Abstract
Modulation of host immunity is one of the proposed benefits of the consumption of probiotics. Nonetheless, comparative studies on the immunological properties that support the selection of strains of the same species for specific health benefits are limited. In this study, the ability of different strains of Bifidobacterium longum to induce cytokine production by peripheral blood mononuclear cells (PBMCs) has been evaluated. Live cells of all B. longum strains greatly stimulated regulatory cytokine interleukin (IL)-10 and proinflammatory cytokine tumour necrosis factor (TNF)-α production. Strains of the same species also induced specific cytokine patterns, suggesting that they could drive immune responses in different directions. The probiotic strain B. longum W11 stimulated strongly the production of T helper 1 (Th1) cytokines while B. longum NCIMB 8809 and BIF53 induced low levels of Th1 cytokines and high levels of IL-10. The effects of cell-surface components obtained by sonication of B. longum strains overall confirm the effects detected by stimulation of PBMCs with live cells, indicating that these components are important determinants of the immunomodulatory activity of B. longum. Genomic DNA of some strains stimulated the production of the Th1 and pro-inflammatory cytokines, interferon (IFN)-γ and TNF-α, but not that of IL-10. None of the cell-free culture supernatants of the studied strains was able to induce TNF-α production, suggesting that the proinflammatory component of these strains is associated mainly with structural cell molecules. The results suggest that despite sharing certain features, some strains can perform a better functional role than others and their careful selection for therapeutic use is desirable.
Keywords: Bacteria, Bifidobacterium, cytokines/interleukins, gut immunology/disease
Introduction
Probiotics are defined as live microorganisms that when ingested in appropriate numbers exert a beneficial effect on the host health, including the prevention and treatment of some pathologies [1]. It is generally accepted that a strain must satisfy a number of requirements to qualify as probiotic, such as the ability to survive the gastrointestinal transit, adhere to mucus and exhibit anti-microbial and immunostimulatory properties [2, 3]. The Food and Agriculture Organization/World Health Organization (FAO/WHO) have generated guidelines for evaluation and selection of probiotics. These include in vitro assessment of the above-mentioned desirable traits as well as in vivo demonstration of the biological effects of the potential probiotic strains by clinical trials [2]. Nevertheless, consensus on the methodology and criteria that should be applied for that purpose has not been reached [4]. The determination of appropriate methodology entails certain difficulties, including variability among strains, the lack of solid correlations between in vitro and in vivo test results and limited knowledge on their mechanisms of action [5]. According to the new European Union (EU) regulations on health claims of functional foods, probiotic-containing products should address specific benefits rather than general ones [6]. While this requirement implies a rational selection of strains for specific uses, thus far probiotic products for multiple applications seem to be common on the market [7].
It is recognized that each strain has unique and different properties and that the probiotic effects of a specific strain must not be extrapolated to other strains [4, 8]. In vitro evaluation tests indicated that properties such as tolerance to gastric pH and bile, resistance to antibiotics, production of anti-microbial compounds and adhesion ability vary depending on the considered strain [9, 10]. Strains of different species have usually been evaluated in a comparative manner, but those belonging to the same species have rarely been compared [11]. Modulation of host immunity is one of the most commonly proposed benefits of the consumption of probiotics. Lactobacillus and Bifidobacterium strains used as probiotics have been acknowledged for their role in preventing and treating acute gastrointestinal infections, allergy and atopic diseases and inflammatory bowel diseases [12–14]. The beneficial effects of these strains are based partly on their ability to regulate differentially the production of anti- and proinflammatory cytokines and the T helper 1 (Th1)/Th2 balance. Nevertheless, comparative studies on the immunological traits of strains of the same species that support a rational selection of probiotic strains for specific immunomodulatory benefits are limited [15, 16]. Moreover, adverse effects of the indiscriminate use of probiotics in early life and in patients at risk remain a possibility [5, 17]. The immune effects of probiotics can be exerted directly by live microbial cells, as the probiotic definition requires, but also by structural cell components (peptidoglycan, lipoteichoic acids and DNA) and secreted compounds [18–21]. This has led to questioning the requirement of viability in probiotic strains, although the effects of live cells tend to be greater than those of dead cells [22].
The aim of this study was to compare the immunological properties of strains of the same species, Bifidobacterium longum, including commercialized probiotics, human commensals and reference strains in order to progress on the criteria used for their evaluation and selection for clinical applications. The effects of structural cell components and secreted molecules of these strains were also evaluated to determine their relative contributions to the overall immune potential of each strain.
