Abstract
Notch signaling regulates multiple developmental processes and is implicated in various human diseases. Through use of the Notch transcriptional co-activator mastermind, we conducted a screen for Notch signal modifiers using the Exelixis collection of insertional mutations, which affects ∼50% of the Drosophila genome, recovering 160 genes never before associated with Notch, extending the previous roster of genes that interact functionally with the Notch pathway and mastermind. As the molecular identity for most recovered genes is known, gene ontology (GO) analysis was applied, grouping genes according to functional classifications. We identify novel Notch-associated GO categories, uncover nodes of integration between Notch and other signaling pathways, and unveil groups of modifiers that suggest the existence of Notch-independent mastermind functions, including a conserved ability to regulate Wnt signaling.
NOTCH signaling is one of a handful of basic signaling mechanisms controlling metazoan development. Its highly pleiotropic action includes involvement in cell differentiation, proliferation, survival, and migration (Artavanis-Tsakonas et al. 1999; Baron et al. 2002; Bray 2006; Louvi and Artavanis-Tsakonas 2006). Notch pathway mutations are also associated with several human pathologies, including cancer. Canonical Notch signaling involves ligand binding to the Notch receptor, triggering its cleavage and release of the intracellular domain into the cytoplasm, which traffics to the nucleus and complexes with Suppressor of Hairless [Su(H)] and Mastermind (Mam) to regulate transcriptional targets (Artavanis-Tsakonas et al. 1999; Baron et al. 2002; Bray 2006; Louvi and Artavanis-Tsakonas 2006).
Genetic screens have been invaluable in elucidating Notch pathway elements (Xu and Artavanis-Tsakonas 1990; Lambie and Kimble 1991; Fortini and Artavanis-Tsakonas 1994; Go and Artavanis-Tsakonas 1998; Chen et al. 2004; Katic et al. 2005), but a systematic experimental approach examining the set of Notch-interacting genes at a genomic level, i.e., the genetic circuitry of the Notch pathway, has been hindered by inherent limitations of these approaches. For instance, an EMS chemical mutagenesis theoretically interrogates 100% of the genome and is truly genomewide; however, the subsequent identification of all or even a large subset of interacting gene products is prohibitively laborious and therefore generally less informative. As a result, the nature and extent of probing a genetic circuitry remains narrowly defined. The Exelixis collection (materials and methods), a set of 15,500 transposon-induced mutations disrupting an estimated 53% of Drosophila coding regions (Parks et al. 2004; Thibault et al. 2004), overcomes many of these limitations. Importantly, as insert sites for nearly all transposons have been sequenced, screens using this collection have allowed rapid gene assignments of Notch signal modifiers at an unprecedented scale. Two inherent limitations of the collection should be kept in mind. First, in many instances the modifying transposon has inserted between genes such that an unambiguous gene assignment cannot be made instantaneously, notwithstanding the immediate ability to narrow down the candidate gene to a very small number of possibilities. Second, without further experimentation, it is unknown whether disrupted loci represent loss- or gain-of-function mutations, thereby limiting our ability to determine precise epistatic relationships between Notch (N) and interacting loci. The collection nonetheless permits a novel perspective on the complexity of N and mam activities and on the nature of genes and the biological processes with which they integrate.
MATERIALS AND METHODS
Drosophila stocks:
Flies were cultured on standard media. Crosses were carried out at 25°. The UAS-MamN construct and the C96-GAL4, UAS-MamN (C96-MamN) stock were previously described (Helms et al. 1999). The following strains were used: mam2 (Lehmann et al. 1983), mamsAX8 (Verheyen et al. 1996), N55e11 (Heiztler and Simpson 1991), N54l9 (Grimwade et al. 1985), nd3, sm1, sm05338, smKG03875, msi1, msi2, orbdec, orb2BG02373, CG17838 (Grumbling and Strelets 2006), lark1 (provided by R. Jackson), NAx16 (Kelley et al. 1987), dx152 (provided by K. Matsuno), spenAH393, and spenXFM911 (provided by I. Rebay). Matthew Freeman provided the GMR-ArmS44Y and GMR-ArmS56F strains (Freeman and Bienz 2001).
The Exelixis collection:
The collection is composed of 15,500 transposon-induced gene disruptions, resulting in mutations in ∼53% of the Drosophila genome (Parks et al. 2004; Thibault et al. 2004) (http://drosophila.med.harvard.edu/). Each insertion is derived from one of four vector types, three piggyBac-derived (PB, RB, and WH), and a fourth, a P-element variant (XP) (Parks et al. 2004; Thibault et al. 2004). Currently, the collection is 22% PB, 20% RB, 35% WH, and 23% XP. There are two classes of disruption events: those leading to inactivation of loci and those driving expression of downstream genes when exposed to GAL4 due to the presence of UAS sequences within the insertional transposon (WH and XP elements) (Parks et al. 2004; Thibault et al. 2004). In the presence of GAL4, UAS-containing insertions could theoretically inactivate loci even if oriented to drive expression, for example, by generating antisense RNA products. This could be investigated genetically by screening the collection in a background containing a loss-of-function mam or Notch allele without any GAL4 driver. WH insertions also contain splice acceptor sites (splice traps), permitting normal transcription of tagged genes, but are designed such that WH, rather than endogenous, splice acceptors are used, allowing for a piece of the piggyBac transposon to be spliced into the final transcript, thereby disrupting translation. In the presence of GAL4, UAS-containing WH and XP insertions may represent hypomorphic, hypermorphic, neomorphic, or antisense alleles. In contrast, PB and RB insertions lack UAS sequences and are likely to represent null or hypomorphic alleles. Therefore, screening in a genetic background containing a GAL4-dependent phenotype allows one to exploit the full potential of the collection and to recover interactors representing both classes of insertional events.
Screening:
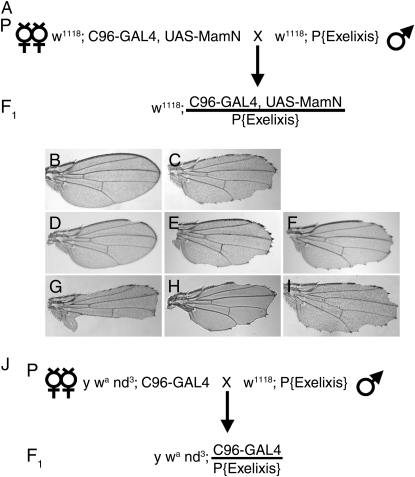
The Exelixis collection (Artavanis-Tsakonas 2004; Parks et al. 2004; Thibault et al. 2004) was screened for genes that dominantly modify wing-nicking phenotypes resulting from C96-GAL4-directed MamN expression using the C96-GAL4, UAS-MamN (C96-MamN) strain (Helms et al. 1999). C96-MamN individuals exhibit fully penetrant, dosage-sensitive, dominant wing phenotypes (Figure 1C) caused by loss of Notch signaling (Smoller et al. 1990; Helms et al. 1999). The strength of the C96-MamN interaction for each modifying insertion was scored in retests using an arbitrary scale of 1–5: 1 represents a strong interaction and 5 represents a weak interaction [supplemental Table 1 at http://www.genetics.org/supplemental/ (MamN column)]. A total of 274 modifying transposons received a value of ≤3.
Figure 1.—
Screen designs and primary screen validation. (A) Primary screen for insertions that modify the C96-GAL4, UAS-MamN (C96-MamN) wing phenotype. C96-MamN individuals exhibit a fully penetrant, dosage-sensitive, dominant wing phenotype (Helms et al. 1999), similar to those associated with loss-of-function mutations in Notch and wingless (Smoller et al. 1990; Helms et al. 1999). Exelixis P-element or piggyBac males were crossed to C96-MamN females. F1 cultures exhibiting C96-MamN wing-notching enhancement or suppression were selected and retested: 219 enhancers and 385 suppressors were isolated. Three insertions caused lethality and three additional inserts could not be characterized. (B) Control w1118 and (C) C96-MamN wings. D–J provide examples of the range of C96-MamN modifier phenotypes (materials and methods). Shown are female wings trans-heterozygous for C96-MamN; interacting loci, stock IDs, and identified genes are listed. Strong, moderate, and weak C96-MamN suppressors [(D) d05358 and CG14709; (E) c06331 and CG8090; and (F) c02035, respectively] and enhancers [(G) c06428 and CG14767; (H) d00059 and klumpfuss (klu); and (I) d07432 and Sin3A, respectively] are shown. (J) Secondary screens for Notch-pathway specific (NIs) and MamN-specific (MSIs) C96-MamN interactors. [y wa nd3; C96-GAL4], [N55e11/FM6; C96-GAL4], [y w Ax16; C96-GAL4], or [w dx152; C96-GAL4] females were crossed to males as in A (nd3 illustrates the strategy used for each allele). In a C96-GAL4 background, F1 cultures exhibiting enhancement or suppression of nd3, N55e11, Ax16, and dx152 wing phenotypes were noted (supplemental Table 1 at http://www.genetics.org/supplemental/).
