Abstract
OTHER ARTICLE PUBLISHED IN THIS MINI-REVIEW SERIES ON IMMUNODEFICIENCY Recognition, clinical diagnosis and management of patients with primary antibody deficiencies: a systematic review. Clin Exp Immunol 2007; 149: 10.1111/j.1365-2249.2007.03432.x
Common variable immunodeficiency (CVID) is a primary immunodeficiency that typically affects adults and is characterized by abnormalities of quantative and qualitative humoral function that are heterogeneous in their immunological profile and clinical manifestations. The recent identification of four monogenic defects that result in the CVID phenotype also demonstrates that the genetic basis of CVID is highly variable. Mutations in the genes encoding the tumour necrosis factor (TNF) superfamily receptors transmembrane activator and calcium-modulating ligand interactor (TACI) and B cell activation factor of the TNF family receptor (BAFF-R), CD19 and the co-stimulatory molecule inducible co-stimulator molecule (ICOS) all lead to CVID and illustrate the complex interplay required to co-ordinate an effective humoral immune response. The molecular mechanisms leading to the immune defect are still not understood clearly and particularly in the case of TACI, where a number of heterozygous mutations have been found in affected individuals, the molecular pathogenesis of disease requires further elucidation. Together these defects account for perhaps 10–15% of all cases of CVID and it is highly likely that further genetic defects will be identified.
Keywords: CVID, co-stimulation, hypogammaglobulinaemia, TACI
Introduction
Primary immunodeficiencies are a highly heterogeneous group of monogenic diseases that range in the severity of clinical symptoms and in the degree of disease prevalence. Over the last 20 years, elucidation of the underlying genetic defect has led to major advances in understanding the molecular and cellular basis of disease pathogenesis and has also led to improvements in genetic diagnosis, prediction of disease prognosis and the development of gene therapy for specific conditions. Moreover, these diseases have been enormously informative in understanding the role played by specific molecules in the immune system and have guided our interpretation of the complex interplay present in many immunological systems.
Of the many primary immunodeficiencies, antibody deficiency syndromes are among the most common and yet it is only very recently that gene defects in these conditions have been identified. Specific IgA deficiency (IgAD) has a prevalence of ∼1:600 individuals in the western world and symptomatic individuals often have deficiency of IgG subclasses and failure of specific responses to carbohydrate antigens in addition to defects of IgA production [1]. Common variable immunodeficiency (CVID) has a prevalence of approximately 1:25 000 in Europeans and is defined clinically and immunologically by low serum immunoglobulin concentrations of one or more isotypes [2], defective specific antibody responses and clinically increased susceptibility to bacterial infections [3]. Approximately 20% are complicated by autoimmune manifestations and lymphoproliferation with splenomegaly seen in one-third of patients. Cellular and immunological defects in CVID are variable and include abnormalities of B cell survival, decreased frequency of circulating CD27+ memory B cells, failure of isotype switching to IgA and IgG and defective B cell activation [4,5]. Further studies highlight the inability to mount responses to polysaccharide antigens, which may be related to the absence of marginal zone IgM memory B cells in a subgroup of CVID patients [6].
Genetic inheritance of CVID
In keeping with the heterogeneous cellular and immunological defects, the inheritance of CVID is also very variable. Although the majority of cases are sporadic, familial patterns of inheritance are seen in ∼10–20% of cases [7]. Most multiplex CVID families show an autosomal dominant pattern of inheritance, but autosomal recessive inheritance is also seen in a significant minority. In a number of families, IgAD and CVID co-exist and furthermore some individuals with IgAD progress to a CVID phenotype, suggesting that at least in some pedigrees, the two conditions share a common genetic aetiology [8]. Initial genetic studies which combined analysis of patients with both CVID and IgAD identified the existence of a susceptibility locus in the MHC region of chromosome 6 [9,10]. Further fine-mapping studies on IgAD families confirmed more clearly the existence of a susceptibility locus at chromosome 6p termed IGAD1[11]. A subsequent study by the same group suggested that the major susceptibility determining locus was at human leucocyte antigen D-related (HLA DQ-DR), although this same study identified non-MHC susceptibility loci on chromosomes 4p, 12p and 14q [7]. The heterogeneity of the genetic basis of CVID is highlighted further by more recent studies which have identified susceptibility loci at 16q, and 4q in a number of different multiplex families [12,13].
CVID arising from defects in genes associated with other primary immunodeficiency diseases (PIDs)
The genetics of CVID is complicated further by a number of observations which demonstrate that the phenotype associated with mutations in certain known genes can, itself, be very variable. Mutations in Btk result typically in complete agammaglobulinaemia with a total lack of peripheral B cell development and hence classical X-linked agammaglobulinaemia (XLA), but in a small number of cases less severe B cell defects arise from Btk mutations and may mimic the CVID phenotype [14]. Similarly, defects in the intracellular adapter molecule SAP lead to X-linked lymphoproliferative disease, which can present in a number of ways including hypogammaglobulinaemia [15]. Other gene defects which may lead to a CVID phenotype in a small number of phenotypic variants include the genes associated with hyper-IgM syndromes and defects in class-switching recombination (CSR); CD40, CD40L, activation-induced cytidine deaminase (AID) and uracil-N-glycosylase (UNG). Studies in which systematic analysis of CVID patients has been undertaken do, however, suggest that the incidence of defects in these genes is likely to be low and can in some cases also be discerned through a characteristic pedigree or family history [16,17].
Over the last 5 years, four monogenic defects leading specifically to the CVID phenotype have been identified. These defects illustrate the multiple complex pathways involved in the co-ordination of an effective humoral immune response and emphasize the crucial roles played by these specific molecules. These recent developments also suggest that as more genetic causes are defined, CVID will no longer be a specific disease entity but will exist as a group of monogenic defects sharing similar immunological and clinical phenotypes.