Materials and methods
Bacterial strains and culture conditions
The strains of the species B. longum used in the present study and their suppliers (in brackets) were the following: B. longum BB536 (Morinaga Milk Industry Co, Zama, Japan; provided by Comercial Química Massó SA, Barcelona, Spain); B. longum NCC 2705 (Nestlé SA, Lausanne, Switzerland); B. longum W 11 (Zirfos, Bologna, Italy); B. longum NCIMB 8809; B. longum American Type Culture collection (ATCC) 15707 and B. longum BIR 324 and BIF 53 (previously isolated from faeces) [23]. Briefly, strain isolation was carried out in MRSC agar containing 80 μg/ml mupirocin. Presumptive bifidobacteria were identified by genus-specific polymerase chain reaction (PCR) [24] and partial 16S rDNA sequencing with Y1 and Y2 primers, as described previously [25]. Strains were grown routinely in de Man, Rogosa and Sharpe (MRS) broth (Scharlau Chemie SA, Barcelona, Spain) supplemented with 0·05% (w/v) cysteine (Sigma, St Louis, MO, USA) (MRS-C) and incubated at 37°C under anaerobic conditions (AnaeroGen; Oxoid, Basingstoke, UK) for 22 h. Cells were harvested by centrifugation (6000 g for 15 min) during stationary growth phase, washed two times in phosphate-buffered saline (PBS, 130 mM sodium chloride, 10 mM sodium phosphate, pH 7·4), and resuspended in PBS containing 20% glycerol. Aliquots of these suspensions were frozen in liquid nitrogen and stored at −80°C until used. The number of live cells after freezing and thawing was determined by colony-forming unit (CFU) counting on MRS-C agar after 48 h incubation. For all strains tested, more than 90% cells were alive upon thawing and no significant differences were found during storage time (4 months). One fresh aliquot was thawed for every new experiment to avoid variability in the cultures between experiments. These samples constituted the live-cell suspensions. The obtained culture supernatants were filter-sterilized (0·22-μm pore size filter, Millipore, Bedford, MA, USA) to eliminate the possible presence of viable cells and their pHs were adjusted to 7·2 with 1 N NaOH. Aliquots of cell-free culture supernatants were kept at −80°C until used.
Extraction of cell-surface components
In order to prepare samples containing cell-surface components the cell suspensions, obtained as described above and adjusted to 1 × 106 CFU/ml, were submitted to five sonication cycles for 15 min each, with 2-min intervals on ice in a Braun-sonic sonicator 1510 (Braun Biotech Inc., Allentown, PA, USA) at 100 W output. After this treatment the absence of viability losses was confirmed by plate counting as described above. Cells were separated by centrifugation (10 000 g, 10 min) and the supernatants were stored at −80°C until used. The protein concentration of surface protein fractions was determined by Lowry's method using a total protein kit (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer's procedure. These fractions were used at a final concentration of 100 μg protein/ml for peripheral blood mononuclear cells (PBMCs) stimulation assays.
Preparation of genomic DNA
Isolation of genomic DNA from B. longum strains was performed using the QIAamp DNA stool kit (Qiagen, Barcelona, Spain) according to the manufacturer's procedure. Concentration and purity of all DNA preparations were confirmed on 1% agarose gels using standards of DNA concentration (Lambda DNA; Invitrogen, Barcelona, Spain).
Isolation and stimulation of PBMCs
PBMCs were isolated from heparinized peripheral blood of four healthy volunteers (median age 30 years, range 24–40 years) after written consent was obtained. The study protocol was approved by the CSIC Committee on Ethical Practice. Briefly, PBMCs were isolated by centrifugation over a Ficoll density gradient (Amersham Biosciences, Piscataway, NJ, USA), washed with RPMI-1640 (Cambrex, New York, USA), and adjusted to 1 × 106 cells/ml in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) (Gibco, Barcelona, Spain), 2 mM l-glutamine, 100 μg/ml streptomycin and 100 U/ml penicillin (Sigma).
PBMCs were incubated in 24-well flat-bottomed polystyrene microtitre plates (Corning, Madrid, Spain) in the presence or absence of different stimulants at 37°C under 5% CO2 for 24 h. Final concentrations of either 1 × 106 CFU/ml live-cell suspensions, l50 μl cell-free culture supernatants, 100 μg cell-surface components or 2 μg/ml genomic DNAs from each B. longum strain were used as stimulants. Purified lipopolysaccharide (LPS) from Escherichia coli O111:B4 (Sigma) was used to stimulate PBMCs at a concentration of 1 μg/ml as a positive control. Non-stimulated PBMCs were also evaluated as controls of basal cytokine production. All reagents were tested by the E-toxate test for LPS (Sigma) and shown to be below the limit of detection (2 pg/ml). Every fraction used as stimulant was assayed in duplicate, and each experiment was performed with PBMCs from four donors. Cell culture supernatants were collected by centrifugation, fractionated in aliquots and stored at −20°C until cytokines were analysed.
