Abstract
Undesired side products of DNA transfections are usually discarded. However, here, we show that such products may provide insight into mutational events that are also a major driving force in protein evolution. While studying the small heat-shock protein αA-crystallin, we transfected the hamster αA-crystallin gene into a mouse muscle cell line. One of the stable transfected cell lines expressed, in addition to the expected normal αA- and alternatively spliced αAins-crystallins, two slightly larger, immunologically cross-reacting proteins. These proteins were found to be encoded by a mutant αA-crystallin gene with a large intragenic duplication, arisen by illegitimate recombination at two CCCAT homologies, ≈1.8 kilobases apart in the normal hamster αA-crystallin gene. As a consequence, a tandem-duplicated exon 3 sequence is present in the mature mRNA of this gene, resulting in a 41-residue repeat in the translated proteins. Cells expressing the elongated αA-crystallins have normal growth characteristics and the usual diffuse cytoplasmic distribution of immunoreactive αA-crystallin. Size-exclusion chromatography of cell extracts indicated that the mutant proteins are readily incorporated into the normal large water-soluble αA-crystallin complexes, showing that the insert does not disturb the integrity of these complexes. This viable αA-crystallin mutant thus mimics the origins and effects of exon duplication, which is a common consequence of exon shuffling in mammalian genome evolution.
Keywords: α-crystallin, exon duplication, illegitimate recombination, protein evolution, small heat-shock proteins
It is known that during passage of plasmid DNA in mammalian cells, all sorts of mutations may spontaneously occur in the transfected DNA (1). Therefore, when transfection of a gene into eukaryotic cells yields aberrant products, the logical reaction is to ignore and discard those cells. However, it should be realized that transfection and other recombinant DNA technology simply exploit the mutational processes normally occurring in living organisms. Here, we show that paying attention to “undesired” by-products of transfections may be rewarding in terms of increasing our understanding of mutational and evolutionary mechanisms.
To compare the intracellular properties of the various mammalian small heat-shock proteins (sHsps), we routinely transfect the corresponding genes into different cell lines. Among the six known mammalian sHsps (2), a special position is taken by the α-crystallins (3). α-Crystallin was first discovered as a major structural eye-lens protein. It occurs as large globular complexes of up to 700 kDa, composed of two related types of 20-kDa subunits, αA- and αB-crystallin (4). In rodents and some other mammals, a minor α-crystallin subunit is present, resulting from alternative splicing of the αA-crystallin gene transcript (5–7). This process yields, in addition to the normal 173-residue αA-crystallin, the elongated αAins-crystallin, with an insertion of 23 amino acid residues. The insert is encoded by the optional exon 2, which is skipped in 80–90% of mRNA because of the presence of an adjacent nonconsensus GC 5′ splice site (6, 8). Like other members of the sHsp family, α-crystallins have in vitro chaperone-like activity, suppressing the aggregation of denaturing proteins (9). The most conspicuous in vivo feature of α-crystallins is their ability to confer thermotolerance on overexpression in different cell types (10, 11).
Although αB-crystallin occurs in many tissues outside the lens, notably in heart and striated muscle, αA-crystallin is restricted essentially to the lens (12–14). To determine whether αA-crystallin has diverged functionally from αB-crystallin in the course of evolution, it is of interest to establish how αA-crystallin behaves in cell types that are normally the domain of αB-crystallin. Expressing the αA-crystallin gene in a muscle cell line might help to answer this question. Doing so, we observed that one of the stably transfected cell lines expressed, in addition to the expected αA- and αAins-crystallins, two immunologically cross-reacting proteins at high levels. It seemed informative to identify those proteins and see how viable they are in terms of interfering with normal cellular functioning, complex formation, and chaperone-like capacities. Characterizing these products might shed light on the recombinational processes taking place during passage of the introduced DNA in the mammalian cells, as well as informing us about the properties of spontaneously arising mutants of αA-crystallin. Our analyses identified a mutant αA-crystallin gene with a duplicated exon 3. This finding is in accord with the hypothesis of exon shuffling (15), a theory that has recently been tested experimentally by Gilbert et al. (16).