Inducible co-stimulator molecule (ICOS) deficiency
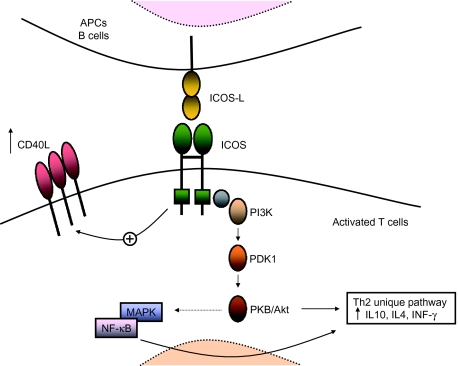
Activation of immature T cells into effector T cells through the T cell antigen receptor (TCR) complex requires the binding of cell surface co-stimulatory proteins such as CD28 and CTLA-4 binding to surface B7 receptors CD80 (B7.1) and CD86 (B7.2) on antigen-presenting cells (APCs). ICOS belongs to the CD28 family of immunoglobulin-like co-stimulatory surface molecules. ICOS is expressed only on activated T cells and is involved in the release of cytokines interleukin (IL)-4, IL-5, IL-6, tumour necrosis factor (TNF)-α, interferon (IFN)-γ and granulocyte–macrophage colony-stimulating factor (GM-CSF), but in particular in the superinduction of IL-10, which leads to terminal differentiation of B cells to memory and plasma cells [18] (Fig. 1). ICOS lacks the B7 binding MYYPPPY motif present in CD28 and CTLA-4 and binds a unique receptor, ICOS-L (B7RP-1, B7h, GL50, LICOS or B7-H2) which is expressed constitutively as a monomer on APC, including naive B cells [19].The ICOS:ICOSL signalling pathway plays a pivotal role in T helper 1 (Th1) and Th2 cell activation and in T cell–B cell co-operation. During early T cell activation by antigens, ICOS regulates Th2 cell differentiation by enhancing NFATc1 expression and initial IL-4 production which, in turn, regulates c-Mac expression [20]. In vivo studies show that ICOS is highly expressed within the T cell zones of secondary lymphoid organs and in the apical light zone of germinal centres (GCs), where T cells induce differentiation of B cells into memory cells and Ig secreting plasma cells [18,21].
Fig. 1.
Inducible co-stimulator molecule (ICOS):ICOS-L signalling can results in multiple pathways. Like CD28, ICOS can bind to the p85 subunit of phosphatidylinositol 3-kinase (PI3K) resulting in activation of lipid kinase (PDK1, PKB/Akt). ICOS can activate mitogen-activated protein kinases (MAPKs) and nuclear factor-kappa B (NF-κB) resulting in increased transcription of interleukin (IL)-10, IL-4 and interferon (IFN)-γ, prevention of cell death and up-regulation of cell metabolism. ICOS can also up-regulate CD40-L.
Evidence that ICOS:ICOSL is a unique receptor–ligand pair is given by the fact that both ICOS−/− and ICOS-L−/− mice show the same defects in T helper cell response and impaired humoral immunity. In particular, after secondary immunization, both ICOS−/− and ICOS-L−/− mice show reduced levels of serum IgE and IgG1 and defects in GCs formation [22–26].
Human ICOS deficiency was first described in 2003 by Grimbacher et al. [27]. Four patients from two unrelated families were found to carry a genomic homozygous deletion of exons 2 and 3 of the human ICOS gene resulting in a frameshift and a truncated protein of 28 amino acids, and resulting in complete loss of ICOS protein expression on the patients' activated T cells. All four patients were diagnosed with CVID and were hypogammaglobulinaemic with a decreased amount of all Ig classes resulting in recurrent bacterial infections. In these patients B cell numbers were reduced, with a considerable decrease in CD27+IgM–IgD– switched memory B cells and naive CD27–IgM+IgD+. Subsequent analysis of five further individuals including children revealed that total peripheral B cells, as well as naive B cells, were normal in number in children but showed a decrease in adults. Switched memory B cells were diminished in both children and adults. The phenotype and function of CD4+ T cells were normal, but the secretion of IL-10 and IL-17 was impaired [28]. These data strengthen the role of ICOS on activated T cells as an important regulator of late B cell differentiation, class switching, B cell memory formation and Ig production. Analysis of the few switched B cells in ICOS-deficient patients shows, however, that somatic hypermutation is relatively well preserved, suggesting that ICOS is necessary for T–B cell-mediated class switch recombination but not for somatic hypermutation.
In total, nine patients from four families have been identified with ICOS mutations and all carry the same large homozygous deletion [29,30]. ICOS deficiency is therefore an autosomal recessive disorder, and all four families are most probably descendent from a common founder, as all ICOS-deficient patients carry the same homozygous haplotype at the D2S2289 locus close to ICOS. The geographical location of all nine affected individuals also suggests a common founder and migration along the river Danube. Overall, the incidence of ICOS deficiency has been estimated in approximately 5%.
More recent studies confirm that ICOS plays a critical role as a co-factor for B cell help during T dependent antibody responses. As in ICOS−/− mice, in ICOS-deficient patients the formation of GCs is impaired and the GC reaction abrogated [28]. This may be due to the low secretion of IL-10 by ICOS-deficient CD4+ T cells, resulting in severely reduced numbers of CD27+ memory B cells and lack of plasma cells in the peripheral lymphoid organs of ICOS-deficient patients [28]. In particular, the GC-specific CD57+CXCR5+ T cell subpopulation normally producing elevated levels of IL-10 was absent in ICOS-deficient patients, and the number of circulating CXCR5+ T cells was also reduced compared to healthy donors [31]. In addition, Rasheed et al. [32] describe a subpopulation of terminally differentiated CXCR5+ follicular helper T (TFH) cells characterized by high levels of expression of CXCR5 and ICOS, named ICOShi CXCR5hi T cells. Taken together, these studies suggest a pivotal role of ICOS in the development of the GC TFH cells explaining the impaired formation of GCs in human ICOS-deficiency, as well as the reduced number of switched memory B cells and the lack of plasma cells in the lymphoid organs of these patients.