Cytokine assays
Cytokine concentrations of supernatants were measured by human Th1/Th2 enzyme-linked immunosorbent assay (ELISA) Ready SET Go! Kit (BD Bioscience, San Diego, CA, USA), including interleukin (IL)-2 and interferon (IFN)-γ as Th1-type and IL-4 and IL-10 as Th2-type cytokines. The proinflammatory cytokine TNF-α was also measured using the ELISA kit from e-Bioscience (BD Biosciences). The detection procedures were performed according to the manufacturer's instructions. The sensitivity of assays for each cytokine was as follows: 4 pg/ml for IL-2, IFN-γ and tumour necrosis factor (TNF)-α, and 2 pg/ml for IL-4 and IL-10.
Statistical analyses
The differences between means of produced cytokines in response to different bacterial stimuli were determined by applying the least significant difference (LSD) test using the StatGraphics software (Manugistics, Rockville, MD, USA). Differences were considered to be statistically significant at P < 0·05. Data are expressed as mean and standard deviation (s.d.) of duplicates measures determined in four independent experiments.
Results
Effects of live bacteria on cytokine production
The results of the effects of live bacteria of different B. longum strains on cytokine production by PBMCs are shown in Fig. 1. Strain-dependent effects were detected in the stimulation of production of all tested cytokines except for the Th2 cytokine IL-4, which was not induced by any of the assayed strains. Live cell suspensions of all strains induced the production of significantly higher amounts (P < 0·01) of the regulatory cytokine IL-10 than the non-stimulated PBMCs. The stimulation of PBMCs by the strains B. longum BIR 324 and W11 induced the production of IL-10 levels similar to those obtained by LPS stimulation. The strains B. longum ATCC 15707 and NCC 2705 showed a significantly higher (P < 0·01) ability to induce the production of IL-10 than the remaining strains, followed by B. longum BIF53, NCIMB 8809 and BB536 (P < 0·05). In contrast, B. longum W11 was the only strain that induced the production of significantly higher levels (P < 0·05) of both Th1-type cytokines, IL-2 and IFN-γ, compared to the non-stimulated and LPS-stimulated cells. In addition, B. longum BB536 and NCC 2705 induced the production of higher amounts (P < 0·05) of IFN-γ, and B. longum BIR324 and ATCC 15707 induced the production of higher amounts (P < 0·05) of IL-2 than the non-stimulated and LPS-stimulated cells. All strains induced the production of significantly higher amounts (P < 0·01) of TNF-α than the non-stimulated and LPS-stimulated cells. The production of this cytokine was particularly high upon stimulation with B. longum BIF53, ATCC 15707, NCC 2705 and, secondly, with B. longum NCIMB 8809 and BB536. In general, live cells of all B. longum strains were potent stimulants of IL-10 and TNF-α production, suggesting a role in immune-regulation and promotion of innate defences. Specific strains also induced specific cytokine responses at the assayed concentrations. Thus, the probiotic B. longum W11 stimulated strongly the production of Th1 cytokines (IL-2 and IFN-γ), while showing the lowest ability to induce the regulatory and Th2 cytokine IL-10. B. longum BB536 induced a similar cytokine profile regarding IFN-γ and IL-10 production. In contrast, B. longum NCIMB 8809 and BIF53 were among the strains inducing the production of the lowest levels of the Th1 cytokines (IL-2 and IFN-γ), while inducing higher IL-10 production. B. longum ATCC 15707 induced a similar cytokine profile regarding IFN-γ and IL-10. Therefore, different strains of the same species are potentially able to drive immune responses into opposite directions in vitro, suggesting different immune potentials in clinical practice in vivo.
Fig. 1.
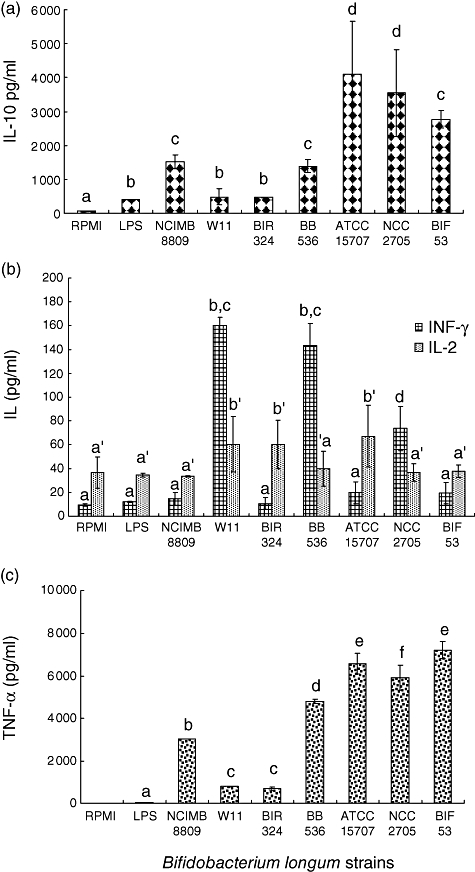
Cytokine production by peripheral blood mononuclear cells (PBMCs) stimulated with live bacteria of different Bifidobacterium longum strains. Purified lipopolysaccharide (1 μg/ml) from Eschericia coli was used as a positive control. Non-stimulated PBMCs were also evaluated as controls of basal cytokine levels. Results are expressed as mean ± s.d. of duplicate measures determined in four independent experiments. Significant differences among samples were established by using the least significant difference test. Means in the same graphic with different letters (a–f or a′–f′) were significantly different (P < 0·05).