MATERIALS AND METHODS
Plasmid Construction and Transfection.
To construct the eukaryotic expression vector for αA-crystallin, an 8.1-kilobase FspI–KpnI fragment containing the hamster αA-crystallin gene under the vimentin promoter (pVim-αA) was taken from a Puc-18 vector (17). The pVim-αA fragment was ligated into the XbaI and SalI sites of a pCMV-neo vector, which contains a neomycin-resistance cassette (18). By using the restriction enzymes XbaI and SalI, the cytomegalovirus (CMV) promoter was deleted from the pCMV-neo plasmid. All ligations were made blunt-end.
Mouse skeletal muscle cells (C2C12) were grown as monolayers in DMEM with glutamax (GIBCO), supplemented with 20% (vol/vol) FCS, penicillin G (100 units⋅ml−1), and streptomycin (100 ml⋅ml−1). Cells were transfected based on the method developed by Graham and van der Eb (19) and modified by Wigler et al. (20), and stable cell lines were obtained by selection with gentamicin (GIBCO).
SDS/PAGE, Two-Dimensional Electrophoresis, and Immunoblotting.
Samples for electrophoresis were prepared by lysing the transfected cells in SDS sample buffer. The protein content of the samples was estimated with the bicinchoninic acid protein reagent (Pierce). One-dimensional SDS/PAGE was performed according to Laemmli’s method (21), and two-dimensional gel electrophoresis was performed essentially as described by O’Farrell (22). After one- and two-dimensional gel electrophoresis, Western blotting was performed according to the method of Towbin et al. (23). A polyclonal rabbit antiserum against bovine lens αA-crystallin and monoclonal antibody Cr.I-1 (7) against rat αA-crystallin were used.
PCR Amplification and DNA Sequencing.
Total RNA was extracted from transfected cells by using Trizol (GIBCO). cDNA was made by 3′ rapid amplification of cDNA ends with the aid of a 5′/3′ rapid amplification of cDNA ends kit (Boehringer Mannheim). A degenerated forward primer was designed on the basis of a conserved exon 1 sequence in the hamster and mouse αA-crystallin gene (5′-CCA-TTC-AGC-ACC-CTT-GGT-TYA-ARC-G-3′) and used in combination with an oligo(dT) and an anchor primer (included in the kit). The PCR product was ligated into the pGEM-T vector (Promega) and sequenced. After sequence determination, the molecular masses and isoelectric points of deduced proteins could be calculated with the aid of the computer program generunner.
Chromosomal DNA was obtained from transfected cells by using the extraction procedure of Blin and Stafford (24). To amplify the genomic region of interest, a forward primer at the 3′ end of exon 3 (5′-GGC-AAG-CAC-AAT-GAG-AGG-CAG-3′) was combined with a reverse primer at the 5′ end of exon 3 (5′-CTT-GTC-CCG-GTC-AGA-TCG-GAC-3′). The amplification cycle used denaturation at 94°C for 2 min, annealing at 65°C for 1 min, and extension at 72°C for 70 s. The PCR product was again ligated into a pGEM-T vector and sequenced subsequently.
Size-Exclusion Chromatography.
The molecular mass of αA-crystallin complexes present in the transfected cell lines was estimated by size-exclusion chromatography. Cells were scraped from culture flasks in lysis buffer, and after centrifugation the soluble fraction was applied onto a Superose 6 HR 10/30 prepacked size-exclusion column (Pharmacia). The buffers used for lysis and elution have been described by Lavoie et al. (25). Elution was performed at a flow rate of 0.5 ml/min, and 15% of each 1-ml fraction was analyzed by SDS/PAGE and immunoblotting with the monoclonal antibody. Total calf lens protein was used for calibration.