CD19 deficiency
B cell development and differentiation is critically dependent upon signal transduction through the B cell antigen receptor (BCR). Co-receptors associated with the BCR can modulate BCR signal transduction positively or negatively and in so doing influence B cell fate decisions at multiple stages of development. The CD19 molecule, together with CD21, CD81 and CD225, forms a BCR co-receptor complex that functions to lower the threshold for BCR signalling following antigen engagement [33,34]. This complex also acts as a link between the innate and adaptive immune systems and a mechanism by which complement-bound antigen can direct an adaptive T cell-dependent response. The CD21 component of the co-receptor complex contains a highly conserved extracellular motif that binds to C3d(g) bound antigen. In contrast, the CD19 molecule has two Ig-like extracellular domains and a cytoplasmic tail with motifs capable of binding the phosphotyrosine kinase, Lyn and PI3 kinase [34]. In this way CD21 serves as the ligand binding subunit that then links the recognition of antigen by complement to the signalling function of CD19. In CD19−/− mice a variety of defects of B cell development and function have been identified [35]. CD19−/− show normal B cell development in the bone marrow, but exhibit marked abnormalities in B-1, marginal zone (MZ) and germinal centre B cells. CD19−/− B cells also show a reduction in serum immunoglobulin production and a profound defect in response to T cell-dependent protein antigens. More recent data highlight the importance of the CD19 cytoplasmic signalling domain and in particular the role of PI3 kinase. Transgenic mice expressing a CD19 molecule with mutations in the cytoplasmic tail tyrosines 482 and 513 (Y482/Y513) [the binding sites of the p85α unit of phosphatidylinositol-3 kinase (PI3K)], show abnormalities of germinal centre B cell differentiation, maturation and proliferation [36]. These defects lead to a failure of production of specific antibodies after a primary response and a failure of high-affinity antibody production after TD antigen boosting. These defects were localized further to the follicular dendritic cell (FDC) zone of the germinal centre and suggest disruption at an early stage in GC B cell differentiation.
Four individuals from two unrelated consanguineous pedigrees (three individuals were siblings from the same family) have now been identified with mutations in CD19 [37]. All four presented in childhood with recurrent bacterial infections and on investigation were found to be hypogammaglobulinaemic with abnormal isohaemagglutinin production. Peripheral B cells were detected at normal numbers by surface expression of CD20, but in contrast CD19 expression was undetectable in one patient and barely detectable in the other three. Sequencing of the CD19 gene in one patient (P1) showed a single base pair (bp) insertion causing a frameshift and premature stop codon in the proximal region of the intracellular domain, thereby removing the majority of the intracellular domain. In the other three patients (patients 2, 3, 4), a homozygous 2 bp deletion again caused a frameshift mutation, and a premature stop codon again deleting a major part of the intracellular domain. Importantly, both mutations led to the production of proteins lacking the C-terminal tyrosine residues critical for CD19 signal transduction. Expression of intracellular CD19 protein was absent or decreased significantly, thereby confirming the flow cytometric expression results. Decreased CD19 expression led to a concomitant reduction in CD21 levels, although the expression of the other members of the co-receptor complex, CD81 and CD225, were normal.
Analysis of B cell compartments showed relatively normal numbers of B cells in the marrow with a normal distribution of precursor populations and normal B cell numbers in the periphery. All four patients had decreased numbers of CD5 B cells and reduced CD27+ memory B cells in comparison to age-matched healthy controls. Germinal centre formation was normal and in contrast to the murine knock-out model, somatic hypermutation in the small memory B cells showed patterns that were comparable to controls. However, B cells from CD19 patients exhibited defective calcium fluxes following IgM stimulation and P1 demonstrated poor proliferation following SAC (Staphyllococcus aureus Cowan I) stimulation. Rabies vaccination in all patients showed a significantly decreased secondary antibody response with poor avidity antibody formation.
In summary, the data suggest that CD19 mutations in these patients lead to relatively normal B cell development but the lack of CD19 signal transduction results in a poor response to antigenic stimuli and an inability to mount an effective humoral response. Unlike transmembrane activator and calcium-modulating ligand interactor (TACI) defects (see below), no autoimmune features or signs of lymphoproliferation were evident in either the four patients described or in murine CD19−/− strains [35,38].
TACI deficiency
Differentiation of mature B cells into effector cells capable of specific humoral immunity is strictly regulated. Tumour necrosis factor receptor superfamily (TNFRSF) members play important and diverse roles in the regulation of activation and apoptosis for specific cells of the immune system. CD40 (TNFRSF5) has important roles in B cell proliferation, differentiation and immunoglobulin isotype switching and mutations in CD40 lead to an autosomal recessive form of hyper-IgM syndrome [39]. CD95 or Fas (TNFRSF6) is a pro-apoptotic molecule which, if mutated, leads to autoimmune lymphoproliferative syndrome (ALPS) [40]. It is now evident that the survival and function of transitional and mature B cells is dependent upon another group of TNFRSF members. TACI [transmembrane activator and calcium modulator and cyclophilin ligand (CAML) interactor] and BCMA (B cell maturation antigen; TNFRSF17) are both expressed on B cells and interact with the ligands BAFF (B cell activation factor of the TNF family receptor) and APRIL (a proliferation-inducing ligand). BAFF also interacts with a unique receptor, BAFF-R (TNFRSF13C) [41,42].