Effects of cell-surface components on cytokine production
The effects of cell-surface components obtained by sonication of B. longum strains were also evaluated, as cell surface components are potentially responsible for the immune functions of probiotic strains (Fig. 2). The stimulation of IL-10 production was the highest in the case of cell-surface components of B. longum NCC 2705 ATCC 15707 and BIF53 according to the results obtained with live cell suspensions. Cell-surface components of B. longum ATCC 15707 and BIF53 were those that induced the highest production (P < 0·05) of both Th1-type cytokines, IL-2 and IFN-γ, which was not in agreement with the results obtained by live cell stimulation. Cell-surface components of B. longum NCC 2705 induced significantly higher (P < 0·05) IFN-γ production when compared with non-stimulated and LPS-stimulated cells, according to the effects of live cells. The surface-protein fractions of B. longum BIF53, ATCC 15707 and NCC 2705 showed the highest effects (P < 0·05) on TNF-α production, in agreement with the results obtained when PBMCs were stimulated by live cells of the same strains. Although to a lesser extent, the effects of cell-surface components of B. longum NCIMB 8809 on TNF-α were also higher (P < 0·05) than in non-stimulated and LPS-stimulated cells, in agreement with the results obtained by stimulation of live cells. The probiotic strain B. longum BB536 was the only exception to this trend.
Fig. 2.
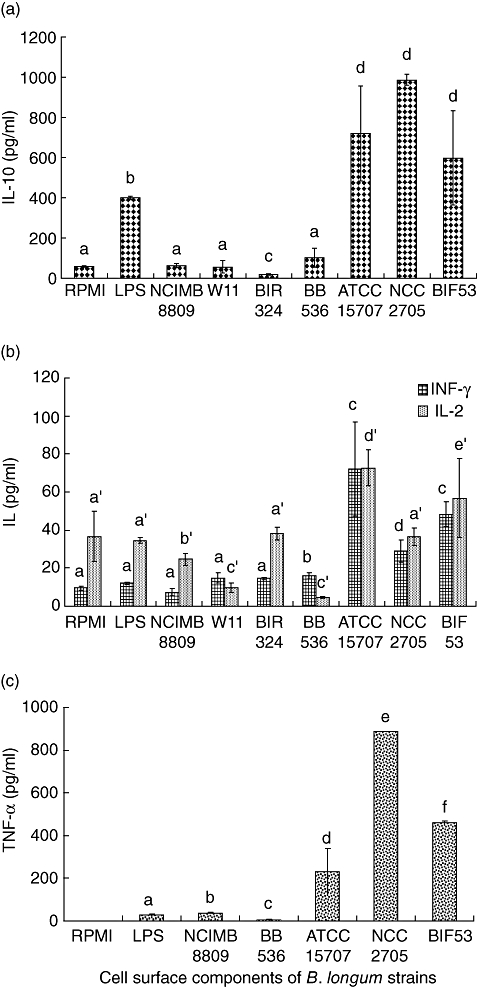
Cytokine production by peripheral blood mononuclear cells stimulated with cell-surface components of different Bifidobacterium longum strains (See Fig. 1 for details).
Effects of cell-free culture supernatants on cytokine production
The immune effects of compounds secreted to cell-free culture supernatants on cytokine production are shown in Fig. 3. All supernatants of B. longum cultures induced the production of significantly higher amounts of IL-10 than those found in the non-stimulated cells, but not superior to the amounts found in the LPS-induced cells. The supernatant of B. longum ATCC 15707 and BIR-324 significantly induced (P < 0·05) IFN-γ production and that of B. longum NCC 2705 ATCC 15707 IL-2 production, when compared with non-stimulated and LPS-stimulated cells. These effects did not correspond to those detected with live cells, suggesting that compounds secreted to the extracellular medium other than those associated with the cell-surface could mediate part of the immune effects. Induction of TNF-α by cell-free culture supernatants of the studied strains was not detected in any case.
Fig. 3.