Secondary screens:
To identify genes with Notch pathway functions, all interacting transposons were crossed to nd3, N.255e11, Ax16, and dx152 mutations in a C96-GAL4 background and examined for synergistic wing effects. A total of 31 genes isolated from the screen are known Notch pathway genetic interactors (Table 1). Of these 31 genes, 12 modified four of these alleles, 6 modified three of these alleles (Beadex, kekkon-1, kuzbanian, mastermind, osa, and Ras85D), and 13 of these genes modified no more than two of the secondary test alleles (echinoid, extra macrochaete, frizzled, inscuteable, kismet, net, nemo, Neurotactin, the X-linked gene rugose, shaggy, sprouty, split ends, and Ultrabithorax.) As several alleles of documented N-interacting genes failed to modify all four alleles tested (including an allele of Delta), defining Notch interactors (NIs) as interacting with all tested alleles seemed too stringent. As most known Notch interactors modified at least three of four tested secondary mutations, we arbitrarily defined autosomal and X-linked NIs as insertions modifying wing phenotypes of at least three of the aforementioned mutations or as modifying wing phenotypes of either N55e11 or NAx16 heterozygotes, respectively. MamN-specific interactors (MSIs) failed to interact with any allele in secondary tests.
TABLE 1.
Identification of known Notch pathway genetic interactors
| Gene | N | mam | Dl/Ser | C96-MamN | C96-GAL4 |
|---|---|---|---|---|---|
| Beadex (Bx) | NA | + | + | E/S | +/NE |
| Delta (Dl) | + | + | + | E/E/E/S | NE/+/NE/+ |
| echinoid (ed) | + | NA | + | S | NE |
| extra macrochaete (emc) | + | NA | + | S | NE |
| escargot(esg) | +* | + | NA | E | + |
| eyegone (eyg) | + | NA | NA | E | + |
| fringe (fng) | + | NA | + | E/E | +/+ |
| frizzled (fz) | + | NA | + | S | NE |
| hephaestus (heph) | + | NA | + | S | NE |
| inscuteable (insc) | + | NA | NA | S/S | NE/NE |
| kekkon-1 (kek1) | NA | + | NA | E | + |
| kismet (kis) | + | NA | NA | E | NE |
| kuzbanian (kuz) | + | NA | + | S | NE |
| mastermind (mam) | + | + | + | E/S | NE/NE |
| net | NA | NA | + | E | NE |
| nemo (nmo) | + | NA | + | S | NE |
| Neurotactin (Nrt) | + | NA | NA | E | NE |
| numb | + | NA | NA | E/E | NE/NE |
| osa | + | NA | NA | E | NE |
| pointed (pnt) | + | NA | NA | E/E | NE/NE |
| polychaetoid (pyd) | + | NA | + | Other | NE |
| puckered (puc) | + | NA | NA | E/S/S | +/NE/NE |
| Ras85D | + | + | NA | E | NE |
| rugose (rg) | NA | NA | + | S | NE |
| scabrous (sca) | + | NA | + | E | NE |
| shaggy (sgg) | + | + | NA | E | NE |
| sprouty (sty) | +* | NA | +* | S | NE |
| split ends (spen) | + | NA | NA | S | NE |
| string (stg) | + | NA | NA | E | NE |
| Ultrabithorax (Ubx) | + | NA | NA | S | NE |
| wingless (wg) | + | + | + | Other | + |
A list of 31 genes previously associated with Notch-signaling activity (Grumbling and Strelets 2006) identified as C96-MamN modifiers. Of these 31 genes, 24 are known to interact genetically with N, and, of these 24, 15 were defined as NIs and are underlined. Known interactors of other Notch pathway members defined as novel NIs are double underlined. Interactions of genes with Notch (N), mastermind (mam), Delta or Serrate (Dl/Ser), C96-MamN, or C96-GAL4 are listed. NA, not available; NE, no effect; +, interaction; +*, interaction from G. D. Hurlbut, M. W. Kankel and S. Artavanis-Tsakonas, unpublished results; E, enhances; S, suppresses. Multiple entries for a given gene under the “C96-MamN” and “C96-GAL4” columns indicate multiply recovered alleles and different entries refer to the type of interaction for each allele.
Control screens:
Two types of control screens were performed:
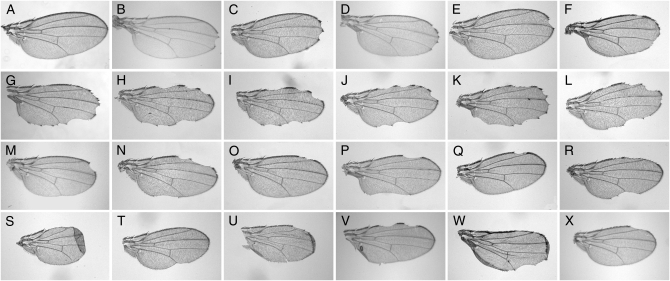
All 610 modifying transposons were crossed to the C96-GAL4 strain to identify insertions that cause MamN-independent wing phenotypes when mis-expressed. Several wing phenotypes are shown in Figure 3. Only two interacting strains exhibited a wing phenotype prior to outcrossing [independently of GAL4-mediated expression (c02134 and c00454, which disrupt the CG3262 and bonus genes, respectively)]. A total of 40 elicited MamN-independent phenotypes (see Secondary screens; Figure 3), including several known Notch pathway interactors (fringe, Figure 3D; Beadex, Figure 3E; kekkon-1, Figure 3P; escargot, Figure 3S; eye gone, Figure 3X; and Delta and wingless, data not shown). Moreover, several modifying insertions phenocopy loss of Notch function wing defects in this background (Figure 3, B–F): two of these, fringe and Beadex, are established pathway interactors (Grumbling and Strelets 2006). Therefore, other genes identified in this manner may function analogously and may include potential novel Notch genetic elements. d00627 inserted ∼10 kb away from both CG15166 and CG15167. Hence, Notch-like phenotypes for d00627/+; C96-GAL4/+ individuals may likely result from GAL4-induced misexpression of one or both of these genes (Figure 3).
Since the C96-MamN phenotype is dosage sensitive (Helms et al. 1999), we expected that mutations in general regulators of transcription would modify C96-GAL4 expression. We took advantage of the dosage-sensitive rough-eye phenotype associated with glass multiple reporter (GMR)-directed GAL4 expression (Kramer and Staveley 2003) to identify potential transcriptional regulators. Thus, putative global transcriptional regulators would modify GMR-GAL4 rough-eye phenotypes by affecting GMR-GAL4 transgene expression while modifying insertions that do not alter the phenotype are presumed to not be involved in global transcription. This test revealed that 294 (48% of 610) insertions modify the GMR-GAL4 eye phenotype, including 139 C96-MamN enhancers and 155 C96-MamN suppressors, several of which are known Notch pathway interactors (e.g., Delta). Of these GMR-GAL4 interactors, 221 enhanced and 51 suppressed the GMR-GAL4 eye phenotypes while 22 (17 enhancers and 3 suppressors) caused GMR-GAL4 lethality. While this class of modifying transposons must include genes that function as general transcriptional regulators, some may simply affect eye imaginal disc development in a Notch-independent or Notch-dependent fashion.