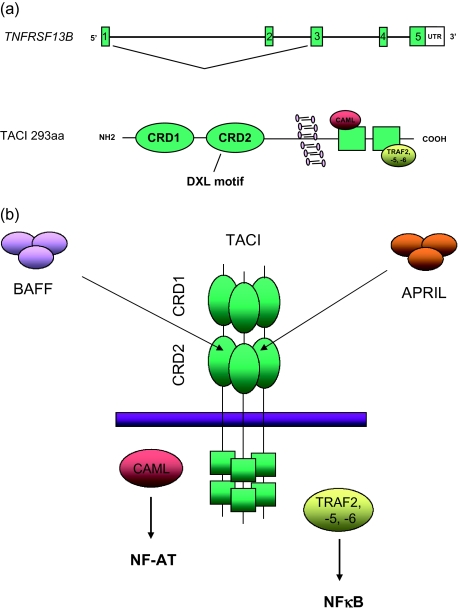
The extracellular domains of BAFF-R, TACI and BCMA contain a cysteine-rich domain (CRD). BAFF-R and BCMA contain one CRD while TACI contains two domains (CRD1 and CRD2), although the N-amino terminal CRD1 of TACI is probably dispensable and CRD2 alone can mediate ligand binding and cell signalling [43] (Fig. 2a,b). Crystal structure analysis shows that the CRDs form a short β-hairpin structure within which is located a common feature termed the DXL motif, a conserved 6-residue sequence, essential for ligand binding. While the hairpin alone is sufficient for BAFF binding, binding to APRIL requires a specific hydrophobic core that is present in TACI and BCMA but not in BAFF-R [43]. Following ligand binding, the intracellular domain of TACI can bind a number of TNF-associated factors (TRAFs) through two consensus TRAF binding sequences (Fig. 2a,b). In experimental systems TACI can bind TRAF-2, -5 and -6 and TRAF activation results in activation of nuclear factor-kappa B (NF-κB) and Jun amino terminal kinases [44]. The carboxy terminal portion of the TACI intracellular domain interacts with CAML up-regulating calcineurin and leading to NF-AT (nuclear factor of activated T cells transcription factor) dephosphorylation and nuclear translocation [45] (Fig. 2b). TACI, like other TNFRSF members such as Fas, requires ligand-induced trimerization or oligomerization for optimal signalling as TRAF molecules bind weakly to a single receptor but bind with high affinity to trimeric TNFRSF receptor complexes (Fig. 2b).
Fig. 2.
(a) Genomic organization and protein structure of transmembrane activator and calcium-modulating ligand (CAML) interactor (TACI). The gene encoding TACI, TNFRSF13B, consists of five exons (represented by boxes and not in scale) and codes for a 293 amino acids protein. TACI contains cysteine-rich domains 1 and 2 (CRD1 and CRD2) in the extracellular domain of TACI and binding sites for tumour necrosis factor (TNF)-associated factors (TRAFs -2, -5 and -6) and calcium modulator and cyclophilin ligand (CAML) in its intracellular domain. The highly conserved DXL motif is located in the CRD2 domain. An alternative splicing can occur, giving rise to a short TACI protein lacking the CRD1. (b) B cell activation factor of the TNF family receptor (BAFF) and a proliferation-inducing ligand (APRIL) bind to the CRD2 of TACI trimers. Intracellular signalling though CAML and TRAFs results in activation of nuclear factor of activated T cells transcription factor (NF-AT) and nuclear factor-kappa B (NF-κB).
TACI is expressed on all human peripheral B cells and is expressed preferentially on late transitional B cells and marginal zone B cells. Although the functions of the various receptors and ligands do overlap, in vitro studies and analysis of knock-out murine models allows us to attribute non-redundant functions to specific molecules (summarized in Fig. 3). In BAFF/BAFF-R/TACI/APRIL/BCMA knock-out mice, early B cell development in the bone marrow is unaffected but on exiting the bone marrow, transitional T1 and T2 splenic B cells are dependent upon BAFF/BAFF-R-mediated survival, as deletion of either of these genes results in a significant reduction of mature B cells in the circulation and in lymphoid organs [46–48]. In contrast, TACI-deficient mice show increased numbers of B cells and develop autoimmune manifestations with systemic lupus erythematosus (SLE)-like symptoms, lymphoproliferation with splenomegaly and development of lymphoma in 15% of mice, suggesting that TACI acts as a negative regulator of B cell development [38,49,50]. Total antibody production is mildly impaired in BAFF-R-deficient mice but T cell-independent type II (TI–II) responses are retained, whereas this response is impaired severely in TACI-deficient mice, suggesting that TI–II responses are mediated predominantly by TACI. Isotype class-switching, especially to IgA, in response to APRIL is abnormal in TACI-deficient mice and identifies a role for the APRIL–TACI interaction in IgA class-switch recombination [38]. The dependence of IgA CSR on an APRIL/TACI signal is supported further by defective IgA CSR in APRIL-deficient mice [51] and by elevated serum levels of IgA in APRIL transgenic mice [52]. These murine data are corroborated by studies on human B cells in which TACI expression was down-regulated by TACI-specific siRNA, and which again show that TACI/APRIL signalling is necessary for IgA production but also highlights the synergistic role played by APRIL binding to heparin sulphate proteoglycans [53]. BCMA-deficient mice, by contrast, show no significant defects in B cell production or antibody production [54]. These defects of B cell function and homeostasis, Ig production, class-switch recombination and impaired T independent responses in BAFF-R- and TACI-deficient mice are similar to the immunological abnormalities observed in CVID. Further, the autoimmune and lymphoproliferative manifestations seen in TACI-deficient mice are also seen in a subset of CVID patients and have thus led to the further study of these molecules in this disease.