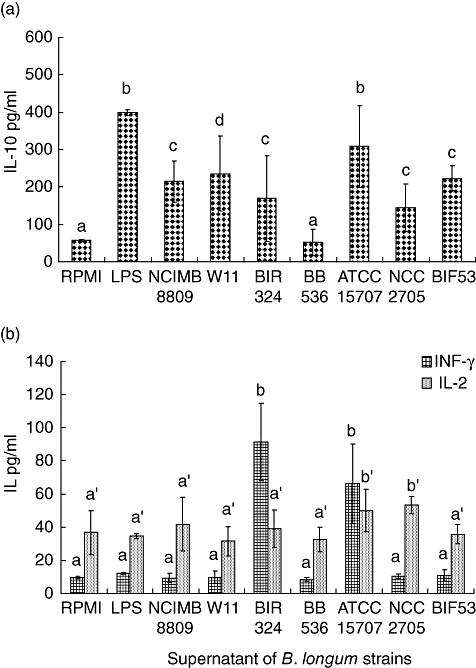
Cytokine production by peripheral blood mononuclear cells stimulated with the supernatant of different Bifidobacterium longum strains (See Fig. 1 for details).
Effects of DNA on cytokine production
The effects of pure genomic DNA of different B. longum strains on stimulation of cytokine production by PBMCs are shown in Table 1. The production of the Th1 cytokine IL-2 and the Th2 cytokine IL-4 were not induced by PBMCs stimulated with the DNA of any B. longum strain. DNA from B. longum NCIMB 8809 and the probiotic strain B. longum W11 showed a significantly higher (P < 0·05) ability to stimulate the production of the Th1-type cytokine IFN-γ and the cytokine levels were also higher than those found in LPS-stimulated PBMCs. None of the DNA from B. longum strains was able to induce IL-10 production significantly. In fact, compared to non-stimulated cells, the production of IL-10 was lower in all cases. This can be a consequence of an insufficient concentration of stimulant that, unexpectedly, seemed to affect negatively basal cytokine levels. DNA from B. longum NCC 2705, the probiotic strain B. longum BB536 and the laboratory isolate B. longum BIR 324 showed a significantly higher (P < 0·05) ability to induce the production of the proinflammatory cytokine TNF-α, but B. longum BB536 induced the highest values when compared with the other strains.
Table 1.
Cytokine production by peripheral blood mononuclear cells (PBMCs) stimulated with genomic DNA from Bifidobacterium strains.
| Cytokine production† | |||
|---|---|---|---|
| Strains | IFN-γ (pg/ml) | TNF-α (pg/ml) | IL-10 (pg/ml) |
| RPMI-1640 | 9·5 ± 1·0a | n.d. | 57·7 ± 2·6a |
| LPS | 11·9 ± 0·5a | 27·8 ± 3·1a | 398 ± 7·8b |
| NCIMB 8809 | 92·1 ± 7·2b | n.d. | 20·46 ± 3·1c |
| W11 | 131·0 ± 13·7b | n.d. | 19·56 ± 2·6c |
| BIR 324 | 33·0 ± 7·1c | 49·1 ± 18·2b | 13·85 ± 1·3d |
| BB 536 | 38·4 ± 8·6c | 308·5 ± 32·9e | 10·23 ± 0·9d |
| ATTC 15707 | 33·3 ± 3·0c | 17·1 ± 5·9a | 10·34 ± 0·8c |
| NCC 2705 | 35·8 ± 7·1c | 122·3 ± 2·6d | 9·33 ± 0·9d |
| BIF 53 | 23·7 ± 0·7c | n.d. | 12·04 ± 1·0d |
Data are expressed as means ± s.d. of duplicate measures determined in four independent assays. Significant differences among samples were established by using the least significant difference test. Means in the same column showing different letters (a–e) were significantly different (P < 0·05). IFN: interferon; IL: interleukin; LPS: lipopolysaccharide; n.d.: non-detectable; TNF: tumour necrosis factor.
Discussion
In this study, a large variation in the ability of different strains of the same Bifidobacterium species to modulate the in vitro production of cytokines by PBMCs has been detected, suggesting different functional roles. A few studies on the species-specific effects of bifidobacteria on immunocompetent cells have been carried out previously [8, 15]. None the less, to our knowledge, this is the first comparative study of the immunoproperties of different strains of the same species. Here, a strain-specific pattern of cytokine production, particularly of IL-10, IFN-γ and TNF-α, has been demonstrated by comparing reference strains, human isolates and commercial probiotics of B. longum. A common feature of the tested strains was their ability to induce IL-10 production, although to a different extent, which was not the case for IL-4, as reported in other studies for different species of lactic acid bacteria [16]. Bifidobacterium strains that were good inducers of IL-10 production were also good inducers of TNF-α production, as indicated in previous studies [22, 26].
In the present study, the induction of production of the Th1 cytokine IFN-γ seemed to be correlated inversely with the production of IL-10 in the strain B. longum W11 and to a lesser extent in BB536, while the opposite trend was observed in the strains B. longum ATCC15707, 8809 and BIF53. Numerous studies support the view that IL-10 exerts a strong suppressive effect on Th1 lymphocytes, antigen-presenting cells and the production of inflammatory mediators [27, 28]. Thus, IL-10 can counteract the production of other cytokines such as IFN-γ or IL-4. Considering our results, strains such as B. longum W11 and BB536 could provide protection against the early stage of infection via Th1 and TNF-α production [29]. In fact, B. longum W11 is claimed to help in the treatment and prevention of gastrointestinal infections and reduce bacterial overgrowth in irritable bowel syndrome (IBD) [26, 30]. In addition, B. longum BB536 was reported to help in fighting pathogens and alleviating symptoms of allergy and atopic diseases by promoting Th1- and counteracting Th2-immune responses [29]. In contrast, strains showing a regulatory cytokine profile characterized by stronger induction of IL-10, such as B. longum ACTT 15707, NCIMB 8809, BIF53 and NCC 2705, could play important regulatory roles against Th1- biased immune response in IBD patients, for example.
