Abstract
We have previously described biological model systems for studying tumor suppression in which, by using H-1 parvovirus as a selective agent, cells with a strongly suppressed malignant phenotype (KS or US) were derived from malignant cell lines (K562 or U937). By using cDNA display on the K562/KS cells, 15 cDNAs were now isolated, corresponding to genes differentially regulated in tumor suppression. Of these, TSAP9 corresponds to a TCP-1 chaperonin, TSAP13 to a regulatory proteasome subunit, and TSAP21 to syntaxin 11, a vesicular trafficking molecule. The 15 cDNAs were used as a molecular fingerprint in different tumor-suppression models. We found that a similar pattern of differential regulation is shared by activation of p53, p21Waf1, and the human homologue of Drosophila seven in absentia, SIAH-1. Because SIAH-1 is differentially expressed in the various models, we characterized it at the protein and functional levels. The 32-kDa, mainly nuclear protein encoded by SIAH-1, can induce apoptosis and promote tumor suppression. These results suggest the existence of a common mechanism of tumor suppression and apoptosis shared by p53, p21Waf1, and SIAH-1 and involving regulation of the cellular machinery responsible for protein folding, unfolding, and trafficking.
Spontaneous reversion of malignant cells to a more normal phenotype is an extremely interesting process that may provide important clues toward the elucidation of tumor suppression mechanisms. However, even if reversion occurs with reasonable frequency in occasional cancer cells, such cells are very hard to identify. This problem can be overcome through the use of the H-1 parvovirus, which kills preferentially tumor cells while sparing their normal counterparts (1–3). Alternatively, it remains possible that besides killing the malignant cells, the H-1 parvovirus by itself also induces a suppression of the malignant phenotype. The method of selecting daughter cells with a suppressed malignant phenotype of a parental population of tumor cells enables the use of comparative molecular approaches. Using this method, we have previously described (4) the KS cells derived from the K562 human erythroleukemia cell line. KS cells have a suppressed transformed phenotype; unlike parental K562, they reexpress wild-type p53 (4). In parallel, we used the same technology for the human monocytic U937 cell line to generate the US clones, which express constitutively elevated p21Waf1 (5). Like KS, US cells have a strongly suppressed malignant phenotype, and some of them completely fail to form tumors when injected into scid/scid mice (5). Using cDNA display on the K562-KS system, we now describe the isolation of 15 differentially expressed cDNA clones. Many of these clones are also differentially expressed in other tumor-suppression models. The identity of some of these genes suggests a link between tumor suppression and protein folding, degradation, and trafficking.
We previously identified a series of genes differentially regulated after wild-type p53 induction in the M1-LTR6 cells (6–9). Among them (6), the vertebrate (siah-1b) homologue (10, 11) of the Drosophila seven in absentia (sina) (12) was selectively up-regulated during the first hours of apoptosis induction by wild-type p53. In Drosophila, sina (12) codes for a metal-binding nuclear protein downstream of the sevenless receptor. It plays a major role in the specification of the R7 photoreceptor cells during Drosophila eye development (12). It has been demonstrated that sina binds PHYL and TTK 88, which is a transcriptional repressor of neuronal cell fate (13, 14). By interacting with the ubiquitin–proteasome pathway, sina regulates degradation of TTK 88 (13, 14). The human homologue of sina, SIAH-1, was identified as a p53-p21Waf1 inducible gene activated in its expression during physiological apoptosis and various model systems of tumor suppression (5). SIAH-1 binds BAG-1, and this binding is responsible for the inhibition of the growth arrest effect of p53 (15). BAG-1 targets Hsp70–Hsc70 to SIAH-1, inducing conformational changes that directly or indirectly abrogate its growth-regulatory function (16). SIAH also binds DCC (deleted in colorectal cancer) and regulates its degradation via the ubiquitin–proteasome pathway (17). SIAH’s N-terminal RING-finger domain is required for proteolysis, whereas the C-terminal cystein-rich region is required for the binding to target proteins (18).
In the present study we demonstrate that SIAH-1 has common downstream effectors with p53 and p21Waf1 and that it can induce apoptosis and tumor suppression.