Fig. 3.
Interactions of B cell activation factor of the TNF family receptor (BAFF) and a proliferation-inducing ligand (APRIL) with their receptors BAFF-R, transmembrane activator and calcium-modulating ligand (CAML) interactor (TACI) and B cell maturation antigen (BCMA) control B cell development and homeostasis. BAFF binds to BAFF-R, TACI and BCMA, while APRIL binds only to TACI and BCMA. The functional outcomes of BAFF and APRIL interactions with their receptors are listed. Simultaneous binding of APRIL to TACI and heparan sulphate proteoglycans mediates IgA production.
Two studies have identified mutations in TACI in two different cohorts of patients with CVID and IgAD [55,56]. As suggested previously, the same mutation within a pedigree led to CVID or IgAD in different individuals and suggests that the two immunological phenotypes are variants of the same gene defect. The inheritance pattern in the patients and pedigrees analysed was complex. Simple heterozygous, compound heterozygous and homozygous mutations have all been identified, with simple heterozygous changes the most frequent, implying both autosomal dominant and recessive patterns of inheritance. The mechanism by which simple heterozygous mutations exert a pathogenic effect is unclear. Assembly of trimeric TACI complexes is necessary for effective signalling and the recruitment of specific TRAFs to the TACI-cytoplasmic domain [44]. The presence of a mutant TACI allele may lead to the formation of trimeric receptor complexes containing both wild-type and mutant receptors, with the mutant exerting a dominant negative effect, although at present there are no data to support this model. Heterozygote TACI+/– mice show a normal phenotype but are not a good comparative model, as in these mice the TACI mutation results in complete loss of mutant protein expression. The most common CVID mutation identified is a 310T→C nucleotide substitution resulting in a C104R (cysteine 104→arginine) substitution in TACI exon 3 [55,56]. This mutation, found in the extracellular domain, disrupts a disulphide bond with C93 that is required for the formation of the CRD2 of TACI. In B cells from a patient with a homozygous C104R mutation, BAFF/APRIL binding was shown to be abolished, whereas in heterozygotes, binding of APRIL was preserved. Expression of the mutant in heterologous cell lines demonstrates clearly that the mutant form is stably expressed. The functional consequences of this mutation in the heterozygous state are therefore unclear, but are likely to relate to defective assembly of wild-type/mutant receptor complexes that interfere with normal binding and abrogated downstream signalling. Indeed, transfection of wild-type and mutant TACI forms into 293T cells to generate the heterozygous phenotype suggests that trimeric complexes containing both species can form and bind ligand but are unable to generate appropriate downstream signals [57].
Within our own patient cohort we have also identified a number of mutations not described previously that may allow us to dissect TACI function in more detail. In one individual, a single nucleotide insertion after nt572 leads to a premature stop and disruption of the amino acid sequence of the intracellular TACI domain and is predicted to abrogate intracellular signalling through TRAF and CAML binding domains. We have also identified two missense mutations in a compound heterozygote. The Y79C (tyrosine 79→cysteine) mutation destabilizes the hairpin structure of CRD2 and would be predicted to abolish both APRIL and BAFF binding. The second mutation (I87N; isoleucine 87→asparagine) is potentially more informative, in that I87 forms part of the hydrophobic core unique to TACI which is essential for binding to APRIL [43]. This mutation is functionally unique among all those described so far in that it would be predicted to preserve BAFF binding but abrogate APRIL association, and could thus highlight the importance of the APRIL/TACI interaction.
Data accumulated from the analysis of large multi-centre CVID cohorts (over 500 patients analysed) now suggest that mutations in TACI are found in ∼8–10% of CVID patients (unpublished data, Bacchelli et al., ESID, October 2006). Simple heterozygous mutations account for the vast majority of changes, with the C104R and A181E missense mutations the most common. A critical factor in determining the pathogenicity of these changes is their frequency in the normal healthy population. Our analysis and those of other groups suggest that these changes are found in healthy controls, but at a significantly decreased frequency for C104R. Data on the A181E change are conflicting, with data from our initial study showing an incidence in the normal population that is equivalent to that found in CVID patients, while a larger study combining European and American cohorts suggests that the incidence of A181E is greater in the CVID population and therefore may be pathogenic [58]. Interestingly, in this latter study both the C104R and A181E changes were not found at a significantly increased frequency in individuals with selective IgAD when compared to controls. The finding of these heterozygous changes in normal individuals questions whether these heterozygous TACI mutations/changes on their own are truly pathogenic or if, indeed, they act as susceptibility or disease-modifying mutations that act in co-operation with other gene defects. Further functional data from expression of these mutants in appropriately relevant in vitro and in vivo models are necessary to answer these clearly important issues. It is also worth noting that the C104R mutation has been found only in the homozygous form in CVID patients and no controls carrying a homozygous C104R defect have been identified, again suggesting that this variant is disease-causing, at least in homozygous form.
While TACI−/− mice showed increased B cell numbers and B cell hyperreactivity, normal B cell numbers were observed in TACI-deficient CVID patients. Analysis of the B cell phenotype in initial studies suggested a decrease in CD27+ switched memory B cells, but further analysis of large numbers of patients does not show any distinct B cell phenotype and no evidence of a genotype/phenotype correlation. Other characteristic features of the TACI-deficient murine model are the tendency to lymphoma and autoimmunity. Lymphoproliferative disease is seen in TACI-deficient patients with benign splenomegaly, tonsillar hyperplasia and follicular nodular hyperplasia of the gastrointestinal tract. However, these features are also seen in CVID patients without TACI mutations, and so comparative data from larger numbers of patients are required before we can determine whether this is a TACI-specific phenotype. The high incidence of severe SLE-like autoimmune symptoms observed in TACI−/− mice [38] is not seen in humans, although patients did show a tendency to autoimmune disease such as hypothyroidism, pernicious anaemia and autoimmune thrombocytopaenia. Again, data from large cohorts are required to determine whether these defects are specifically associated with the TACI mutations.