RESULTS
Stable Transfection of the Hamster αA-Crystallin Gene Yields Elongated By-Products.
pVim-αA was transfected into the mouse skeletal muscle cell line C2C12 by using the calcium phosphate precipitation procedure. Among 12 stably transfected lines, 11 showed—to different extents—expression of αA- and αAins-crystallin, as assayed by Western blotting (not shown). Surprisingly, one of the positive cell lines, pVim-αA7, expressed, in addition to αA- and αAins-crystallin, two somewhat larger immunologically cross-reacting proteins (Fig. 1A). This cell line and a line with high expression of αA- and αAins-crystallin (pVim-αA11) were used for further comparison and will be referred to as lines 7 and 11, respectively.
Figure 1.
αA-crystallin immunoreactive proteins present in mouse C2C12 cells after stable transfection with the hamster αA-crystallin gene. (A) Immunoblot after SDS/PAGE of total extracts from transfected cells with a high expression level of normal αA- and αAins-crystallin with masses of 19.8 and 22.5 kDa, respectively (line pVim-αA11; left lane) and transfected cells expressing two additional αA-crystallin-like proteins: super αA-crystallin and super αAins-crystallin (line pVim-αA7; right lane). Detection was achieved with a polyclonal anti-αA-crystallin antiserum. (B) Immunoblot after two-dimensional electrophoresis of cell extract from line pVim-αA7. Detection was achieved with a monoclonal anti-αA-crystallin antiserum. Arrowheads indicate presumed monophosphorylation products. The different ratios between the four αA-crystallin-positive proteins in A and B resulted from the use of different antisera and staining conditions.
Two-dimensional gel electrophoresis of line 7 cell extract, followed by immunoblotting, showed that the isoelectric points of the two additional proteins lie between those of αA- and αAins-crystallin (Fig. 1B). Their positions and proportions relative to normal αA- and αAins-crystallin led us to name them super αA-crystallin and super αAins-crystallin. From the Western blots in Fig. 1 (see also Fig. 3B), it is difficult to estimate the amounts of the super forms relative to normal αA- and αAins-crystallin. It is clear, however, that both are expressed at considerable levels. Interestingly, and at variance with earlier αA-crystallin transfections in HeLa cells (26), conspicuous phosphorylation of the expressed proteins seems to occur (arrowheads in Fig. 1B).
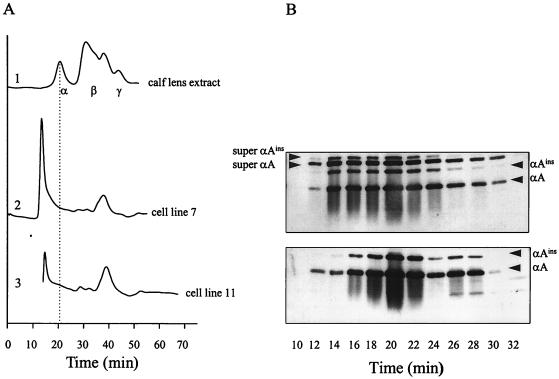
Figure 3.
Complex size of α-crystallin expressed in transfected mouse C2C12 cells. (A) Elution profiles after size-exclusion chromatography (Superose 6) of (i) calf lens soluble proteins, (ii) extract of cell line 7, and (iii) extract of cell line 11. Calf lens α-crystallin complexes have a mass of ≈700 kDa. (B) Western blotting after SDS/PAGE of Superose 6 fractions from cell line 7 (Upper) and cell line 11 (Lower). Lanes are numbered according to the elution times shown in A. Blots are stained with a monoclonal anti-αA-crystallin antiserum.
As for the viability of the transfected cells, it turned out that line 7 cells, harboring the super forms, grew and divided as readily as cells expressing only the normal αA- and αAins-crystallins. Also, immunofluorescence studies of cell line 7, as compared with cell line 11, showed no differences in pattern or distribution of αA-crystallin immunoreactivity. In both cell lines, the antiserum gave a similar homogeneous cytoplasmic staining (data not shown).
Sequence Determination Identifies Exon Duplication by Illegitimate Recombination.