The effects of cell-surface components obtained from B. longum strains by sonication overall confirm the effects of live cells on cytokine production, indicating that surface structures are important in determining immunostimulating activity of probiotic bacteria [31, 32]. The differential effects detected in some cases between the use of live cells and cell-surface components as stimulants might be due to differences in the efficiency of the sonication procedure to extract bioactive components among strains. In addition, compounds other than those obtained by sonication could have been involved in immune stimulation. TNF-α production was only slightly induced by cell-surface components in four of the seven B. longum strains. The production of this cytokine in response to bacteria has been related to tertiary configuration of their peptidoglycan and surface proteins, which could also have been disturbed during extraction by sonication [33].
It has also been demonstrated that compounds released to culture supernatants by some of the studied B. longum strains could play an additional role in immunomodulation. Soluble factors of probiotic or commensal bacteria have been demonstrated to have immune functions, but there are still few reports on the identification of those bioactive compounds [21]. An interesting finding of this work is that none of the supernatants were able to induce TNF-α, indicating that the proinflammatory component of B. longum is associated only with structural cell components.
The immunostimulatory effects of DNA from different B. longum strains led to the production of the proinflammatory cytokines IFN-γ and TNF-α. DNA extracted from pure cultures of the probiotic mixture VSL#3 and from human faeces collected after probiotic ingestion influence cytokine production by PBMC decreasing IL-1β and increasing IL-10 [18]. The ability of the probiotic mixture VSL#3 to attenuate experimental colitis was also shown to be mediated by DNA via TLR9 signalling [34]. It has been suggested that the high guanine cytosine (GC) content of the Bifidobacterium genus (58–61%) could favour IL-10 production. The differential effects exerted by the studied B. longum strains could be due to differences in the presence or redundancy of CpG motifs in their DNAs, as it is demonstrated that some of these motifs exert a more pronounced immunomodulatory effect than others [35]. In contrast to what has been reported in other studies, the induction of IL-10 production upon stimulation of PBMCs with genomic DNA of B. longum strains was not higher than that induced by LPS, although lower concentrations of stimuli were used in this study [18]. The oligodeoxynucleotide BL07S identified in B. longum BB536 has been reported to exert anti-allergy effects in vitro and in vivo by increasing Th1 and decreasing Th2 cytokine production [36, 37]. In this study the DNA from this strain was, however, not exerting the strongest immunostimulatory effect on Th1-type cytokine production compared to the DNA from other tested strains. Although the study of individual traits of a strain is not enough to predict its functionality, the results presented highlight the need for conducting a comparative evaluation of strains of the same species for an improved selection of probiotics for specific uses.
Overall, B. longum strains have shown to divert immune responses into different directions in vitro, either towards a proinflammatory or a regulatory profile. This suggests that different strains may have different functional roles and applications in diverse pathological conditions. The fact that scientific publications suggest that similar effects (alleviation of lactose intolerance, improvement of defences against acute infections, etc.) are produced by different strains must not lead to generalizations about probiotic effects. Some strains may have functional traits in common but not all do, and studies on specific strains are needed [5]. Even sharing certain features, the obtained results suggest that some strains can perform a functional role better than others and so a careful selection of strains for therapeutic use is desirable.
Acknowledgments
This work was supported by grant AGL2005-05788-C02-01 from the Spanish Ministry of Science and Education and grant 200570F0091 from CSIC (Spain).