Defects in BAFF-R
The finding of defects in TACI, together with the understanding of the role of BAFF/BAFF-R signalling in B cell homeostasis and function, has led naturally to the hunt for BAFF-R defects in CVID patients. Despite the analysis of large cohorts only one patient, a 60-year-old male with hypogammaglobulinaemia, has been identified with a BAFF-R defect [59]. This individual had a 24-bp homozygous deletion in exon 2, which codes for the transmembrane region of the receptor. Limited information is published at present, but B cells from the patient expressed no BAFF-R and detailed B cell phenotyping revealed a block at the transitional B cell stage and is in keeping with the specific role of BAFF and BAFF-R in peripheral B cell survival. Other studies have detected heterozygous sequence variations in the BAFF-R gene, but these changes were also detected at a similar frequency in the normal population and are therefore unlikely to be pathogenic [60].
Summary
CVID has been characterized in many ways, and despite the variety of clinical, cellular and immunological defects described, the inability to make specific antibody responses remains a consistent hallmark of the disease phenotype. The identification of molecular defects in ICOS, CD19, TACI and BAFF-R is entirely consistent with this immunological phenotype, although each molecule disrupts B cell maturation, function and differentiation at a different stage. TACI and BAFF-R are critical to the maintenance of B cell homeostasis and have important roles in isotype class-switching. CD19 deficiency highlights the importance of antigen receptor signalling, while ICOS deficiency illustrates the absolute need for T:B cell interaction in co-ordinating an effective secondary humoral response. The defects identified in patients with CVID are supported by remarkably similar findings in the respective murine knock-out models, highlighting the important conserved roles of these molecules across species. Although four molecular defects have now been identified, it is likely that these abnormalities account for only ∼10–15% of all cases of CVID. Our understanding of the other molecules in B cell function and maturation including co-stimulatory molecules, cytokines and their receptors and other mediators of B cell homeostasis suggests that CVID will eventually (and probably not before too long) be categorized according to a variety of different molecular defects.
References
- 1.Cunningham-Rundles C. Physiology of Iga and Iga deficiency. J Clin Immunol. 2001;21:303–9. doi: 10.1023/a:1012241117984. [DOI] [PubMed] [Google Scholar]
- 2.Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies) Clin Immunol. 1999;93:190–7. doi: 10.1006/clim.1999.4799. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin Immunol. 1999;92:34–48. doi: 10.1006/clim.1999.4725. [DOI] [PubMed] [Google Scholar]
- 4.Di Renzo M, Pasqui AL, Auteri A. Common variable immunodeficiency: a review. Clin Exp Med. 2004;3:211–17. doi: 10.1007/s10238-004-0027-2. [DOI] [PubMed] [Google Scholar]
- 5.Warnatz K, Denz A, Drager R, et al. Severe deficiency of switched memory B cells (CD27(+) IgM(-) IgD(-) in subgroups of patients with common variable immunodeficiency: a new approach to classify a heterogeneous disease. Blood. 2002;99:1544–51. doi: 10.1182/blood.v99.5.1544. [DOI] [PubMed] [Google Scholar]
- 6.Kruetzmann S, Rosado MM, Weber H, et al. Human immunoglobulin M memory B cells controlling Streptococcus pneumoniae infections are generated in the spleen. J Exp Med. 2003;197:939–45. doi: 10.1084/jem.20022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kralovicova J, Hammarstrom L, Plebani A, Webster AD, Vorechovsky I. Fine-scale mapping at IGAD1 and genome-wide genetic linkage analysis implicate HLA-DQ/DR as a major susceptibility locus in selective IgA deficiency and common variable immunodeficiency. J Immunol. 2003;170:2765–75. doi: 10.4049/jimmunol.170.5.2765. [DOI] [PubMed] [Google Scholar]
- 8.Burrows PD, Cooper MD. IgA deficiency. Adv Immunol. 1997;65:245–76. [PubMed] [Google Scholar]
- 9.Volanakis JE, Zhu ZB, Schaffer FM, et al. Major histocompatibility complex class III genes and susceptibility to immunoglobulin A deficiency and common variable immunodeficiency. J Clin Invest. 1992;89:1914–22. doi: 10.1172/JCI115797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder HW, Jr, Zhu ZB, March RE, et al. Susceptibility locus for IgA deficiency and common variable immunodeficiency in the HLA-DR3-B8-A1 haplotypes. Mol Med. 1998;4:72–86. [PMC free article] [PubMed] [Google Scholar]
- 11.Vorechovsky I, Cullen M, Carrington M, Hammarstrom L, Webster AD. Fine mapping of IGAD1 in IgA deficiency and common variable immunodeficiency: identification and characterization of haplotypes shared by affected members of 101 multiple-case families. J Immunol. 2000;164:4408–16. doi: 10.4049/jimmunol.164.8.4408. [DOI] [PubMed] [Google Scholar]
- 12.Finck A, Van der Meer JW, Schaffer AA, et al. Linkage of autosomal-dominant common variable immunodeficiency to chromosome 4q. Eur J Hum Genet. 2006;14:867–75. doi: 10.1038/sj.ejhg.5201634. [DOI] [PubMed] [Google Scholar]