Figure 3.—
Mam-independent wing phenotypes. Wing phenotypes of C96-MamN interactors in a mam+ background identified loci that interact with Notch pathway elements. Except for B, all panels show wings prepared from genotypes containing C96-GAL4. (A) w1118 control strain. (B) A null N allele, N54l9. (C) d01997, fringe, or CG9119. (D) d08885, fringe. (E) d01047, Beadex. (F) d00627, CG15167. (G) C96-MamN. (H–J) d11666, d10223, d03908, cp309. (K) d09084, CG17390. (L) d04807, lark. (M) d00992, smooth. (N) d04790, CG17836. (O) d03727, CG4612. (P) d03841, kekkon-1. (Q) d04745, cropped. (R) d05894, dorsal. (S) d05415, escargot. (T) d05123, CG7443, or CG9603. (U) d08364, Ecdysone inducible protein 75B. (V) d11052, reaper. (W) d11677, distal antenna. (X) d01629, eyegone. All genotypes are trans-heterozygous for the Exelixis insertion.
Mapping:
An Exelixis database contains genomic sequence flanking all 15,500 inserts and provides chromosomal locations for all modifiers (supplemental Table 1 at http://www.genetics.org/supplemental/). Using this database, along with BLAST and visual inspection, we focused on Exelixis transposons within 2 kb of a single gene model (based on Drosophila melanogaster R5.1 annotations) for which a unique FBti (FlyBase transposon insertion identifier) site was determined according to a prerelease version FB2007_01 (provided by Rob Kulathinal at FlyBase). Using these criteria, 408 gene products (accounting for multiple alleles) were identified for all 610 recovered, modifying insertions with high confidence (supplemental Table 1). For some strains, assignment of a unique molecular identity requires further experimentation, due to insertions for which no sequence data are available, multiple inserts (37 strains), or ambiguous effects of transposon insertion between adjacent genes. No single candidate gene could be identified for 37 strains with multiple inserts. Extrapolation of the 37 strains with multiple inserts indicates that ∼6% of all 15,500 strains contain multiple inserts. For the remaining 146 modifying insertions, the transposon inserted between two different genes such that we could not unambiguously distinguish between disrupted loci. Most of the 610 C96-MamN-interacting transposons are single-allele modifiers. However, at least 35 complementation groups are represented by two alleles, including 13 complementation groups represented by three or more alleles. Complementation groups were defined as interacting insertions that disrupted the same gene as determined by our mapping criteria (see above).
Mounting of wings:
Adult wings were removed, dehydrated in isopropanol, and mounted in a 3:1 dilution of CMCP-10 mounting media (Masters, Wood Dale, IL) and lactic acid. Images were taken on a Zeiss Axioplan Microscope with a ×5 objective using IP Lab software (Scanalytics) and assembled using Adobe Photoshop and Microsoft PowerPoint.
Eye microscopy:
All eye images are from female adults using standard techniques. Images were taken on a Wild M10 dissecting microscope at ×25 using a ProgRes 3008 digital camera and ProgRes 5.0 software and assembled using Adobe Photoshop and Microsoft PowerPoint.
Gene ontology analysis:
For the gene ontology (GO) analysis, we focused on inserts within 2 kb of a single gene for which a unique FBti (FlyBase transposon insertion) site was determined according to a prereleased β version of FB2007_01 (provided by R. Kulathinal at FlyBase). Using these criteria, we could readily determine the identity of 3606 disrupted genes within the Drosophila genome. Although our analysis assessed the entire collection, which covers 53% of the genome, in our GO analysis we included only transposons for which unique gene assignments could be made with high confidence. Each disrupted gene was assigned a unique refseq protein ID and put into the Database for Annotation, Visualization and Integrated Discovery (DAVID) to provide a measure of term co-occurrence probability (Dennis et al. 2003). GO analysis of these 3606 genes identified statistically significant overrepresentation of GO terms among identified interactors (Ashburner et al. 2000). Categories with low membership were penalized and P < 0.05 is used for all terms classified as overrepresented. Terms identified by GO analysis from these 3606 genes were used as a baseline for comparisons to identify novel GO terms associated with the following subgroups: (1) known Notch genetic interactors described at FlyBase represented by the collection (ExFBNGInts), (2) all NIs, novel NIs, (4) all MSIs, and (5) 610 C96-MamN-interacting transposons, which include 408 identified genes. The genes/computed or curated genes (CGs) for all five subgroups are listed in supplemental Table 6 at http://www.genetics.org/supplemental/.
Statistics:
We tested for enrichment of known N genetic interactors and other specific subgroups using the Yates corrected χ2 test. We assumed that the D. melanogaster genome consists of 14,000 genes and focused on the 3606 insertions where highly confident gene assignment was possible as described (see Gene ontological analysis).
Luciferase reporter assay:
293T cells were seeded on 24-well plates at 100,000 cells/well 1 day before transfection and transfected with appropriate combinations of expression plasmid DNA corresponding to a final amount of 1 μg of DNA. Total plasmid amounts were maintained constant by adding appropriate amounts of empty vectors. Transfected cells were harvested 36 hr post-transfection. Luciferase activities were measured using the luciferase reporter assay system (Promega, Madison, WI). Luciferase values were corrected for transfection efficiency by normalizing to Renilla activity.
RESULTS
A screen for mastermind modifiers:
Previous characterization of the Exelixis collection affords rapid identification of affected genes or, in cases of ambiguity, the narrowing down of gene identity to very few candidates. As many insertions are GAL4 responsive, we screened in a background allowing for recovery of potential gain-of-function mutations through mis-expression. We screened all 15,500 insertions within the collection (Figure 1A) for mutations that dominantly modify a fully penetrant wing-margin phenotype associated with GAL4-dependent expression (Brand and Perrimon 1993) of a dominant-negative, C-terminal Mam truncation [C96-GAL4, UAS-MamN (C96-MamN)] (Figure 1C) (Helms et al. 1999; Wu et al. 2000; Kitagawa et al. 2001). Notably, the collection disrupts 87 of the 193 (45.1%) genes known to interact genetically with N, mam, or both (Grumbling and Strelets 2006) (Figure 2), serving as a control for sensitivity. C96-MamN competes with wild-type Mam, reducing its function to <50%, a level of activity not associated with dominant phenotypes of endogenous alleles (Helms et al. 1999); affects expression of the Notch downstream targets cut, vestigal, and wingless in this context (Helms et al. 1999; Yedvobnick et al. 2001); and is sensitive to mutations in all core Notch pathway components tested (Helms et al. 1999). We expect modifiers to include bona fide Notch pathway genes, genes affecting wing development or transgene expression, other nonspecific enhancers and suppressors, and, importantly, those involved in Notch-independent mam functions.
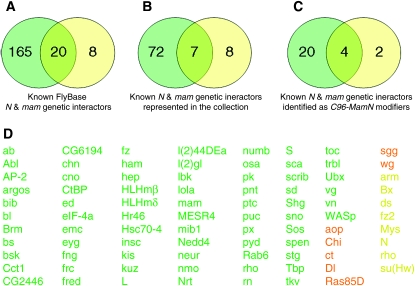
Figure 2.—
Numbers of known Notch and mastermind genetic interactors. Venn diagrams showing the number of genes associated genetically with Notch and mam. The number of genes known to interact genetically with N (green), mam (yellow), or both (orange) as described in FlyBase (A) and disrupted within the Exelixis collection (B) or (C) identified as C96-MamN modifiers from the Exelixis collection. (C) Of the 87 genes (B), 26 (29.9%) within the collection previously known to be associated genetically with N, mam, or both were isolated as C96-MamN modifiers, representing a statistically significant enrichment for pathway modifiers (Yates' χ2 = 28.8, P = 8.14 × 10−08). Identified C96-MamN interactors were enriched for known genetic interactors of N [24 of 79 genes (30.4%)] (Yates' χ2 = 27.3, P = 1.70 × 10−07) and mam [6 of 15 genes (40.0%)] (Yates' χ2 = 9.7, P = 1.90 × 10−03). (D) Known Notch and mastermind genetic interactors described by FlyBase and represented within the Exelixis collection. Green, yellow, and orange fonts indicate Notch, mastermind, and overlapping genetic interactors, respectively.