References
- 1.Havenaar R, Huis in't Veld JHJ. Probiotics: a general view. In: Wood BJB, editor. The lactic acid bacteria vol. 1. The lactic acid bacteria in health and disease. London: Chapman & Hall; 1992. pp. 209–24. [Google Scholar]
- 2.Food and Agriculture Organization of the United Nations and World Health Organization (FAO/WHO). Guidelines for the evaluation of probiotics in food. London/Ontario/Canada: FAO/WHO.; [Google Scholar]
- 3.Tuomola E, Crittenden R, Playne M, Isolauri E, Salminen S. Quality assurance criteria for probiotic bacteria. Am J Clin Nutr. 2001;73:393–8. doi: 10.1093/ajcn/73.2.393s. [Review]. [DOI] [PubMed] [Google Scholar]
- 4.Pineiro M, Stanton C. Probiotic bacteria: legislative framework − requirements to evidence basis. J Nutr. 2007;137:850–3. doi: 10.1093/jn/137.3.850S. [DOI] [PubMed] [Google Scholar]
- 5.Daniel C, Poiret S, Goudercourt D, Dennin V, Leyer G, Pot B. Selecting lactic acid bacteria for their safety and functionality by use of a mouse colitis model. Appl Environ Microbiol. 2006;72:5799–805. doi: 10.1128/AEM.00109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Wright A. Regulating the safety of probiotics − the European approach. Curr Pharm Des. 2005;11:17–23. doi: 10.2174/1381612053382322. [Review]. [DOI] [PubMed] [Google Scholar]
- 7.Gueimonde M, Salminen S. New methods for selecting and evaluating probiotics. Dig Liver Dis. 2006;38:242S–7S. doi: 10.1016/S1590-8658(07)60003-6. [DOI] [PubMed] [Google Scholar]
- 8.Boyle RJ, Robins-Browne RM, Tang ML. Probiotic use in clinical practice: what are the risks? Am J Clin Nutr. 2006;83:1256–64. doi: 10.1093/ajcn/83.6.1256. [Review]. [DOI] [PubMed] [Google Scholar]
- 9.Dunne C, O'Mahony L, Murphy L, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001;73:386–92. doi: 10.1093/ajcn/73.2.386s. [Review]. [DOI] [PubMed] [Google Scholar]
- 10.Huis in't Veld J, Havenaar R, Marteau P. Establishing a scientific basis for probiotic. R&D Trends Biotechnol. 1994;12:6–8. doi: 10.1016/0167-7799(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 11.Haller D, Colbus H, Gänzle MG, Scherenbacher P, Bode C, Hammes WP. Metabolic and functional properties of lactic acid bacteria in the gastro-intestinal ecosystem: a comparative in vitro study between bacteria of intestinal and fermented food origin. Syst Appl Microbiol. 2001;24:218–26. doi: 10.1078/0723-2020-00023. [DOI] [PubMed] [Google Scholar]
- 12.Ishida Y, Nakamura F, Kanzato H, et al. Effect of milk fermented with Lactobacillus acidophilus strain L-92 on symptoms of Japanese cedar pollen allergy: a randomized placebo-controlled trial. Biosci Biotechnol Biochem. 2005;69:1652–60. doi: 10.1271/bbb.69.1652. [DOI] [PubMed] [Google Scholar]
- 13.Ohno H, Tsunemine S, Isa Y, Shimakawa M, Yamamura H. Oral administration of Bifidobacterium bifidum G9-1 suppresses total and antigen specific immunoglobulin E production in mice. Biol Pharm Bull. 2005;28:1462–6. doi: 10.1248/bpb.28.1462. [DOI] [PubMed] [Google Scholar]
- 14.Gionchetti P, Lammers KM, Rizzello F, Campieri M. VSL#3: an analysis of basic and clinical contributions in probiotic therapeutics. Gastroenterol Clin North Am. 2005;34:499–513. doi: 10.1016/j.gtc.2005.05.003. [Review]. [DOI] [PubMed] [Google Scholar]
- 15.Young SL, Simon MA, Baird MA, et al. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin Diagn Lab Immunol. 2004;11:686–90. doi: 10.1128/CDLI.11.4.686-690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niers LE, Timmerman HM, Rijkers GT, et al. Identification of strong interleukin-10 inducing lactic acid bacteria which down-regulate T helper type 2 cytokines. Clin Exp Allergy. 2005;35:1481–9. doi: 10.1111/j.1365-2222.2005.02375.x. [DOI] [PubMed] [Google Scholar]
- 17.Boyle RJ, Tang ML. The role of probiotics in the management of allergic disease. Clin Exp Allergy. 2006;36:568–76. doi: 10.1111/j.1365-2222.2006.02472.x. [Review]. [DOI] [PubMed] [Google Scholar]
- 18.Lammers KM, Brigidi P, Vitali B, et al. Immunomodulatory effects of probiotic bacteria DNA: IL-1 and IL-10 response in human peripheral blood mononuclear cells. FEMS Immunol Med Microbiol. 2003;38:165–72. doi: 10.1016/S0928-8244(03)00144-5. [DOI] [PubMed] [Google Scholar]
- 19.Pena JA, Versalovic J. Lactobacillus rhamnosus GG decreases TNF α production in lipopolysaccharide-activated murine macrophages by a contact-independent mechanism. Cell Microbiol. 2003;5:277–85. doi: 10.1046/j.1462-5822.2003.t01-1-00275.x. [DOI] [PubMed] [Google Scholar]