- 13.Schaffer AA, Pfannstiel J, Webster AD, Plebani A, Hammarstrom L, Grimbacher B. Analysis of families with common variable immunodeficiency (CVID) and IgA deficiency suggests linkage of CVID to chromosome 16q. Hum Genet. 2006;118:725–9. doi: 10.1007/s00439-005-0101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanegane H, Tsukada S, Iwata T, et al. Detection of Bruton's tyrosine kinase mutations in hypogammaglobulinaemic males registered as common variable immunodeficiency (CVID) in the Japanese Immunodeficiency Registry. Clin Exp Immunol. 2000;120:512–7. doi: 10.1046/j.1365-2249.2000.01244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nistala K, Gilmour KC, Cranston T, et al. X-linked lymphoproliferative disease: three atypical cases. Clin Exp Immunol. 2001;126:126–30. doi: 10.1046/j.1365-2249.2001.01599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weston SA, Prasad ML, Mullighan CG, Chapel H, Benson EM. Assessment of male CVID patients for mutations in the Btk gene: how many have been misdiagnosed? Clin Exp Immunol. 2001;124:465–9. doi: 10.1046/j.1365-2249.2001.01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eastwood D, Gilmour KC, Nistala K, et al. Prevalence of SAP gene defects in male patients diagnosed with common variable immunodeficiency. Clin Exp Immunol. 2004;137:584–8. doi: 10.1111/j.1365-2249.2004.02581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutloff A, Dittrich AM, Beier KC, et al. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;397:263–6. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 19.Carreno BM, Collins M. The B7 family of ligands and its receptors: new pathways for costimulation and inhibition of immune responses. Annu Rev Immunol. 2002;20:29–53. doi: 10.1146/annurev.immunol.20.091101.091806. [DOI] [PubMed] [Google Scholar]
- 20.Nurieva RI, Duong J, Kishikawa H, et al. Transcriptional regulation of th2 differentiation by inducible costimulator. Immunity. 2003;18:801–11. doi: 10.1016/s1074-7613(03)00144-4. [DOI] [PubMed] [Google Scholar]
- 21.Beier KC, Hutloff A, Dittrich AM, et al. Induction, binding specificity and function of human ICOS. Eur J Immunol. 2000;30:3707–17. doi: 10.1002/1521-4141(200012)30:12<3707::AID-IMMU3707>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 22.Dong C, Juedes AE, Temann UA, et al. ICOS co-stimulatory receptor is essential for T-cell activation and function. Nature. 2001;409:97–101. doi: 10.1038/35051100. [DOI] [PubMed] [Google Scholar]
- 23.McAdam AJ, Greenwald RJ, Levin MA, et al. ICOS is critical for CD40-mediated antibody class switching. Nature. 2001;409:102–5. doi: 10.1038/35051107. [DOI] [PubMed] [Google Scholar]
- 24.Tafuri A, Shahinian A, Bladt F, et al. ICOS is essential for effective T-helper-cell responses. Nature. 2001;409:105–9. doi: 10.1038/35051113. [DOI] [PubMed] [Google Scholar]
- 25.Mak TW, Shahinian A, Yoshinaga SK, et al. Costimulation through the inducible costimulator ligand is essential for both T helper and B cell functions in T cell-dependent B cell responses. Nat Immunol. 2003;4:765–72. doi: 10.1038/ni947. [DOI] [PubMed] [Google Scholar]
- 26.Nurieva RI, Mai XM, Forbush K, Bevan MJ, Dong C. B7h is required for T cell activation, differentiation, and effector function. Proc Natl Acad Sci USA. 2003;100:14163–8. doi: 10.1073/pnas.2335041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimbacher B, Hutloff A, Schlesier M, et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat Immunol. 2003;4:261–8. doi: 10.1038/ni902. [DOI] [PubMed] [Google Scholar]
- 28.Warnatz K, Bossaller L, Salzer U, et al. Human ICOS deficiency abrogates the germinal center reaction and provides a monogenic model for common variable immunodeficiency. Blood. 2006;107:3045–52. doi: 10.1182/blood-2005-07-2955. [DOI] [PubMed] [Google Scholar]
- 29.Salzer U, Maul-Pavicic A, Cunningham-Rundles C, et al. ICOS deficiency in patients with common variable immunodeficiency. Clin Immunol. 2004;113:234–40. doi: 10.1016/j.clim.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Ohm-Laursen L, Schjebel L, Jacobsen K, Permin H, Svejgaard A, Barington T. Normal ICOS, ICOSL and AID alleles in Danish patients with common variable immunodeficiency. Scand J Immunol. 2005;61:566–74. doi: 10.1111/j.1365-3083.2005.001603.x. [DOI] [PubMed] [Google Scholar]
- 31.Bossaller L, Burger J, Draeger R, et al. ICOS deficiency is associated with a severe reduction of CXCR5+CD4 germinal center Th cells. J Immunol. 2006;177:4927–32. doi: 10.4049/jimmunol.177.7.4927. [DOI] [PubMed] [Google Scholar]
- 32.Rasheed AU, Rahn HP, Sallusto F, Lipp M, Muller G. Follicular B helper T cell activity is confined to CXCR5(hi) ICOS(hi) CD4 T cells and is independent of CD57 expression. Eur J Immunol. 2006;36:1892–903. doi: 10.1002/eji.200636136. [DOI] [PubMed] [Google Scholar]
- 33.Carter RH, Fearon DT. CD19: lowering the threshold for antigen receptor stimulation of B lymphocytes. Science. 1992;256:105–7. doi: 10.1126/science.1373518. [DOI] [PubMed] [Google Scholar]
- 34.Fearon DT, Carroll MC. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu Rev Immunol. 2000;18:393–422. doi: 10.1146/annurev.immunol.18.1.393. [DOI] [PubMed] [Google Scholar]