We identified 610 insertions that modify the C96-MamN wing-nicking phenotype. Examples of the range of modification are shown in Figure 1, D–I. As each modifying insert may affect multiple genes, definitive gene assignments necessitate reversion and rescue studies to provide unambiguous gene identity. However, in most instances a single candidate gene was apparent such that 408 gene assignments were determined with high confidence for the 610 interacting transposons (see materials and methods; supplemental Tables 1 and 6 at http://www.genetics.org/supplemental/), including 31 known Notch pathway genetic interactors (Table 1 and Figure 2). This represents a significant enrichment for known Notch pathway modifiers among the 408 interacting transposons compared to their prevalence among identifiable genes covered by the collection (P = 1.83 × 10−09), supporting the relevance of novel candidates for the Notch pathway. Not all documented N interactors represented in the collection were recovered.
In a control screen (materials and methods) designed to reveal MamN-independent wing modifiers, we crossed all insertions to the C96-GAL4 strain. This test revealed that only 40 of the 610 (6.6%) recovered modifying transposons elicited MamN-independent phenotypes (Figure 3), suggesting that the interactions observed for most of the modifying insertions are unlikely to result from additive effects.
Functional analysis of modifiers:
We sought to distinguish Notch-dependent from Notch-independent C96-MamN-interacting transposons in secondary genetic assays by testing for interactions with Notch (N55e11, nd3, NAx16) and/or deltex [dx (dx152)], a ubiquitin ligase that functions in the cytoplasm to modulate Notch activity (supplemental Table 1 at http://www.genetics.org/supplemental/). Biologically, these alleles have different effects on pathway activity, leading to a ligand-dependent gain (NAx16) or loss (N55e11, nd3, and dx152) of Notch function. In a C96-GAL4 background, these alleles gave highly penetrant, modifiable wing-nicking (N55e11 and nd3) or venation (dx152 and NAx16) phenotypes (Figure 4, row 1). Ultimately, classifying modifiers on the basis of their relationship to these alleles may provide insights into epistatic relationships and the nature of the recovered interactions.
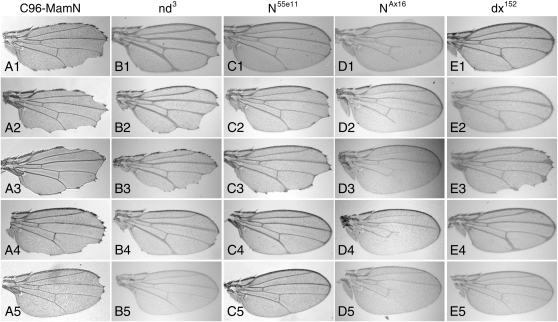
Figure 4.—
Validation of the secondary screens. C96-MamN NIs modify wing phenotypes resulting from mutations in Notch pathway genes. Efficacy of secondary genetic assays is illustrated for two NI enhancers [scabrous (sca) and klumpfuss (klu)] and two NI suppressors (CG8090 and CG7370). Notch/sca interactions have been reported (Mlodzik et al. 1990); other interactions define novel Notch-signaling modulators. Wings from genotypes contain the following Notch pathway mutations: (A1–A5) C96-MamN. (B1–B5) y w nd3. (C1–C5) w N55e11. (D1–D5) y wa NAx16. (E1–E5) w dx152. Strains in columns are mated with (row 2) scad09400, (row 3) klud00059, (row 4) CG8090c06331, and (row 5) CG7370f06222. All phenotypes are heterozygous for C96-GAL4. Each mutation fails to produce wing phenotypes when combined with C96-GAL4 (not shown). (A1) C96-MamN wing nicking is dominantly enhanced by (A2) scad09400 and (A3) klud00059 and suppressed by (A4) CG8090c06331 and (A5) CG7370f06222 heterozygotes. (B1) nd3 combined with C96-GAL4 causes a highly penetrant wing-nicking phenotype. (B2) scad09400 and (B3) klud00059 heterozygotes increase the number and severity of nd3 wing notching. (B4) nd3; CG8090c06331 trans-heterozygous wings were nearly suppressed to wild type while (B5) CG7370f06222 suppresses nd3 wings to wild type. (C1) Distal wing notching exhibited by N55e11 heterozygotes increased in (C2) scad09400 and (C3) klud00059 backgrounds and was dominantly suppressed to wild type at high penetrance by (C4) CG8090c06331 and (C5) CG7370f06222. (D1) L4 and L5 longitudinal wing-vein shortening in hemizygous NAx16 males. (D2) scad09400 suppresses L4, but not L5 phenotypes. (D3) klud00059, (D4) CG8090c06331, and (D5) CG7370f06222 cause additional L4 and L5 longitudinal wing-vein shortening. (E1) dx152 hemizygotes display distal wing-vein thickening with occasional wing notching, which is dominantly enhanced by (E2) scad09400, (E3) klud00059, and (E4) CG8090c06331. For klud00059, the degree and penetrance of wing notching is also more severe. (E5) CG7370f06222 completely suppresses dx152 vein defects.
On the basis of the results from this assay, we chose to define two separate categories for further study: NIs and MSIs. We defined NIs by their ability to modify at least three of these alleles (materials and methods) whereas MSIs were defined as failing to modify any allele in this assay. As a group, the NIs consist of 235 modifying insertions or 175 genes (accounting for multiple alleles) (Figure 4; supplemental Tables 1, 2, and 6 at http://www.genetics.org/supplemental/) and are enriched for known Notch pathway genetic interactors compared to their prevalence among identifiable genes covered by the collection (P = 1.6 × 10−08) (Grumbling and Strelets 2006). Importantly, this group also includes 160 genes never before associated with Notch (supplemental Tables 2 and 6), nearly doubling the roster of genes known to interact genetically with Notch. Not all of the 31 known Notch pathway genetic interactors defined at FlyBase are included within the NIs subgroup; however, 15 genes known to interact genetically with N are included (Table 1).
The second group, the MSIs, failed to modify any of the secondary test alleles and are of special interest, as they may reveal Notch-independent mam functions and are composed of 118 modifiers or 79 genes (supplemental Tables 3 and 6 at http://www.genetics.org/supplemental/). The relatively large size of this group may suggest a broader-than-anticipated role for mam in development. Failure to interact with these alleles does not rigorously preclude possible MSI interactions with other Notch pathway components. Given the context dependency of Notch signaling, it is important to note that our data do not exclude the possibility that the MSIs might interact with Notch in other contexts. However, each of the 31 known pathway interactors uncovered, including 15 defined as NIs, modified at least one secondary test allele.
Gene ontology:
To further assess the modifiers uncovered by the screen, GO analyses were performed to reveal overrepresentation of specific functionally or structurally related groups. Our approach was based on comparing the prevalence of specific GO terms among modifier subgroups to their prevalence in the Exelixis collection, which served as the baseline for these analyses. We examined several modifier subgroups (shown in supplemental Table 6 at http://www.genetics.org/supplemental/), which include the entire set of NIs (175 genes), the novel set of NIs (160 genes), and finally, the MSIs (79 genes). These three subgroups were compared to the 79 known Notch genetic interactors identified by FlyBase represented in the collection (ExFNGInts) (supplemental Table 6). Novel, statistically significant GO categories unique to each subgroup were determined (materials and methods; supplemental Table 4). Figure 5 shows the degree of overlap between different categories defined for each subgroup relative to ExFNGInts, which are summarized in Figure 6.
Figure 5.—
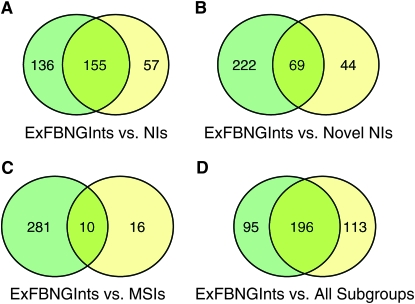
Overlapping functional categories of C96-MamN modifiers relative to the FlyBase Notch genetic interactors represented within the Exelixis collection. Venn diagrams show the number of overlapping, statistically significant (Fisher's exact test, P < 0.05) functional categories between (A) the FlyBase Notch genetic interactors represented within the Exelixis collection (ExFBNGInts) and the entire set of NIs. (B) the ExFBNGInts and the novel set of NIs. (C) The ExFBNGInts and the MSIs. (D) The ExFBNGInts and the combined list of functional categories identified in the screen (all C96-MamN modifiers, the entire set of NIs, the novel set of NIs, and the MSIs). In A–D, the ExFBNGInts are shown in green, the overlap in yellow-green, and the other subgroups in yellow.