To establish the nature of the two additional proteins, total RNA was extracted from cell line 7. A 3′ rapid amplification of cDNA ends was performed with a forward primer corresponding to nucleotide positions 11–26 in exon 1 of the αA-crystallin gene (see arrowhead a in Fig. 2A), an oligo(dT) reverse primer, and an anchor primer. The amplification products were ligated into a pGEM-T vector and transformed into an Escherichia coli DH5α host strain. Restriction analyses identified four different amplification products. Sequence determination confirmed the presence of normal hamster αA- and αAins-crystallin mRNAs, whereas the two other products contained a duplicated exon 3 (named 3* in Fig. 2A). No endogenous mouse αA-crystallin sequences were encountered in the products. Translation of the duplicated exon 3 should result in an additional 41 amino acid residues in otherwise normal αA- and αAins-crystallins. The length and composition of the duplicated sequence correspond well with the observed apparent masses and isoelectric points of the super proteins. The calculated molecular masses are 19.8, 22.5, 24.7, and 27.4 kDa, and the isoelectric points are 6.00, 6.67, 6.02, and 6.52 for normal αA-, αAins-, super αA-, and super αAins-crystallin, respectively.
Figure 2.
Exon duplication in the super αA-crystallin gene. (A) Schematic representations of the super α-crystallin mRNAs as deduced from cDNA sequencing. Exons are numbered below the mRNAs. Exon 2 is retained only in the alternatively spliced αAins-crystallin mRNA. The duplicated exon 3 sequence is indicated as 3*. Numbers above the mRNAs give the amino acid residue positions corresponding to the 3′ end of each exon and the total coding region to indicate the lengths of the mutant proteins and inserted regions. Arrowheads point to the positions of the forward primer (a), used to obtain cDNA, and the forward (b) and reverse (c) primers used for sequencing the mutant gene in the genomic DNA. (B) Nonhomologous recombination between two copies of the transfected hamster αA-crystallin gene (top and middle genes). Introns are numbered above the genes. Recombination has occurred at the position of the vertical dotted line, apparently facilitated by the CCCAT homologies in exon 2 and intron 3 (arrowheads). As a result, the super αA-crystallin gene (bottom gene) contains a 1.7-kilobase duplication resulting in a truncated intron 3 (indicated as 3−) and exon 2 (2−), as well as an additional intron 2 (2*) and exon 3 (3*). Because the intron splice site between pseudointron 3− and pseudoexon 2− is lost, these sequences are combined with intron 2* to form the large intron 3+ between the duplicated exons 3 and 3*. The double arrow beneath the super gene indicates the region that was amplified by using primers b and c in Fig. 2A. The thicker part of this line indicates the position of the sequence presented in Fig. 2C. (C) The sequence around the illegitimate recombination junction in intron 3+ of the super αA-crystallin gene is aligned with the 3′ end of intron 3 and the 5′ end of exon 2 of the normal hamster αA-crystallin gene. The switch from the intron 3 to the exon 2 sequence occurs at the CCCAT homology at positions 596–600. Numbers above the sequence correspond to the position in the hamster αA-crystallin intron 3 and exon 4.
At this stage, it could be concluded that the mutant αA-crystallin gene had originated from nonhomologous or illegitimate recombination between two copies of the transfected hamster gene and that the mutant mRNA was alternatively spliced like the normal rodent αA-crystallin mRNA. Although utterly unlikely in recA− E. coli, we verified whether the recombination might have occurred already in the bacteria before DNA isolation and transfection. Thorough PCR analyses of the isolated plasmid gave no indication for such a recombination. To reconstruct how the elongated mutant gene had arisen, we analyzed the structure of this gene in the chromosomal DNA from cell line 7. Because the site of illegitimate recombination should be located between the duplicated exons 3 and 3*, we performed PCR with a forward primer complementary to the 3′ end of exon 3 and a reverse primer annealing to the 5′ end of exon 3* (arrowheads b and c in Fig. 2A). The resulting amplification product was ≈1.7 kilobases long. As shown in Fig. 2B (see the line under bottom gene), sequence analyses identified, in this order, the expected 5′ 21 bp of exon 3; an almost complete intron 3 (3−), which, at the very end, changed into a 5′ truncated exon 2 (2−); a complete intron 2 (2*); and finally, the expected 3′ 21 bp of the next exon 3 (3*).