METHODS
Antibodies and Western Blot Analysis.
Polyclonal antibodies against the first 16 aa of SIAH-1 were generated in chickens and affinity-purified. Protein extraction and sample processing for detection with the anti-SIAH-1 polyclonal antibody was performed by using standard conditions. Signals were detected by using a secondary antibody coupled to peroxidase. Subcellular fractionation was performed by using differential centrifugation as described (19).
Cells and Transfectants.
K562/KS, (4) U937/US (5), and the p21Waf1 transfectants of U937 cells (20) were previously described. Transfection of U937 cells with human SIAH-1 was performed by using Lipofectin. The cDNA corresponding to the coding region was subcloned in pBK-RSV (Stratagene), and the transfection was followed by selection with 1.5 mg/ml of G418 (Sigma) for 3 weeks.
Flow Cytometry.
For both the propidium iodide DNA content profile and terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) assay, cells were processed as described before (8).
Tumorigenicity Analysis.
Injection into scid/scid mice was done as described (13). Cells (107) were injected per site (20 sites). The Mann–Whitney U test was used for the statistical analysis.
Differential cDNA Display and Northern Blot Analysis.
cDNA display was performed on mRNA derived from K562 and KS cells. The modifications of the original protocol were previously described, and the same combination of primers was used (6). Northern blot analysis was performed by using 2 μg of mRNA. TSAP9–22 and TSIP3 were used as random primed 32P-labeled probes.
Rapid Amplification of cDNA Ends–PCR.
The cDNA clones isolated by differential display of mRNA were expanded by rapid amplification of cDNA ends–PCR using the Marathon cDNA amplification kit (CLONTECH), following the manufacturer’s instructions. The amplified products were cloned by using the TA cloning system (Invitrogen). TSAP13 was extended by using the primers 5′-CTCGGAGTACACAGACTTCAG-3′, derived from the sequence of the differentially expressed cDNA fragment and AP1. TSAP19 was extended by using the following primers: 5′-GCAACATAGAGAATCCGTCTCA-3′ derived from the sequence of the differentially expressed cDNA fragment and AP1. TSAP21 was extended by using two rounds of amplification. The first round used the following primers: 5′-AATTGCATAGCGTAGACCGGATG-3′, derived from the sequence of the differentially expressed cDNA fragment and AP1. The second round used the following anchored primers: 5′-GAGTTCATTCTATTAGCAGATGC-3′ and AP2.
RESULTS
Identification of 15 cDNAs for the Molecular Fingerprinting of Tumor Suppression and Apoptosis.
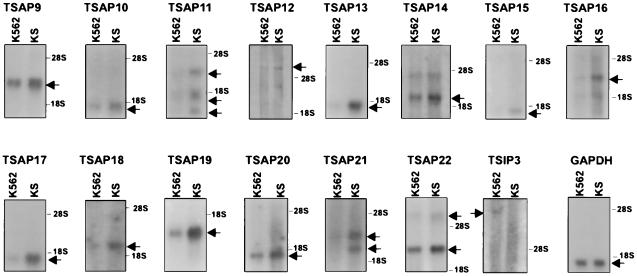
Analysis of differential gene expression in tumor-suppressed, p53-expressing K562 revertants led to the isolation of 15 cDNAs, of which 14 are activated (TSAP, Tumor Suppressor-Activated Pathway) and one inhibited (TSIP, Tumor Suppressor-Inhibited Pathway) (Fig. 1; Table1). Of these, TSAP9 corresponds to a TCP-1 chaperonin (21) and TSAP13 to the regulatory proteasome subunit p40.5/Nas7p (22); TSAP21 is an N-ethylmaleimide-sensitive factor-attachment protein receptor (SNARE) family member, syntaxin 11 (23). The homologies with sequences in the database were only present in the cDNA display fragment corresponding to TSAP9, whereas for TSAP13 and TSAP21, they were obtained by extending the sequence of the cDNA display fragment by using rapid amplification of cDNA ends–PCR (Table1). All of the other cDNAs represent unknown genes, nine of which correspond to expressed sequence tags. Using these 15 cDNAs as probes, we investigated by Northern blot analysis whether they are also differentially expressed in the U937/US model system (5) and in p21Waf1 transfectants (20) and SIAH-1 transfectants of U937 cells (Table 1). It is important to note that all four model systems have in common a suppression of the malignant phenotype and/or activation of programmed cell death. Table 1 indicates that p53 activation as seen in the K562/KS cells, or alternatively p21Waf1 activation as seen in the U937/US cells, both correlate with the triggering of a markedly overlapping set of genes. Strikingly, transfectants of U937 cells with either p21Waf1 or SIAH-1 show a similar gene-expression fingerprint (Table 1).