Figure 6.—
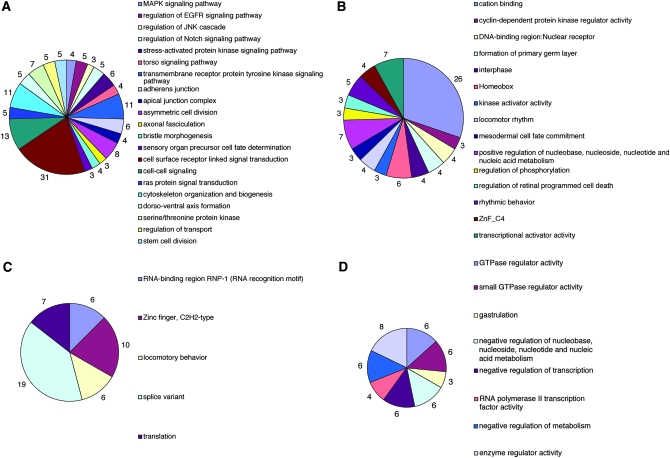
GO analysis. The prevalence of specific GO terms among gene subgroups was compared to their prevalence in the Exelixis collection (materials and methods). Intersubgroup comparisons uncovered novel, statistically significant categories (Fisher's exact test, P < 0.05) unique to each subgroup as identified by DAVID and are summarized for (A) the known FlyBase Notch genetic interactors represented by the Exelixis collection (ExFBNGInts), (B) the 175 NIs, and (C) the 160 novel NIs. (D) Statistically significant functional categories (Fisher's exact test, P < 0.05) uniquely associated with 79 MSIs.
These analyses allowed us to evaluate the significance of recovered modifiers, several aspects of which are noteworthy. Comparisons between significant GO terms represented by ExFNGInts and NIs indicate that the two sets are not identical despite extensive overlap (Figure 5A). This is not surprising, given that the nuclear Notch effector mam was used to generate the phenotypic parameter, which may not be sufficiently sensitive to uncover the full spectrum of known functional categories associated with N. However, the similar distribution of terms represented in both sets suggests that the screen was unbiased and, importantly, validates the significance of novel categories associated with Notch function (Figure 6, B and C; supplemental Table 4 at http://www.genetics.org/supplemental/).
Of particular interest are three such categories that link Notch activity to RNA processing: splice variant, translation, and RNA-binding region RNP-1 proteins [RNA recognition motif (RRM)]. A significant enrichment of RRM genes among all 408 C96-MamN-interacting genes was identified (n = 15, P = 0.000108). RRM genes include components of small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins (hnRNPs), proteins implicated in alternative splicing. The potential importance of RRM genes in regulating Notch signaling is highlighted by hephaestus (Abdelilah-Seyfried et al. 2000), an hnRNP that attenuates Notch signaling by regulating the processing, stability, or translation of one or more Notch pathway mRNAs (Dansereau et al. 2002) and which was identified as an NI. Another RRM gene, lark, exhibits phenotypes in adult mechanosensory bristles (McNeil et al. 2001) that require Notch signaling during development. Recovery of smooth, an hnRNP heph complex member, further implicates RRM genes in the regulation of Notch activity. Moreover, transcript levels of numerous RRM genes are regulated in response to activated Notch in the Drosophila embryo (G. D. Hurlbut, M. W. Kankel and S. Artavanis-Tsakonas, unpublished results), some of which display genetic interactions with Notch pathway components (Grumbling and Strelets 2006; G. D. Hurlbut, M. W. Kankel and S. Artavanis-Tsakonas, unpublished results).
Although the GO analysis suggests that the interrelationships between Notch signaling and RRM genes appear to be extensive, the importance and specificity of this mutual influence remains unknown. Specific Notch-signaling outputs may rely on proper RNA localization, a notion supported by the recovery of smooth (Arn et al. 2003), orb (Grumbling and Strelets 2006), and orb2 (Cooperstock and Lipshitz 1997), three genes involved in RNA localization. However, given the large number of recovered genes involved in RNA processing, we sought to determine their potential importance in regulating Notch activity by examining whether a subset of these genes could modify N54l9 and/or N55e11 haplo-insufficient wing-nicking phenotypes. We focused on seven genes, including split ends (Powell et al. 2001), which antagonizes Notch signaling (Doroquez et al. 2007) and therefore served as a positive control. No prior genetic links had been documented for the remaining six genes tested. At 25°, N54l9 and N55e11 display wing-nicking phenotypes at 20.4 and 23.6% penetrance, respectively (Table 2). All seven genes tested dominantly altered the N54l9 and/or N55e11 haplo-insufficient wing-nicking phenotypes, displaying qualitative (Figure 7) and quantitative (Table 2) modification. Significantly, the interactions displayed by these alleles, which were generated independently of the collection, serve to corroborate the identity of these insertions identified by the screen.
TABLE 2.
RNA-processing genes modify Notch haplo-insufficient wing phenotypes
| Gene | N54l9 (%) | Total | N55e11 (%) | Total |
|---|---|---|---|---|
| isoA | 20.4 | 54 | 23.6 | 72 |
| sm1 | 95.0 | 20 | 88.1 | 42 |
| sm05338 | 95.2 | 42 | 68.8 | 32 |
| smKG03875 | 54.2 | 48 | 42.5 | 40 |
| spenXFM911 | 0.0 | 36 | 0.0 | 22 |
| spenAH393 | 8.3 | 24 | 2.6 | 78 |
| lark1 | 64.0 | 50 | 67.9 | 28 |
| msi1 | 54.5 | 44 | 70.8 | 48 |
| msi2 | 82.1 | 56 | 50.0 | 34 |
| orbdec | 57.7 | 26 | ND | ND |
| orb2BG02373 | 50.0 | 82 | 46.7 | 30 |
| CG17838 | 66.7 | 42 | 74.2 | 54 |
The smooth (sm), split ends (spen), lark, musashi (msi), orb, orb2, and CG17838 genes were identified as C96-MamN modifiers and tested for their ability to dominantly modify the wing-nicking phenotypes associated with the N54l9 and N55ell null alleles. Specific alleles are listed. Percentages indicate the penetrance of the Notch wing-nicking phenotype for N54l9 and N55ell. For penetrance calculations, wings displaying any margin defect are scored as mutant. “Total” indicates the number of wings scored. ND, not done.
Figure 7.—
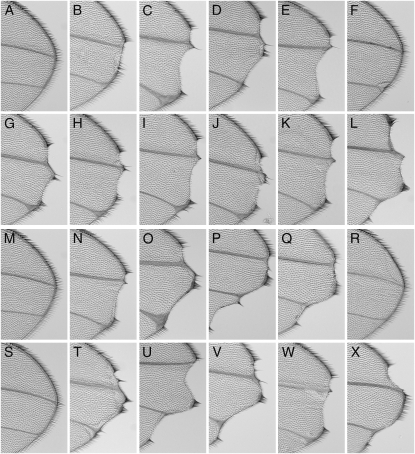
RNA-processing genes modify Notch haplo-insufficient wing phenotypes. Several RNA-processing genes were tested for their ability to modify N54l9 and/or N55e11 wing phenotypes. Shown at ×10 magnification are the distal portion of the wings where typical Notch haplo-insufficiency wing notching is observed. (A and M) Wild-type Drosophila wings. (B) Notch control N54l9/+, mutant phenotype present in 20% of wings. In B–L, wings are heterozygous for both N54l9 and the following alleles: (C) sm1, (D) sm05338, (E) smKG03875, (F) spenAH393, (G) lark1, (H) msi1, (I) msi2, (J) orbdec, (K) orb2BG02373, and (L) CG17838. (N) Notch control N55e11/+, mutant phenotype present in 23% of wings. In O–X, wings are heterozygous for both N55e11 and the following alleles: (O) sm1, (P) sm05338, (Q) smKG03875, (R) spenXFM911, (S) spenAH393, (T) lark1, (U) msi1, (V) msi2, (W) orb2BG02373, and (X) CG17838.