Evidently one hamster αA-crystallin gene has illegitimately recombined at a site located at the 3′ end of its intron 3 (Fig. 2B, middle gene) with a site at the 5′ end of exon 2 of another hamster αA-crystallin gene (Fig. 2B, top gene). The sequence of this transition zone in the mutant gene allows us to identify the position of the site of illegitimate recombination more precisely. Up to position 600, this sequence (presented as intron 3+ in Fig. 2C) is identical to the normal intron 3 (top sequence), and from position 596 onward it is identical to the normal exon 2 (bottom sequence). The overlapping 5-bp sequence CCCAT (bold and underlined) is present both in the normal exon 2 and in the normal intron 3 (see arrowheads in Fig. 2B) and must have provoked the illegitimate recombination between two copies of the transfected αA-crystallin gene. The recombination event leaves a 5′ truncated second exon 2 in the mutant gene (2− in Fig. 2B), which thus becomes part of the third intron of that gene (3+).
The Super Forms Are Normally Incorporated into the Large α-Crystallin Complexes.
One might imagine that a large insertion, such as the 41 amino acid residues encoded by the duplicated exon 3, would interfere with normal folding, stability, and complex formation of αA-crystallin. To assess whether super αA-crystallin and super αAins-crystallin are indeed incorporated into the large and soluble complexes normally formed by αA-crystallin and αAins-crystallin, soluble cell extracts from lines 7 and 11 were analyzed by size-exclusion chromatography. After scraping the cells in lysis buffer, the super α-crystallins were found in the soluble fraction, together with the normal forms. The elution profiles of the extracts of cell lines 7 and 11 were very similar (Fig. 3A, profiles 2 and 3, respectively). SDS/PAGE of the eluted fractions followed by Western blotting with αA-crystallin antiserum (Fig. 3B) showed that, for both cell line 7 and 11, the immunoreactive proteins had elution times similar to control calf lens α-crystallin, between 12 and 30 min (compare profile 1 in Fig. 3A). Notably, no later eluting smaller complexes or monomers were observed in either cell line.
The complexes containing super αA-crystallin and super αAins-crystallin start to elute earlier than the αA-/αAins-crystallin complexes (Fig. 3B, Upper vs. Lower). This fact may be attributed to the mass of the 41-residue insert in the super subunits. Also, the α-crystallin complexes in cell line 7 elute over a broader range than those in line 11, which indicates a greater dispersal of the former complexes. It also seems, as shown in Fig. 3B (Upper), that the ratios of the four subunits vary with the elution time; the super forms tend to be more prominent in the earlier eluting fractions. However, it may be concluded that even the 64-residue insert in super αAins-crystallin, as compared with normal 173-residue αA-crystallin, constitutes no detectable hindrance for its regular integration into the large and soluble α-crystallin particles.
DISCUSSION
How Did the Super αA-Crystallin Mutant Arise?
Before integration into the genome, transfected circular plasmid vectors are randomly cleaved and may form, by homologous recombination, concatamers in which arrays of monomers are oriented in the same direction (27–29). These concatamers integrate into the chromosomes by illegitimate recombination, doing so preferentially at unstable loci where frequent double-strand breaks and rearrangements occur (28). Joining of transfected and chromosomal DNAs is clearly facilitated by short regions of identity, from 1 to 5 bp, at the junction points of illegitimately recombining DNA molecules (28, 30, 31). Illegitimate recombination requires double-strand breaks, frequently followed by 5′ → 3′ and 3′ → 5′ exonuclease processing, allowing single-strand annealing at short homologous sequences (31). In our case, introduction of the pVim-αA expression vector into mouse C2C12 cells has resulted in illegitimate recombination between two copies of the transfected hamster αA-crystallin gene. The recombination has apparently been triggered by the short homologies CCCAT in exon 2 and intron 3. Interestingly, this sequence also contains a preferred triplet sequence, CAT, for nicking by topoisomerase I, which is known to be directly involved in illegitimate recombination (32). It is difficult to decide whether the illegitimate recombination has occurred before or after the chromosomal integration, although the former seems more likely. In any case, the hamster-specific sequence of the super αA-crystallin gene showed that the endogenous mouse gene had not been involved in the recombination and integration process.