Figure 1.
Differential expression of various mRNA species in K562 and KS cells. Northern blot analysis demonstrates activation of TSAP9–TSAP 22 and inhibition of TSIP 3 in the KS cells. Glyceraldehyde-3-phosphate dehydrogenase is used as a control for equal loading.
Table 1.
Characteristics of differentially expressed cDNA clones
| cDNA clones | mRNA kb | Homology | K562 /KS | U937 /US | U937 p21 | U937 SIAH |
|---|---|---|---|---|---|---|
| SIAH-1 | D | D | D | D | ||
| TSAP9 | 2.6 | Chaperonin | D | D | D | D |
| TSAP10 | 1.6 | EST | D | D | D | D |
| TSAP11 | 2.8 | EST | D | N | N | N |
| TSAP12 | 5.5 | EST | D | N | N | N |
| TSAP13 | 1.8 | Proteasome | D | D | D | D |
| TSAP14 | 2.5 | EST | D | D | D | D |
| TSAP15 | 1.6 | EST | D | D | D | D |
| TSAP16 | 2.5 | No | D | D | N | D |
| TSAP17 | 1.8 | No | D | D | D | N |
| TSAP18 | 2.0 | EST | D | N | N | D |
| TSAP19 | 1.5 | EST | D | D | N | N |
| TSAP20 | 1.7 | No | D | D | D | N |
| TSAP21 | 2.1 | SNARE | D | D | D | D |
| TSAP22 | 2.6 | EST | D | D | D | D |
| TSIP3 | 9.5 | EST | D | D | D | D |
D, differential expression by Northern analysis; N, no differential expression by Northern analysis; EST, expressed sequence tag. Homologies: SIAH-1 from ref. 5; chaperonin, chaperonin containing t complex polypeptide 1 from ref. 21; Proteasome, subunit p40.5 (Nas 7p) from ref. 22; SNARE syntaxin 11 from ref. 23.
SIAH-1 Induces Apoptosis and Tumor Suppression.
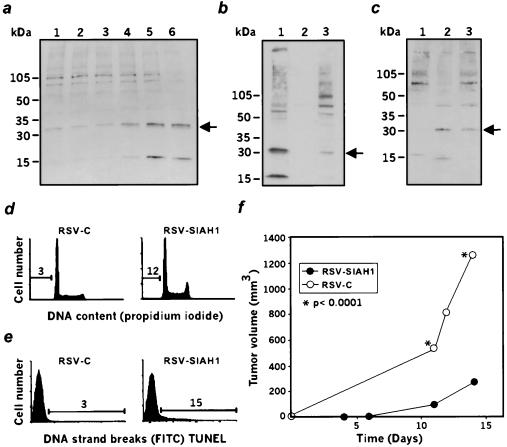
We raised polyclonal antibodies against the 16 first residues of SIAH-1 (5) and looked at the protein expression in M1-LTR6 cells (9). Shifting the LTR6 cells to 32°C induces wild-type p53 function (9). We previously reported a strong induction of siah 1b mRNA after p53 activation (6). Fig. 2a shows that a 32-kDa polypeptide, weakly expressed in LTR-6 cells at 37°C, is induced on wild-type p53 activation at 32°C. Subcellular fractionation reveals that the majority of the protein is found in the nuclear fraction, with a small percentage detected in the cytoplasmic fraction (Fig. 2b). This is consistent with the previous nuclear localization of the Drosophila seven in absentia (12) and of transfected SIAH-1(15). We next investigated the expression of SIAH-1 protein in the U937/US system and in U937 transfectants. This 32-kDa band is undetectable in U937 cells transfected with control vector but is highly expressed in the suppressed US revertants of U937 (Fig. 2c), confirming earlier data obtained by using Northern blot analysis (5). The same band is also reexpressed in U937 cells stably transfected with a SIAH-1 expression vector (Fig. 2c). Fluorescence-activated cell sorting analysis of these transfectants indicates that a significant fraction of the cells reexpressing SIAH-1 exhibit sub-G1 DNA content (Fig. 2d) and TUNEL positivity (Fig. 2e), unlike control cells. Thus, SIAH-1 overexpression induces apoptosis. Finally, injection of the U937- SIAH-1 transfectants into scid/scid mice reveals a significant suppression of their tumorigenic potential (Fig. 2f).