Novel genetic links between Notch and the cell cycle and other signaling pathways:
Many novel NIs fall within GO categories previously associated with Notch function, such as proliferation. The link between Notch signals and proliferation is well documented but poorly understood (Artavanis-Tsakonas et al. 1999; Bray 2006). Identification of taranis (tara) as an NI potentially links Notch activity to the cell cycle machinery (CDKs). TARA is homologous to a family of mammalian proteins, including human p34SEI-1/TRIP-Br1 (Calgaro et al. 2002), which regulate the transcriptional activation of CDKs involved in cell cycle regulation (Sugimoto et al. 1999; Hsu et al. 2001). This potential link is strengthened through our observed interaction of N and Cyclin A (Cyc A) and the ability of TARA-related mammalian TRIP-Br proteins to interact in vitro with Cyc A (Hsu et al. 2001). Finally, wing defects observed for viable tara alleles phenocopy those seen with certain Cyclin E alleles (Duronio et al. 1998), a CDK isolated as a novel NI.
In addition to these new links to the cell cycle, several novel NIs link N to other signaling pathways. For example, crosstalk between Notch and EGFR signaling is detected through the transcription factor klumpfuss (klu) (Figure 4, A3–E3) whereas mirror (mirr) provides a potential node that integrates Notch signaling with the TGF-β's Hh or WNT pathways. mirr, araucan (ara), and caupolican (caup) encode the homeodomain proteins of the Iroquois complex, which affects wing-vein formation and proneural gene activity through regulation of achaete-scute genes (Gomez-Skarmeta et al. 1996), known Notch targets. Hh and TGF-β signaling induce, while wingless (wg) represses, Iroquois complex gene expression (Gomez-Skarmeta et al. 1996). We link Mirr activity with mam and N in the wing and note that Notch activation downregulates ara (Nagaraj et al. 1999) and caup transcription (G. D. Hurlbut, M. W. Kankel and S. Artavanis-Tsakonas, unpublished results).
Functional classification of MSIs:
Of the 408 genes identified in the screen, 79 genes interacted only with C96-MamN (MSIs). The existence of this class of interactors raises the possibility that Mam performs a more prominent biological role that includes Notch-independent functions, which could, notably, include noncanonical, Su(H)-independent Notch signaling (e.g., Klein et al. 2000; Ehebauer et al. 2006). GO analysis was performed to determine if MSIs share functions that might also be mam specific. Identified GO terms were compared to those derived from all other subgroups to identify overrepresented terms specific to MSIs. Several of these classes were identified, including PRC1 complex and Small GTPase regulator activity (summarized in Figure 6D; supplemental Table 4E at http://www.genetics.org/supplemental/). The broad overlap between functional categories enriched in the NIs and the novel NIs helps validate MSI-specific categories as representing possible Notch-independent mam functions.
As Mam participates in the Notch transcriptional complex (Bray 2006), 12 MSIs with a transcriptional role identified in the GO analysis are of interest [pita, CG31716, caudal, Sex comb on midleg (Scm), corto, knirps-like, simjang (simj), defective proventriculus, brain tumor, pterin-4A-carbinolamine dehydratase, deadpan and Peptidoglycan recognition protein LC]. Nearly half are known negative transcriptional regulators (Grumbling and Strelets 2006), suggesting that Mam can either activate or suppress target gene expression, consistent with observed polytene chromosome colocalization of Mam and the Groucho suppressor (Bettler et al. 1996).
Identification of the MamN-specific GO category PRC1 complex also suggests a more general involvement of Mam in gene regulation. This multimeric protein complex, consisting of several Polycomb group genes, directly antagonizes ATP-dependent remodeling of nucleosomal arrays to maintain a transcriptionally repressed state. C96-MamN-specific interactions observed for Scm and corto, two physically interacting PRC1 complex components (Salvaing et al. 2003), underscore the Mam/PRC1 complex link and raise the possibility that Mam binds to the PRC1 complex to repress target gene expression independently of Notch.
One functional category that distinguished MSIs from other subgroups was Small GTPase regulator activity, which, given the pleiotropic action of this gene group, implicates Notch-independent Mam activity in numerous processes. Because of their involvement in carcinogenesis, RAS/GAP Nf1 and CG32560, a small GTPase linked to the MAPKKK cascade (Grumbling and Strelets 2006), are of particular interest. Using Homophila, a database of Drosophila homologs of human-disease-related genes (Reiter et al. 2001; Chien et al. 2002), 136 C96-MamN modifiers, including 50 NIs and 16 MSIs (supplemental Table 5 at http://www.genetics.org/supplemental/), had disease-associated homologs (BLAST E-value of 1 × 10−10 or lower), suggesting a broad relationship between mam activity and disease gene homologs.
Uncovering Notch-independent Mastermind functions:
Although recent reports support the notion that Mam functions independently of Notch as a co-activator for β-catenin/T cell factor (TCF) (Alves-Guerra et al. 2007), Myocyte Enhancer Factor 2c (Shen et al. 2006), p53 (Zhao et al. 2007), and Xenopus neural gene expression (Katada et al. 2006), the full extent and in vivo significance of such independent functions remain unknown. To probe the validity of Notch-independent mam functions, we examined the MSI armadillo (arm), the Drosophila β-catenin homolog, and asked whether this relationship remains valid in the developmental context of the eye. GMR-GAL4-directed mis-expression of either Drosophila equivalent of the two mammalian β-catenin oncogenic point mutations (S44Y and S56F) produced malformed eyes (Freeman and Bienz 2001) (Figure 8, A and F). However, consistent with previous observations (Freeman and Bienz 2001), N haplo-insufficiency had little, if any, effect (Figure 8, D and E and I and J); halving mam gene dosage in Y55 and F76 strains enhanced the phenotype, increasing eye glossiness and producing smaller and rougher eyes (Figure 8, B and C and G and H). As N haplo-insufficiency, unlike heterozygosity for mam, can result in a rough-eye phenotype, modifications of activated arm by mam may reflect functional differences between mam and N in regulating arm activity. Thus, mam can function independently of N to affect arm function in both the wing and eye of Drosophila.
Figure 8.—
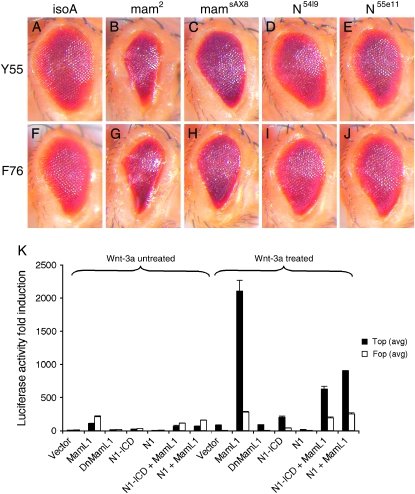
Mastermind enhances activated Armadillo eye phenotypes. Modification of the eye phenotype resulting from mis-expression of activated Arm. All eyes are from females heterozygous for (A–E) GMR-ArmS44Y (Y55) or (F–J) GMR-ArmS56F (F76). Eyes from (A) Y55 and (F) F76 individuals exhibit roughness and are smaller than wild type (not shown). In reducing mam levels, (B and G) mam2 and (C and H) mamsAX8 strongly enhance Y55 and F76 phenotypes while haplo-insufficiency for Notch (D) and (I) N54l9 and (E and J) N55e11 exert little, if any, effect. Genotypes are as follows: (A) GMR-ArmS44Y/+, (B) GMR-ArmS44Y/mam2, (C) GMR-ArmS44Y/mamsAX8, (D) N54l9/+; GMR-ArmS44Y/+, (E) N55e11/+; GMR-ArmS44Y/+, (F) GMR-ArmS56F/+, (G) GMR-ArmS56F/mam2, (H) GMR-ArmS56F/mamsAX8, (I) N54l9/+; GMR-ArmS56F/+, and (J) N55e11/+; GMR-ArmS56F/+. (K) In the absence of the Wnt-3A ligand, transfection of 293T cells with MamL1, dominant-negative MamL1 (DnMamL1), full-length and activated human Notch 1 (Fl-N1 and N1-ICD, respectively) fails to activate the TOP-FLASH reporter (K, untreated). However, MamL1, but not DnMamL1, Fl-N1, or N1-ICD, potentiates Wnt-3A-induced activation of Wnt signaling (∼20 times) as measured by the TOP-FLASH assay (K, Wnt-3A treated). Moreover, this ability of MamL1 to activate Wnt signaling is suppressed by the presence of Fl-N1 and N1-ICD. MamL1, DnMamL1, Fl-N1, and N1-ICD had no effect on the FOP-FLASH luciferase reporter, which contains multiple copies of the mutant form of TCF-binding sites, indicating that the observed effect is specific. Normalized luciferase activities for untreated and Wnt-3A-treated cells are shown.