- 20.Matsuguchi T, Takagi A, Matsuzaki T, et al. Lipoteichoic acids from Lactobacillus strains elicit strong tumor necrosis factor alpha-inducing activities in macrophages through Toll-like receptor 2. Clin Diagn Lab Immunol. 2003;10:259–66. doi: 10.1128/CDLI.10.2.259-266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoarau C, Lagaraine C, Martin L, Velge-Roussel F, Lebranchu Y. Supernatant of Bifidobacterium breve induces dendritic cell maturation, activation, and survival through a Toll-like receptor 2 pathway. J Allergy Clin Immunol. 2006;117:696–702. doi: 10.1016/j.jaci.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 22.Miettinen M, Vuopio-Varkila J, Varkila K. Production of human tumor necrosis factor alpha, interleukin-6, and interleukin-10 is induced by lactic acid bacteria. Infect Immun. 1996;64:5403–5. doi: 10.1128/iai.64.12.5403-5405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collado MC, Gueimonde M, Hernandez M, Sanz Y, Salminen S. Adhesion of selected Bifidobacterium strains to human intestinal mucus and the role of adhesion in enteropathogen exclusion. J Food Prot. 2005;68:2672–8. doi: 10.4315/0362-028x-68.12.2672. [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl Environ Microbiol. 1997;63:1268–73. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gueimonde M, Delgado S, Mayo B, Ruas-Madiedo P, Margolles A, De Los Reyes-Gavilan C. Viability and diversity of probiotic Lactobacillus and Bifidobacterium populations included in commercial fermented milks. Food Res Int. 2004;37:839–50. [Google Scholar]
- 26.Gackowska L, Michalkiewicz J, Krotkiewski M, Helmin-Basa A, Kubiszewska I, Dzierzanowska D. Combined effect of different lactic acid bacteria strains on the mode of cytokines pattern expression in human peripheral blood mononuclear cells. J Physiol Pharmacol. 2006;57:13–21. [PubMed] [Google Scholar]
- 27.Feng CG, Kullberg MC, Jankovic D, et al. Transgenic mice expressing human interleukin-10 in the antigen-presenting cell compartment show increased susceptibility to infection with Mycobacterium avium associated with decreased macrophage effector function and apoptosis. Infect Immun. 2002;70:6672–9. doi: 10.1128/IAI.70.12.6672-6679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balcewicz-Sablinska MK, Gan H, Remold HG. Interleukin 10 produced by macrophages inoculated with Mycobacterium avium attenuates mycobacteria-induced apoptosis by reduction of TNF-alpha activity. J Infect Dis. 1999;180:1230–7. doi: 10.1086/315011. [DOI] [PubMed] [Google Scholar]
- 29.Sanz Y, Nadal I, Sánchez E. Probiotics as drugs against human gastrointestinal infections. Recent Patents Anti-infect Drug Discov. 2007;2:148–56. doi: 10.2174/157489107780832596. [DOI] [PubMed] [Google Scholar]
- 30.Collado MC, Gueimonde M, Sanz Y, Salminen S. Adhesion properties and competitive pathogen exclusion ability of bifidobacteria with acquired acid resistance. J Food Prot. 2006;69:1675–9. doi: 10.4315/0362-028x-69.7.1675. [DOI] [PubMed] [Google Scholar]
- 31.Grangette C, Nutten S, Palumbo E, et al. Enhanced anti-inflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–6. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto S, Hara T, Hori T, et al. Probiotic Lactobacillus-induced improvement in murine chronic inflammatory bowel disease is associated with the down-regulation of pro-inflammatory cytokines in lamina propria mononuclear cells. Clin Exp Immunol. 2005;140:417–26. doi: 10.1111/j.1365-2249.2005.02790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timmerman CP, Mattsson E, Martinez-Martinez L, et al. Induction of release of tumor necrosis factor from human monocytes by staphylococci and staphylococcal peptidoglycans. Infect Immun. 1993;61:4167–72. doi: 10.1128/iai.61.10.4167-4172.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rachmilewitz D, Katakura K, Karmeli F, et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. 2004;126:520–8. doi: 10.1053/j.gastro.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Yi AK, Yoon JG, Yeo SJ, Hong SC, English BK, Krieg AM. Role of mitogen-activated protein kinases in CpG DNA-mediated IL-10 and IL-12 production: central role of extracellular signal-regulated kinase in the negative feedback loop of the CpG DNA-mediated Th1 response. J Immunol. 2002;168:4711–20. doi: 10.4049/jimmunol.168.9.4711. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi N, Kitazawa H, Iwabuchi N, et al. Immunostimulatory oligodeoxynucleotide from Bifidobacterium longum suppresses Th2 immune responses in a murine model. Clin Exp Immunol. 2006;145:130–8. doi: 10.1111/j.1365-2249.2006.03111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi N, Kitazawa H, Iwabuchi N, et al. Oral administration of an immunostimulatory DNA sequence from Bifidobacterium longum improves Th1/Th2 balance in a murine model. Biosci Biotechnol Biochem. 2006;70:2013–17. doi: 10.1271/bbb.60260. [DOI] [PubMed] [Google Scholar]