Figure 2.
Characterization of SIHA1. (a) Expression of the murine siah-1b in the course of p53-induced apoptosis, as measured by Western blot analysis with anti-SIAH-1. LTR-6 cells at 37°C (lane 1) and after 4 hr, 7 hr, 9 hr, 16 hr, or 24 hr of incubation at 32°C (lanes 2–6). Arrow indicates the 32-kDa siah-1b protein. (b) Subcellular localization of murine siah-1b in LTR-6 cells determined by Western blot analysis with anti-SIAH-1 antibodies. Lanes 1, 2, 3 represent the nuclear, membrane, and cytoplasmic fractions respectively. (c) Expression of SIAH-1 in human U937 cells. Lane 1, U937 cells transfected with the control vector. Lane 2, US cells derived from U937 cells but displaying a suppressed malignant phenotype. Lane 3, U937 cells stably transfected with SIAH-1. (d and e) Fluorescence-activated cell sorting analysis of DNA content (d) or TUNEL staining (e) in U937 cells transfected with the vector (RSV-C) or transfected with SIAH-1 (RSV-SIAH-1). (f) Tumorigenicity assay in scid/scid mice after injection of U937 cells transfected with either the control vector (○, RSV C) or with SIAH-1 (●, RSV-SIAH-1).
DISCUSSION
In the present work, we identified a series of 15 cDNAs differentially expressed in the K562/KS (4) system. We further used these cDNAs as a fingerprint in the U937/US system (5) and in p21Waf1 (20) and SIAH-1 transfectants of U937 cells. The striking overlap in differential expression of these genes in the different model systems suggests that at least those sharing differential expression may be part of the tumor suppression and programmed cell death process. Between the K562/KS and the U937/US cells both selected by H-1 parvovirus, there is an overlap of 12 of 15 genes. This suggests that various types of tumor cells may use a series of common genes when reverting to a suppressed phenotype. TSAP11, TSAP12, and TSAP18 are differentially expressed only in cells with active p53 but not in p21Waf1 overproducers, suggesting that they are p53-dependent and p21Waf1-independent. The same is true for the transfectants of U937 cells with either p21Waf1 or SIAH-1: 11 of the 15 genes are differentially regulated in both systems.
TSAP9 corresponds to the group of chaperonin containing t-complex of polypeptide 1 (TCP-1) (21), a multisubunit machinery for protein folding and assembly (15). Chaperonins are also linked with other aspects of p53-mediated signal transduction. The chaperone machinery constitutes an essential function in preventing protein aggregation and in refolding misfolded proteins. The induction during tumor suppression of this chaperonin may provide the cell with a “protein repair” function that is not sufficiently active in the cancer cell.
TSAP 13 encodes a protein recently identified as p40.5, the homologue of Nas7p in yeast (22). It is part of the human 26S proteasome, active in degrading ubiquitinated proteins (24). Disruption in yeast of Nas7p causes high sensitivity to heat stress and inability to proliferate at 37°C (22). In general, ubiquitin-dependent proteolysis plays a pivotal role in controlling cell-cycle transition. p53, by binding MDM-2, is degraded by the proteasome system; this may be the main mechanism responsible for regulation of cellular p53 levels (25–26).
