained to some degree in culture during subsequent cell divisions. The mechanism causing clonal heterogeneity in this subset of fragments is uncertain. It is possible that heterogeneity developed slowly over time. Alternatively, heterogeneity may have occurred at the time tissue methylation patterns were established with methylation faithfully replicated subsequently.
Although there is currently little data concerning the extent of clonal variation in DNA methylation at multiple unrelated sites across the genome for cells that are related by lineage, several studies have addressed the issue of clonal variation in the methylation status of specific genes or DNA sequences. In early studies by Shmookler Reis and Goldstein (22), a mass culture of human diploid fibroblasts and eight clones isolated from the mass culture were examined for the methylation patterns of several genes. They found a striking degree of interclonal heterogeneity, with each gene exhibiting a clone-specific pattern of DNA methylation. More recently, Fitzpatrick et al. (23) analyzed IFN-γ and IL-3 gene methylation and expression in primary CD8+ T cells. They observed substantial heterogeneity in the methylation patterns of the two genes between clones. The methylation profile of either gene correlated with gene expression, leading to the conclusion that differential gene methylation among individual T cells is an important feature of the T cell response (23). Our data extend the findings of clonal methylation heterogeneity to NotI sites across the genome in human T cells and provide evidence for the occurrence of methylation heterogeneity before in vitro cell growth.
It should be pointed out that not all fragments with reduced intensity in the clones exhibited methylation heterogeneity at their NotI sites. Some 130 fragments, other than the set of 156 with methylation heterogeneity, had reduced intensities of 0.5 or less, well below expectations for fragments present in two copies in the genome. This subset likely includes fragments derived from the X chromosome, imprinted loci as well as fragments for which partial NotI site methylation is stably maintained through methylation/demethylation. Evidence for the occurrence of sites for which partial methylation is stably maintained in a cell population stems in part from studies by Turker et al. (24). They identified a CpG site with a partial methylation profile located upstream of the mouse adenine phosphoribosyltransferase promoter region and demonstrated that partial methylation of this site could be stably maintained both in vitro as well as in vivo, presumably because of a high rate of methylation/demethylation that maintained stability of partial methylation (24).
NotI sites occur primarily in CpG islands, which are widely distributed across the genome. There is substantial evidence that links NotI sites to CpG islands and genes (25). In a study of 143 genomic clones containing sequences surrounding NotI sites, a majority of clones were found to contain transcribed sequences (26). All four cloned NotI fragments we have analyzed contained a CpG island, and one of the fragments contained a sequence that matched several expressed sequence tags in databases. Methylation at the NotI site of this fragment correlated with expression of the corresponding gene in the cloned T cells. This finding does not establish that methylation at NotI sites necessarily affects expression. However, it is likely that methylation at NotI sites reflects overall methylation of surrounding sequences, which may often be related to gene expression. Transcriptional repression has been shown to depend on CpG methylation density (27).
Our findings indicate that, under normal physiological states, individual cells in a cell population such as peripheral blood T cells exhibit substantial genomic methylation differences. Although these cells may have inherited essentially the same genome from a nucleotide sequence point of view, operationally they may differ considerably because of epigenetic modifications. Such modifications, by affecting the spectrum of genes expressed in a cell, have functional consequences because they also affect the RNA and protein profiles of the cell and, as a result, affect the ability of an individual cell to respond to exogenous factors. We propose the term “operome” to capture the unique make-up of an individual cell at the genome, transcriptome, and proteome levels. Connections between these different levels are not unidirectional. Diverse external stimuli that induce changes in the proteome may impact on the DNA methylation machinery and the DNA methylation profile of a cell. For example, a number of signaling molecules have been implicated in the regulation of 5-methylcytosine DNA transferase levels. Signaling through the ras pathway has been found to result in an increase in DNA demethylation activity (28) and in the induction of 5-methylcytosine DNA transferase (29). 5-methylcytosine DNA transferase is also one of the targets of the protooncogene that functions in a transcription factor network that responds to extracellular stimuli (30). Other enzymes that may affect the methylation status of a cell also may be affected by external stimuli. A mammalian protein with specific demethylase activity for mCpG DNA has recently been identified (28). Some methylation changes, such as those affecting the subset of NotI sites we have detected, may be stably inherited by daughter cells, and others may be more rapidly reversible, all contributing to the unique methylation profile of individual cells. Although extensive epigenetic diversity between cells may have some benefit, it also may contribute to pathologic states.
The identities of the genes associated with the set of fragments that exhibited methylation heterogeneity is currently unknown. One of the long-standing paradoxes in genetic medicine and evolution is that, for many critical enzymes, satisfactory function is possible at a fraction of the normal level. Heterozygotes for biochemical disorders usually perform normally with half the usual amount of the enzyme. Furthermore, many systems, such as Factors VIII, IX, and XI of the coagulation cascade, seem to perform adequately on as little as 25% of the usual enzyme level. A particularly extreme example is provided by the essential enzyme adenosine deaminase, for which reductions to as little as 2–5% of normal adenosine deaminase activity (31) may be tolerated. This seems a rather wide margin of safety that is not without cost to the cell. Given, now, that methylation levels of specific DNA sequences may vary, it may be speculated that one function of this margin is to provide a buffer against unprogrammed methylation.