To extend these observations across species, we tested whether Mam can affect Wnt signaling independently of Notch by examining the human mam ortholog, mastermind-like1 (MamL1), for influence on ligand-induced activation of Wnt signaling (Willert et al. 2003). The TOP-FLASH luciferase reporter, which contains multiple copies of an optimal TCF-binding site, was used (van de Wetering et al. 1997). Transfection of 293T cells with MamL1 in the presence of the Wnt-3A ligand dramatically increases luciferase activity (∼20× times) while it remains unaffected by treatment with a constitutively activated form of human Notch 1 (Figure 8K). The ability of MamL1 to interact with β-catenin function has also been elegantly demonstrated in a ligand-independent assay using HeLa cells (Alves-Guerra et al. 2007). The genetic data indicate that mam activity functions to suppress Wnt output through arm in the Drosophila eye, and the cell culture data suggest that MamL1 behaves as a co-activator to potentiate Wnt signaling. Resolving this discrepancy requires additional mechanistic studies. Nevertheless, both observations clearly point to a role for Mam in regulating Wnt pathway activity independently of Notch and suggest the existence of additional factors that specify the effects of Mam on Wnt. Significantly, these results imply that use of dominant-negative Mam has additional effects beyond Notch signaling and thus should not be considered to inhibit only Notch activity (Proweller et al. 2007).
DISCUSSION
Availability of the Exelixis collection allowed us to perform a genetic screen for the Drosophila Notch-signaling pathway through use of C96-MamN at a breadth comparable to reverse genetic approaches. Although previous screens uncovered key Notch pathway elements (Fortini and Artavanis-Tsakonas 1994; Verheyen et al. 1996; Go and Artavanis-Tsakonas 1998; Armstrong et al. 2005; Katic et al. 2005; Mahoney et al. 2006), they were inevitably of limited genomewide scope as conventional screens cannot escape laborious mapping procedures necessary for identifying gene products for most modifiers. Even for the most comprehensive genetic screen performed to date (Karim et al. 1996), relatively few complementation groups were mapped. Thus, such screens do not assess the genetic circuitry at a near genomewide scale.
Unlike traditional approaches, which tend to generate loss-of-function mutations, the collection allows simultaneous screening of loss- and gain-of-function mutations. Moreover, hypomorphic mutations in genetically redundant genes may not display phenotypes in the screening background and not all genes are equally susceptible to mutagen treatment, precluding identification of a particular gene as a modifier in these cases. Hence, screens that also include mis-expression, such as this one, may overcome these types of limitations, and the relatively large number of recovered interactors may reflect this. Although the collection disrupts ∼50% of the Drosophila genome, this screen has doubled the number of known Notch interactors, suggesting that the complexity of Notch circuitry is greater than previously understood. Ultimately, it will be important to determine whether modifiers represent hyper- or hypomorphic mutations or to test well-characterized, preexisting alleles of genes of interest to establish epistatic relationships between Notch and these genes, thereby positioning interactors within the context of the Notch pathway.
Given the speed at which an Exelixis screen can be completed, it is feasible to study numerous aspects of Notch signaling in this manner. For example, GAL4-directed expression of transgenes inhibiting Notch signals through mutant forms of the Delta ligand, the Notch receptor, the cytoplasmic modulator of Notch activity, dx, and Su(H), a distinct nuclear effector of Notch signals, would undoubtedly broaden the genetic network corresponding to Notch activity but also help to classify the network into categories affecting signaling at distinct cellular levels. Analyses of such scope, using Notch as an example, will afford a unique perspective into the complexity and nature of cellular signaling, setting the stage for comparative studies across tissues and species.
Finally, this study validates the Exelixis collection as a valuable tool. The speed and accuracy of this analysis afford a new experimental paradigm in genetic screening. Such forward-systems analysis, combined with reverse genetic approaches, not only will help us to better understand processes governing development but also will likely uncover links relevant to human disease. Importantly, this analysis not only points to novel functional relationships involving Notch but also provides unprecedented insight into the overall complexity of the genetic circuitry of Notch signaling, providing a novel framework for future studies.
Acknowledgments
We thank Exelixis for donating the collection and the robotics necessary for its maintenance; Exelixis and the Department of Cell Biology at Harvard Medical School for providing continued support; M. Freeman and K. Matsuno for fly stocks; R. Kulathinal (FlyBase) and D. Dimlich for amending insertion annotations; and B. al-Anzi, J. Arboleda-Velasquez, D. Dimlich, L. Houghton, B. Lake, A. Louvi, and B. Obar for manuscript improvements. B.Y. was supported by National Science Foundation grant IBN-0415453. This work was supported in part by a National Institutes of Health (NIH) postdoctoral fellowship (M.W.K.) and NIH grants NS26084 and CA098402 (S.A.T.).
References
- Abdelilah-Seyfried, S., Y. M. Chan, C. Zeng, N. J. Justice, S. Younger-Shepherd et al., 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155: 733–752 (erratum: Genetics 157: 457). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Guerra, M. C., C. Ronchini and A. J. Capobianco, 2007. Mastermind-like 1 is a specific coactivator of β-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 67: 8690–8698. [DOI] [PubMed] [Google Scholar]
- Armstrong, J. A., A. S. Sperling, R. Deuring, L. Manning, S. L. Moseley et al., 2005. Genetic screens for enhancers of brahma reveal functional interactions between the BRM chromatin-remodeling complex and the delta-notch signal transduction pathway in Drosophila. Genetics 170: 1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arn, E. A., B. J. Cha, W. E. Theurkauf and P. M. Macdonald, 2003. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev. Cell 4: 41–51. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., 2004. Accessing the Exelixis collection. Nat. Genet. 36: 207. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., M. D. Rand and R. J. Lake, 1999. Notch signaling: cell fate control and signal integration in development. Science 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler et al., 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, M., H. Aslam, M. Flasza, M. Fostier, J. E. Higgs et al., 2002. Multiple levels of Notch signal regulation (review). Mol. Membr. Biol. 19: 27–38. [DOI] [PubMed] [Google Scholar]
- Bettler, D., S. Pearson and B. Yedvobnick, 1996. The nuclear protein encoded by the Drosophila neurogenic gene mastermind is widely expressed and associates with specific chromosomal regions. Genetics 143: 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Bray, S. J., 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7: 678–689. [DOI] [PubMed] [Google Scholar]
- Calgaro, S., M. Boube, D. L. Cribbs and H. M. Bourbon, 2002. The Drosophila gene taranis encodes a novel trithorax group member potentially linked to the cell cycle regulatory apparatus. Genetics 160: 547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., X. Li and I. Greenwald, 2004. sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans. Genetics 166: 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, S., L. T. Reiter, E. Bier and M. Gribskov, 2002. Homophila: human disease gene cognates in Drosophila. Nucleic Acids Res. 30: 149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperstock, R. L., and H. D. Lipshitz, 1997. Control of mRNA stability and translation during Drosophila development. Semin. Cell Dev. Biol. 8: 541–549. [DOI] [PubMed] [Google Scholar]
- Dansereau, D. A., M. D. Lunke, A. Finkielsztein, M. A. Russell and W. J. Brook, 2002. Hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster. Development 129: 5553–5566. [DOI] [PubMed] [Google Scholar]