TSAP21 encodes a SNARE corresponding to syntaxin 11, which is localized to the post-Golgi complex (23). Syntaxins are regulators of intracellular trafficking, distribution and restriction of molecules to specific membrane compartments, most extensively studied in the presynaptic nerve terminal. It should be stressed that not all TSAPs may be necessarily expressed to promote cell death. In fact, some of them may be activated to protect the cell from apoptosis (27).
Conceivably, the activation of TSAP 9, TSAP 13, and TSAP 21 in the K562/KS and U937/US system could be related to the fact that the KS and US cells still produce active H-1 parvovirus, which in turn may activate a machinery involving protein structuring and vesicular trafficking. However, the fact that these genes are also independently activated in p21Waf1 and SIAH-1 transfectants of U937 cells indicates that the H-1 parvovirus cannot be the only key player in their induction. Because the majority of the 15 genes described here are coordinately regulated in both p53 and p21Waf1 systems, they are most probably not direct transcriptional targets of p53. Rather, they are likely to represent downstream effectors modulated by functional induction of p53 and p21Waf1 in the orchestration of apoptosis and tumor suppression.
In this regard, SIAH-1 is the best example, being activated independently by p53 and p21Waf1. As shown here, transfection of U937 cells with SIAH-1 induces apoptosis and tumor suppression. This finding is in line with the strong induction of SIAH-1 expression in different models of tumor suppression and apoptosis (5, 6), and with the fact that during differentiation of the intestinal epithelium, SIAH-1 is expressed only in cells that have reached the terminal stage and are dying by apoptosis (5). Furthermore, SIAH-1 was recently shown to induce growth arrest (15). The differential induction of either growth arrest or apoptosis is most probably due to the different cell types used (15). Moreover, SIAH-1 binds BAG-1, a protein involved in programmed cell death, and binding of BAG-1 to SIAH-1 inhibits its growth arrest function (15). These experiments implicate once more SIAH-1 in the regulation of cell proliferation versus growth arrest, and together with the data presented here suggest that it is responsible for the modulation of important cell fate decisions. On the other hand, the experiments with both the Drosophila sina (13, 14) and the mammalian SIAH-1 (17, 18) suggest that these proteins may play a direct role in protein ubiquitination and degradation. Intriguingly, SIAH-1 binds BAG1 (15) but does not promote its degradation as it does to DCC. Further identification of molecules interacting with SIAH might help elucidate its precise function in the cell. One clue might be the differential localization of SIAH in various cells. Most studies indicate that both Drosophila sina (12) and SIAH-1 (15) are nuclear proteins. Although our findings are similar, we also detect a small proportion of SIAH-1 in the cytoplasm. Such different localization may allow SIAH to fulfill a different function.
In conclusion, these results suggest that tumor suppression is achieved through the concerted action of a group of genes, many of which are differentially modulated in a similar fashion by both p53-dependent and -independent pathways. Among these genes, SIAH-1 plays a central role through its multiple effects on cell fate. Other putative common components of the tumor suppression machinery such as TSAP9, -13, and -21 are tightly linked to mechanisms that control protein structure and intracellular localization. Altogether, these genes represent molecular targets whose modulation may trigger a tumor suppressive potential that is otherwise dormant in cancer cells.
Acknowledgments
R.B.A. and A.T. dedicate this work to Daniel Cohen. We thank Gilles Thomas and Christian Rebollo for their support. The tumorigenicity assays were performed by Dr. Philippe Genne and Olivier Duchamp at Oncodesign. The cell cultures were performed by Lucien Cazes and Fabienne Dufour. We thank Bernard Boursin and the photography department at the Hôpital St Louis for the photographic work. We are indebted to Cécile Rouzaud and Pascale Villedieu for efficiency and help.