Acknowledgments
Supported by Public Health Service Grants CA26803, AG014783, AR42525, AI42753, and AR08309 and a grant from the Office of Research and Development, Department of Veterans Affairs.
ABBREVIATIONS
- 2-D
two-dimensional
- RT
reverse transcription
Footnotes
To whom reprint requests should be addressed at: University of Michigan Medical Center, 1150 West Medical Center Drive, MSRB I, Room A520C, Ann Arbor, MI 48109-0656. e-mail: shanash@umich.edu.
References
- 1.Bestor T H, Tycko B. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 2.Razin A. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji W, Hernandez R, Zhang X-Y, Qu G, Frady A, Varela M, Ehrlich M. Mutat. Res. 1997. 33–41. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez R, Frady A, Zhang X-Y, Varela M, Ehrlich M. Cytogenet. Cell Genet. 1997. 196–201. [DOI] [PubMed] [Google Scholar]
- 5.Chen R Z, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. Nature (London) 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 6.Jones P A, Gonzalgo M L. Proc Natl Acad Sci USA. 1997;94:2103–2105. doi: 10.1073/pnas.94.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 8.Gonzalgo M L, Jones P A. Mutat Res. 1997;386:107–118. doi: 10.1016/s1383-5742(96)00047-6. [DOI] [PubMed] [Google Scholar]
- 9.Asakawa J, Kuick R, Neel J V, Kodaira M, Satoh C, Hanash S M. Proc Natl Acad Sci USA. 1994;91:9052–9056. doi: 10.1073/pnas.91.19.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatada I, Hayashizaki Y, Hirotsune S, Komatsubara H. Proc Natl Acad Sci USA. 1991;88:9523–9527. doi: 10.1073/pnas.88.21.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, et al. Nat Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa H, Fujiyama A, Nakai K, Inazawa J, Matsubara K. Genomics. 1998;49:237–246. doi: 10.1006/geno.1998.5246. [DOI] [PubMed] [Google Scholar]
- 13.Kuick R, Asakawa J, Neel J V, Satoh C, Hanash S M. Genomics. 1995;25:345–353. doi: 10.1016/0888-7543(95)80032-h. [DOI] [PubMed] [Google Scholar]
- 14.Thoraval D, Asakawa J, Kodaira M, Chang C, Radany E, Kuick R, Lamb B, Richardson B, Neel J V, Glover T, et al. Proc Natl Acad Sci USA. 1996;93:4442–4447. doi: 10.1073/pnas.93.9.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thoraval D, Asakawa J, Wimmer K, Kuick R, Lamb B, Richardson B, Ambros P, Glover T, Hanash S. Genes Chromosomes Cancer. 1996;17:234–244. doi: 10.1002/(SICI)1098-2264(199612)17:4<234::AID-GCC5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. J Immunol. 1988;140:2197–2200. [PubMed] [Google Scholar]
- 17.Richardson B, Liebling M, Hudson J. Clin Immunol Immunopathol. 1990;55:368–381. doi: 10.1016/0090-1229(90)90125-a. [DOI] [PubMed] [Google Scholar]
- 18.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 19.Golbus J, Salata M, Greenwood J, Hudson J, Richardson B C. Clin Immunol Immunopathol. 1988;46:129–140. doi: 10.1016/0090-1229(88)90013-x. [DOI] [PubMed] [Google Scholar]
- 20.Wimmer K, Thoraval D, Kuick R, Lamb B J, Hanash S M. Biochem Soc Trans. 1997;25:262–267. doi: 10.1042/bst0250262. [DOI] [PubMed] [Google Scholar]
- 21.Wimmer K, Thoraval D, Asakawa J, Kuick R, Kodaira M, Lamb B, Fawcett J, Glover T, Cram S, Hanash S. Genomics. 1996;38:124–132. doi: 10.1006/geno.1996.0607. [DOI] [PubMed] [Google Scholar]
- 22.Shmookler Reis R J, Goldstein S. Nucleic Acids Res. 1982;10:4293–4300. doi: 10.1093/nar/10.14.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzpatrick D R, Shirley K M, McDonald L E, Bielefeldt-Ohmann H, Graham F K, Kelso A. J Exp Med. 1998;188:103–118. doi: 10.1084/jem.188.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turker M S, Swisshelm K, Smith A C, Martin G M. J Biol Chem. 1989;264:11632–11636. [PubMed] [Google Scholar]
- 25.Larsen F, Gundersen G, Prydz H. Genet Anal Tech Appl. 1992;9:80–85. doi: 10.1016/1050-3862(92)90002-m. [DOI] [PubMed] [Google Scholar]
- 26.Allikmets R L, Kashuba V I, Pettersson B, Gizatullin R, Lebedeva T, Kholodnyuk I D, Banikov V M, Petrov N, Zakharyev V M, Winberg G. Genomics. 1994;2:303–309. doi: 10.1006/geno.1994.1062. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh C-L. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya S, Ramchandani S, Cervoni N, Mosheszyf N. Nature (London) 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Deng C, Hemati N, Hanash S M, Richardson B C. J Immunol. 1997;159:1303–1309. [PubMed] [Google Scholar]
- 30.Bakin A V, Curran T. Science. 1999;283:387–390. doi: 10.1126/science.283.5400.387. [DOI] [PubMed] [Google Scholar]
- 31.Kredich N M, Hirschfield M J. In: The Metabolic Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1989. p. 1064. [Google Scholar]