Evolutionary Lessons from a Transfection “Artifact.”
Gene transfections make use of the regular DNA repair and rearrangement mechanisms that continuously operate on the mammalian genome. Illegitimate recombination plays a predominant role in these processes (33). However, amazingly little is known about its mechanisms and genetic control (32). Causing deletions, translocations, and amplifications, illegitimate recombination is a rich source of genetic variation, the raw material for evolutionary change. It affects both chromosomal and extrachromosomal DNA. Because extrachromosomal small circular DNAs occur abundantly in eukaryotic cells (30), it is realistic to liken artifacts, such as those described in this paper, to the possible pathways of molecular evolution. Illegitimate recombination is a major innovative force in protein evolution, generating novel combinations of protein domains by exon shuffling (15). Interestingly, although exon shuffling will mostly occur by recombination in introns, our mutant αA-crystallin gene shows that recombination between an exon, albeit a short and optional one, and an intron may occur as well. It has been estimated that at least 19% of all exons have been involved in shuffling events (34). Duplicated exons, too, occur abundantly; at least 6% of all exons in the human genome have recognizably arisen by exon duplication (35). A prerequisite for effective exon duplication is that the involved exons need to be symmetrical with respect to adjacent intron phases. If they are not, the reading frame will be shifted at the junction of the duplicated exons. Also, the viable exon duplication in our super gene has been possible, because the introns flanking exon 3 are both in phase 0, interrupting the reading frame in between two codons. We may conclude that the exon-duplication mutant of αA-crystallin that we have trapped reflects authentic protein evolutionary mechanisms and fits the idea that exon shuffling has a fundamental role in increasing gene diversity (36).
Structural Implications of Exon Duplication in the Super αA-Crystallins.
The haphazard insertion, addition, or duplication of a whole domain in a protein that has been molded during hundreds of millions of years of adaptive evolution is most likely to frustrate the integrity of that protein. Only rarely will such a major recombination be favorable, yielding a “hopeful monster” (37) at the protein level, with novel and useful properties as already predicted by Gilbert (15) in his classic 1978 paper. It will most often lead to “hopeless” or at best “neutral monsters”. Our super αA-crystallins probably belong to the latter category, considering that the duplication of 41 residues does not detectably interfere with their in vivo viability. The mutant proteins, present at levels similar to the wild-type αA-crystallins (Fig. 1), are not rapidly degraded, have a normal cytoplasmic distribution, do not disturb cell growth, and are incorporated into normal α-crystallin complexes. It seems surprising that a gross structural intrusion can be accommodated by αA-crystallin, which is evolutionarily highly conserved, not having accepted even a single amino acid replacement in the murine rodent lineage for at least 60 million years (38).
This paradox resembles the enigmatic evolutionary origins of αAins-crystallin, which must have occurred more than 100 million years ago (7). The 23-residue insert in αAins-crystallin can probably be accommodated as a bulge at the surface or in any internal space of the α-crystallin complex (39). This situation might apply as well for the 41-residue duplication in super αA-crystallin, which is right next to the insert in αAins-crystallin. Cryo-electron microscopy studies of αB-crystallin complexes identify a highly variable and roughly spherical protein shell around a large central cavity (40). A more regular hollow spherical complex composed of 24 subunits is reported for the crystal structure of the sHsp from Methanococcus jannaschii (MjHSP16.5; ref. 41). It would seem, by comparison, that exon 3 of αA-crystallin codes for three β-strands in the Ig-like fold of the conserved C-terminal domain of the sHsp monomer. The more flexible quaternary structure of the α-crystallins might well allow accommodation of the duplicated β-strands of the super αA-crystallin monomer, either in the central cavity or at the surface. Indeed, an alignment of sHsp sequences from very diverse organisms indicates that major length variation is allowed only in specific regions, notably around the site where the inserts in αAins- and super αA-crystallins are located (3).