ABBREVIATIONS
- TSAP
tumor suppressor-activated pathway
- TSIP
tumor suppressor-inhibited pathway
Footnotes
References
- 1.Mousset S, Rommelaere J. Nature (London) 1982;300:537–539. doi: 10.1038/300537a0. [DOI] [PubMed] [Google Scholar]
- 2.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns K I. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Telerman A, Tuynder M, Dupressoir T, Robaye B, Sigaux F, Shaulian E, Oren M, Rommelaere J, Amson R. Proc Natl Acad Sci USA. 1993;90:8702–8706. doi: 10.1073/pnas.90.18.8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemani M, Linares-Cruz G, Bruzzoni-Giovanelli H, Roperch J-P, Tuynder M, Bouguerlet L, Cherif D, Medhioub M, Pasturaud P, Alvaro V, et al. Proc Natl Acad Sci USA. 1996;93:9039–9042. doi: 10.1073/pnas.93.17.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amson R B, Nemani M, Roperch J P, Israeli D, Bougueleret L, Le Gal I, Medhioub M, Linares-Cruz G, Lethrosne F, Pasturaud P, et al. Proc Natl Acad Sci USA. 1996;93:3953–3957. doi: 10.1073/pnas.93.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Israeli D, Tessler E, Haupt Y, Elkeles A, Wilder S, Amson R, Telerman A, Oren M. EMBO J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roperch J P, Alvaro V, Prieur S, Tuynder M, Nemani M, Lethrosne F, Piouffre L, Gendron M-G, Israeli D, Dausset J, et al. Nat Med. 1998;4:835–838. doi: 10.1038/nm0798-835. [DOI] [PubMed] [Google Scholar]
- 9.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Nature (London) 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 10.Della N G, Bowtell D D L, Beck F. Cell Tissue Res. 1995;279:411–419. doi: 10.1007/BF00318499. [DOI] [PubMed] [Google Scholar]
- 11.Della N G, Senior P V, Bowtell D D L. Development. 1993;117:1333–1343. doi: 10.1242/dev.117.4.1333. [DOI] [PubMed] [Google Scholar]
- 12.Carthew R W, Rubin G M. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- 13.Tang A, Neufeld T, Kwan E, Rubin G. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Li Y, Carthew R W, Lai Z-C. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzawa S, Takayama S, Froesch B A, Zapata J M, Reed J C. EMBO J. 1998;17:2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takayama S, Bimston D N, Matsuzawa S, Freeman B C, Aime-Sempe C, Xie Z, Morimoto R I, Reed J C. EMBO J. 1998;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu G, Zhang S, Vidal M, La Baer J, Xu T, Fearon E R. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu G, Fearon E R. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg M E, Edelman G M. Cell. 1983;33:767–779. doi: 10.1016/0092-8674(83)90019-3. [DOI] [PubMed] [Google Scholar]
- 20.Linares-Cruz G, Bruzzoni-Giovanelli H, Alvaro V, Roperch J-P, Tuynder M, Schoevaert D, Nemani M, Prieur S, Lethrosne F, Piouffre L, et al. Proc Natl Acad Sci USA. 1998;95:1131–1135. doi: 10.1073/pnas.95.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubota H, Hynes G, Willison K. Eur J Biochem. 1995;230:3–16. doi: 10.1111/j.1432-1033.1995.tb20527.x. [DOI] [PubMed] [Google Scholar]
- 22.Hori T, Kato S, Saeki M, De Martino G N, Slaughter C A, Takeuchi J, Toh-e A, Tanaka K. Gene. 1998;216:113–122. doi: 10.1016/s0378-1119(98)00309-6. [DOI] [PubMed] [Google Scholar]
- 23.Advanit R J, Bae H- R, Bock J B, Chao D S, Doung Y-C, Prekeris R, Yoo J-S, Schellers R H. J Biol Chem. 1998;273:10317–10324. doi: 10.1074/jbc.273.17.10317. [DOI] [PubMed] [Google Scholar]
- 24.Baumeister W, Walz J, Zühl F, Seemüller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 25.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 26.Kubbutat M H G, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 27.Lassus P, Ferlin M, Piette J, Hibner U. EMBO J. 1996;15:4566–4573. [PMC free article] [PubMed] [Google Scholar]