- Dennis, G., Jr., B. T. Sherman, D. A. Hosack, J. Yang, W. Gao et al., 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4: 3. [PubMed] [Google Scholar]
- Doroquez, D. B., T. L. Orr-Weaver and I. Rebay, 2007. Split ends antagonizes the Notch and potentiates the EGFR signaling pathways during Drosophila eye development. Mech. Dev. 124: 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio, R. J., P. C. Bonnette and P. H. O'Farrell, 1998. Mutations of the Drosophila dDP, dE2F, and cyclin E genes reveal distinct roles for the E2F-DP transcription factor and cyclin E during the G1-S transition. Mol. Cell. Biol. 18: 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehebauer, M., P. Hayward and A. Martinez-Arias, 2006. Notch signaling pathway. Sci. STKE 2006: cm7. [DOI] [PubMed] [Google Scholar]
- Fortini, M. E., and S. Artavanis-Tsakonas, 1994. The suppressor of hairless protein participates in notch receptor signaling. Cell 79: 273–282. [DOI] [PubMed] [Google Scholar]
- Freeman, M., and M. Bienz, 2001. EGF receptor/Rolled MAP kinase signalling protects cells against activated Armadillo in the Drosophila eye. EMBO Rep. 2: 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go, M. J., and S. Artavanis-Tsakonas, 1998. A genetic screen for novel components of the notch signaling pathway during Drosophila bristle development. Genetics 150: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Skarmeta, J. L., R. Diez del Corral, E. de la Calle-Mustienes, D. Ferre-Marco and J. Modolell, 1996. Araucan and caupolican, two members of the novel Iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 85: 95–105. [DOI] [PubMed] [Google Scholar]
- Grimwade, B. G., M. A. Muskavitch, W. J. Welshons, B. Yedvobnick and S. Artavanis-Tsakonas, 1985. The molecular genetics of the Notch locus in Drosophila melanogaster. Dev. Biol. 107: 503–519. [DOI] [PubMed] [Google Scholar]
- Grumbling, G., and V. Strelets, 2006. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34: D484–D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiztler, P., and P. Simpson, 1991. The choice of cell fate in the epidermis. Cell 64: 1083–1092. [DOI] [PubMed] [Google Scholar]
- Helms, W., H. Lee, M. Ammerman, A. L. Parks, M. A. Muskavitch et al., 1999. Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev. Biol. 215: 358–374. [DOI] [PubMed] [Google Scholar]
- Hsu, S. I., C. M. Yang, K. G. Sim, D. M. Hentschel, E. O'Leary et al., 2001. TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. EMBO J. 20: 2273–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim, F. D., H. C. Chang, M. Therrien, D. A. Wassarman, T. Laverty et al., 1996. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143: 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada, T., M. Ito, Y. Kojima, S. Miyatani and T. Kinoshita, 2006. XMam1, Xenopus Mastermind1, induces neural gene expression in a Notch-independent manner. Mech. Dev. 123: 851–859. [DOI] [PubMed] [Google Scholar]
- Katic, I., L. G. Vallier and I. Greenwald, 2005. New positive regulators of lin-12 activity in Caenorhabditis elegans include the BRE-5/Brainiac glycosphingolipid biosynthesis enzyme. Genetics 171: 1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, M. R., S. Kidd, W. A. Deutsch and M. W. Young, 1987. Mutations altering the structure of epidermal growth factor-like coding sequences at the Drosophila Notch locus. Cell 51: 539–548. [DOI] [PubMed] [Google Scholar]
- Kitagawa, M., T. Oyama, T. Kawashima, B. Yedvobnick, A. Kumar et al., 2001. A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol. Cell. Biol. 21: 4337–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, T., L. Seugnet, M. Haenlin and A. Martinez Arias, 2000. Two different activities of Suppressor of Hairless during wing development in Drosophila. Development 127: 3553–3566. [DOI] [PubMed] [Google Scholar]
- Kramer, J. M., and B. E. Staveley, 2003. GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet. Mol. Res. 2: 43–47. [PubMed] [Google Scholar]
- Lambie, E. J., and J. Kimble, 1991. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112: 231–240. [DOI] [PubMed] [Google Scholar]
- Lehmann, R., F. Jimenez, W. Dietrich and J. A. Campos-Ortega, 1983. On the phenotype and development of mutants for early neurogenesis in Drosophila melanogaster. Rouxs Arch. Dev. Biol. 192: 62–74. [DOI] [PubMed] [Google Scholar]
- Louvi, A., and S. Artavanis-Tsakonas, 2006. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7: 93–102. [DOI] [PubMed] [Google Scholar]
- Mahoney, M. B., A. L. Parks, D. A. Ruddy, S. Y. Tiong, H. Esengil et al., 2006. Presenilin-based genetic screens in Drosophila melanogaster identify novel notch pathway modifiers. Genetics 172: 2309–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, G. P., A. J. Schroeder, M. A. Roberts and F. R. Jackson, 2001. Genetic analysis of functional domains within the Drosophila LARK RNA-binding protein. Genetics 159: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik, M., N. E. Baker and G. M. Rubin, 1990. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 4: 1848–1861. [DOI] [PubMed] [Google Scholar]
- Nagaraj, R., A. T. Pickup, R. Howes, K. Moses, M. Freeman et al., 1999. Role of the EGF receptor pathway in growth and patterning of the Drosophila wing through the regulation of vestigal. Development 126: 975–985. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Powell, P. A., C. Wesley, S. Spencer and R. L. Cagan, 2001. Scabrous complexes with Notch to mediate boundary formation. Nature 409: 626–630 (erratum: Nature 410: 718). [DOI] [PubMed] [Google Scholar]
- Proweller, A., A. C. Wright, D. Horng, L. Cheng, M. M. Lu et al., 2007. Notch signaling in vascular smooth muscle cells is required to pattern the cerebral vasculature. Proc. Natl. Acad. Sci. USA. [DOI] [PMC free article] [PubMed]
- Reiter, L. T., L. Potocki, S. Chien, M. Gribskov and E. Bier, 2001. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 11: 1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvaing, J., A. Lopez, A. Boivin, J. S. Deutsch and F. Peronnet, 2003. The Drosophila Corto protein interacts with Polycomb-group proteins and the GAGA factor. Nucleic Acids Res. 31: 2873–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., A. S. McElhinny, Y. Cao, P. Gao, J. Liu et al., 2006. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 20: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller, D., C. Friedel, A. Schmid, D. Bettler, L. Lam et al., 1990. The Drosophila neurogenic locus mastermind encodes a nuclear protein unusually rich in amino acid homopolymers. Genes Dev. 4: 1688–1700. [DOI] [PubMed] [Google Scholar]
- Sugimoto, M., T. Nakamura, N. Ohtani, L. Hampson, I. N. Hampson et al., 1999. Regulation of CDK4 activity by a novel CDK4-binding protein, p34(SEI-1). Genes Dev. 13: 3027–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- van de Wetering, M., R. Cavallo, D. Dooijes, M. van Beest, J. van Es et al., 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88: 789–799. [DOI] [PubMed] [Google Scholar]
- Verheyen, E. M., K. J. Purcell, M. E. Fortini and S. Artavanis-Tsakonas, 1996. Analysis of dominant enhancers and suppressors of activated Notch in Drosophila. Genetics 144: 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert, K., J. D. Brown, E. Danenberg, A. W. Duncan, I. L. Weissman et al., 2003. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423: 448–452. [DOI] [PubMed] [Google Scholar]
- Wu, L., J. C. Aster, S. C. Blacklow, R. Lake, S. Artavanis-Tsakonas et al., 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26: 484–489. [DOI] [PubMed] [Google Scholar]
- Xu, T., and S. Artavanis-Tsakonas, 1990. deltex, a locus interacting with the neurogenic genes, Notch, Delta and mastermind in Drosophila melanogaster. Genetics 126: 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedvobnick, B., W. Helms and B. Barrett, 2001. Identification of chromosomal deficiencies that modify Drosophila mastermind mutant phenotypes. Genesis 30: 250–258. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., R. B. Katzman, L. M. Delmolino, I. Bhat, Y. Zhang et al., 2007. The notch regulator maml1 interacts with p53 and functions as a coactivator. J. Biol. Chem. 282: 11969–11981. [DOI] [PubMed] [Google Scholar]