Of course, the survival of a mutant protein in a heterologous cell line is not directly comparable with the “real life” long-term constraints acting in evolution. It nevertheless means that the mutant has passed a first and crucial test, being stable and able to cope with the intracellular environment. Therefore, the super αA-crystallins form a realistic model for studying the evolutionary latitude of the structure–function relationships of the sHsps. Also, in the case of transfection experiments with other genes, it may be rewarding to take advantage of the natural and preselected mutational creativity of the genes themselves, rather than to ignore and discard any unexpected by-products.
Acknowledgments
We thank Perry Overkamp and Marcel Sweers for technical assistance. This work was supported by European Community-BIOMED (BMH4-CT96-1593).
ABBREVIATIONS
- sHsp
small heat-shock protein
- pVim-αA
hamster αA-crystallin gene under the vimentin promoter
References
- 1.Calos M P, Lebkowski J S, Botchan M R. Proc Natl Acad Sci USA. 1983;80:3015–3019. doi: 10.1073/pnas.80.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boelens W C, van Boekel M A M, de Jong W W. Biochim Biophys Acta. 1998;1388:513–516. doi: 10.1016/s0167-4838(98)00215-5. [DOI] [PubMed] [Google Scholar]
- 3.de Jong W W, Caspers G, Leunissen J A M. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 4.Bloemendal H. Science. 1977;197:127–138. doi: 10.1126/science.877544. [DOI] [PubMed] [Google Scholar]
- 5.Cohen L H, Westerhuis L W, de Jong W W, Bloemendal H. Eur J Biochem. 1978;89:259–266. doi: 10.1111/j.1432-1033.1978.tb20921.x. [DOI] [PubMed] [Google Scholar]
- 6.King C R, Piatigorsky J. Cell. 1983;32:707–712. doi: 10.1016/0092-8674(83)90056-9. [DOI] [PubMed] [Google Scholar]
- 7.Hendriks W, Sanders J, de Leij L, Ramaekers F, Bloemendal H, de Jong W W. Eur J Biochem. 1988;174:133–137. doi: 10.1111/j.1432-1033.1988.tb14072.x. [DOI] [PubMed] [Google Scholar]
- 8.Smulders R H P H, Kokke B P A, Gijsen M L J, de Jong W W. Mol Biol Rep. 1998;25:225–230. doi: 10.1023/a:1006897910253. [DOI] [PubMed] [Google Scholar]
- 9.Horwitz J. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klemenz R, Fröhli E, Steiger R H, Schäfer R, Aoyama A. Proc Natl Acad Sci USA. 1991;88:3652–3656. doi: 10.1073/pnas.88.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den IJssel P R L A, Overkamp P, Knauf U, Gaestel M, de Jong W W. FEBS Lett. 1994;335:54–56. doi: 10.1016/0014-5793(94)01175-3. [DOI] [PubMed] [Google Scholar]
- 12.Bath S P, Nagineni C N. Biochem Biophys Res Commun. 1989;158:319–325. doi: 10.1016/s0006-291x(89)80215-3. [DOI] [PubMed] [Google Scholar]
- 13.Kato K, Shinohara H, Kurobe N, Inaguma Y, Shimizu K, Ohshima K. Biochim Biophys Acta. 1991;1074:201–208. doi: 10.1016/0304-4165(91)90062-l. [DOI] [PubMed] [Google Scholar]
- 14.Kato K, Shinohara H, Kurobe N, Goto S, Inaguma Y, Ohshima K. Biochim Biophys Acta. 1991;1080:173–180. doi: 10.1016/0167-4838(91)90146-q. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert W. Nature (London) 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert W, de Souza S J, Long M. Proc Natl Acad Sci USA. 1997;94:7698–7703. doi: 10.1073/pnas.94.15.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloemendal H, Enzlin J H, van Rijk A F, Jansen H J. Exp Eye Res. 1997;64:1037–1041. doi: 10.1006/exer.1997.0294. [DOI] [PubMed] [Google Scholar]