- 35.Rickert RC, Rajewsky K, Roes J. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature. 1995;376:352–5. doi: 10.1038/376352a0. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Carter RH. CD19 regulates B cell maturation, proliferation, and positive selection in the FDC zone of murine splenic germinal centers. Immunity. 2005;22:749–61. doi: 10.1016/j.immuni.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 37.van Zelm MC, van der Reisli I, et al BM. An antibody-deficiency syndrome due to mutations in the CD19 gene. N Engl J Med. 2006;354:1901–12. doi: 10.1056/NEJMoa051568. [DOI] [PubMed] [Google Scholar]
- 38.Seshasayee D, Valdez P, Yan M, Dixit VM, Tumas D, Grewal IS. Loss of TACI causes fatal lymphoproliferation and autoimmunity, establishing TACI as an inhibitory BLyS receptor. Immunity. 2003;18:279–88. doi: 10.1016/s1074-7613(03)00025-6. [DOI] [PubMed] [Google Scholar]
- 39.Ferrari S, Giliani S, Insalaco A, et al. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc Natl Acad Sci USA. 2001;98:12614–9. doi: 10.1073/pnas.221456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieux-Laucat F, Le Deist F, Hivroz C, et al. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268:1347–9. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 41.Mackay F, Schneider P, Rennert P, Browning J. BAFF and APRIL: a tutorial on B cell survival. Annu Rev Immunol. 2003;21:231–64. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 42.Schneider P. The role of APRIL and BAFF in lymphocyte activation. Curr Opin Immunol. 2005;17:282–9. doi: 10.1016/j.coi.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Hymowitz SG, Patel DR, Wallweber HJ, et al. Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J Biol Chem. 2005;280:7218–27. doi: 10.1074/jbc.M411714200. [DOI] [PubMed] [Google Scholar]
- 44.Bishop GA. The multifaceted roles of TRAFs in the regulation of B-cell function. Nat Rev Immunol. 2004;4:775–86. doi: 10.1038/nri1462. [DOI] [PubMed] [Google Scholar]
- 45.von Bulow GU, Bram RJ. NF-AT activation induced by a CAML-interacting member of the tumor necrosis factor receptor superfamily. Science. 1997;278:138–41. doi: 10.1126/science.278.5335.138. [DOI] [PubMed] [Google Scholar]
- 46.Sasaki Y, Casola S, Kutok JL, Rajewsky K, Schmidt-Supprian M. TNF family member B cell-activating factor (BAFF) receptor-dependent and -independent roles for BAFF in B cell physiology. J Immunol. 2004;173:2245–52. doi: 10.4049/jimmunol.173.4.2245. [DOI] [PubMed] [Google Scholar]
- 47.Miller DJ, Hayes CE. Phenotypic and genetic characterization of a unique B lymphocyte deficiency in strain A/WySnJ mice. Eur J Immunol. 1991;21:1123–30. doi: 10.1002/eji.1830210506. [DOI] [PubMed] [Google Scholar]
- 48.Shulga-Morskaya S, Dobles M, Walsh ME, et al. B cell-activating factor belonging to the TNF family acts through separate receptors to support B cell survival and T cell-independent antibody formation. J Immunol. 2004;173:2331–41. doi: 10.4049/jimmunol.173.4.2331. [DOI] [PubMed] [Google Scholar]
- 49.Yan M, Wang H, Chan B, et al. Activation and accumulation of B cells in TACI-deficient mice. Nat Immunol. 2001;2:638–43. doi: 10.1038/89790. [DOI] [PubMed] [Google Scholar]
- 50.von Bulow GU, van Deursen JM, Bram RJ. Regulation of the T-independent humoral response by TACI. Immunity. 2001;14:573–82. doi: 10.1016/s1074-7613(01)00130-3. [DOI] [PubMed] [Google Scholar]
- 51.Castigli E, Scott S, Dedeoglu F, et al. Impaired IgA class switching in APRIL-deficient mice. Proc Natl Acad Sci USA. 2004;101:3903–8. doi: 10.1073/pnas.0307348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Planelles L, Carvalho-Pinto CE, Hardenberg G, et al. APRIL promotes B-1 cell-associated neoplasm. Cancer Cell. 2004;6:399–408. doi: 10.1016/j.ccr.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 53.Sakurai D, Hase H, Kanno Y, Kojima H, Okumura K, Kobata T. TACI regulates IgA production by APRIL in collaboration with HSPG. Blood. 2007;109:2961–7. doi: 10.1182/blood-2006-08-041772. [DOI] [PubMed] [Google Scholar]
- 54.Xu S, Lam KP. B-cell maturation protein, which binds the tumor necrosis factor family members BAFF and APRIL, is dispensable for humoral immune responses. Mol Cell Biol. 2001;21:4067–74. doi: 10.1128/MCB.21.12.4067-4074.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salzer U, Chapel HM, Webster AD, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. 2005;37:820–8. doi: 10.1038/ng1600. [DOI] [PubMed] [Google Scholar]
- 56.Castigli E, Wilson SA, Garibyan L, et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37:829–34. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 57.Garibyan L, Lobito AA, Siegel RM, Call ME, Wucherpfennig KW, Geha RS. Dominant-negative effect of the heterozygous C104R TACI mutation in common variable immunodeficiency (CVID) J Clin Invest. 2007;117:1550–7. doi: 10.1172/JCI31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan-Hammarstrom Q, Salzer U, Du L, et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat Genet. 2007;39:429–30. doi: 10.1038/ng0407-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warnatz K, Salzer U, Gutenburger S, et al. Finally found: human BAF-R deficiency causes CVID. XIth Meeting of the European Society for Immunodeficiencies. 2004;October [Google Scholar]
- 60.Losi CG, Silini A, Fiorini C, et al. Mutational analysis of human BAFF receptor TNFRSF13C (BAFF-R) in patients with common variable immunodeficiency. J Clin Immunol. 2005;25:496–502. doi: 10.1007/s10875-005-5637-2. [DOI] [PubMed] [Google Scholar]