- 18.Bakker S J, Markowitz S, Fearon E R, Willson J K V, Vogelstein B. Science. 1990;249:912–914. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 19.Graham F L, van der Eb A J. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 20.Wigler M, Sweet R, Sim G K, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Axel R. Cell. 1979;16:777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.O’Farrell P H. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 23.Towbin H, Staehlin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blin N, Stafford D W. Nucleic Acid Res. 1976;3:2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavoie J N, Lambert H, Hickey E, Weber L A, Landry J. Mol Cell Biol. 1995;15:505–516. doi: 10.1128/mcb.15.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den IJssel P R L A, Overkamp P, Bloemendal H, de Jong W W. Biochem Biophys Res Commun. 1998;247:518–523. doi: 10.1006/bbrc.1998.8699. [DOI] [PubMed] [Google Scholar]
- 27.Coffin J M. J Med Virol. 1990;31:43–49. doi: 10.1002/jmv.1890310109. [DOI] [PubMed] [Google Scholar]
- 28.Merrihew R V, Marburger K, Pennington S L, Roth D B, Wilson J H. Mol Cell Biol. 1996;16:10–18. doi: 10.1128/mcb.16.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishop J O. Reprod Nutr Dev. 1996;36:607–618. [PubMed] [Google Scholar]
- 30.Iwasaki T, Ohki R, Kiyama R, Oishi M. FEBS Lett. 1995;363:239–245. doi: 10.1016/0014-5793(95)00325-4. [DOI] [PubMed] [Google Scholar]
- 31.Henderson G, Simons J P. Mol Cell Biol. 1997;17:3779–3785. doi: 10.1128/mcb.17.7.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, Schiestl R H. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth D, Wilson J. In: Genetic Recombination. Kucherlapati R, Smith G, editors. Washington, DC: Am. Soc. Microbiol.; 1988. pp. 621–653. [Google Scholar]
- 34.Long M, Rosenberg C, Gilbert W. Proc Natl Acad Sci USA. 1995;92:12495–12499. doi: 10.1073/pnas.92.26.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fedorov A, Fedorova L, Starshenko V, Filatov V, Grigor’ev E. J Mol Evol. 1998;46:263–271. doi: 10.1007/pl00006302. [DOI] [PubMed] [Google Scholar]
- 36.de Souza S J, Long M, Gilbert W. Genes Cells. 1996;16:493–505. doi: 10.1046/j.1365-2443.1996.d01-264.x. [DOI] [PubMed] [Google Scholar]
- 37.Goldschmidt R. The Material Basis of Evolution. New Haven, CT: Yale Univ. Press; 1940. [Google Scholar]
- 38.Hendriks W, Leunissen J, Nevo E, Bloemendal H, de Jong W W. Proc Natl Acad Sci USA. 1987;84:5320–5324. doi: 10.1073/pnas.84.15.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smulders R H P H, van Geel I G, Gerards W L H, Bloemendal H, de Jong W W. J Biol Chem. 1995;270:13916–13924. doi: 10.1074/jbc.270.23.13916. [DOI] [PubMed] [Google Scholar]
- 40.Haley D A, Horwitz J, Stewart P L. J Mol Biol. 1998;277:27–35. doi: 10.1006/jmbi.1997.1611. [DOI] [PubMed] [Google Scholar]
- 41.Kim K K, Kim R, Kim S. Nature (London) 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]