Abstract
The proportions of mutant and wild-type mtDNA are crucial in determining the severity of mitochondrial diseases. It has been generally considered that deletion-mutant mtDNA has replication advantages and accumulates with time. Here, we examine the tissue-by-tissue proportions of mutant mtDNA with a 4696-bp deletion (ΔmtDNA) and wild-type mtDNA in mitochondrial disease model mice (mito-mice). Comparison of the proportions of ΔmtDNA in each tissue at various ages showed that the rate of accumulation of ΔmtDNA differed among tissues. The heart, skeletal muscles, kidney, liver, testis, and ovary showed increases in the proportion of ΔmtDNA with age, but the pancreas, spleen, brain, and blood showed only a slight or no increase in proportion. In contrast to the somatic tissues, however, the germ cells of female mito-mice and resultant offspring showed a strong decrease in ΔmtDNA with maternal age. The decrease was so acute that some offspring showed complete disappearance of ΔmtDNA, even though their elder brothers and sisters had high proportions of ΔmtDNA. Female germ cells have a machinery that prevents the inheritence of defective mtDNA to the following generation since germ cells are kept for a long time until they are ovulated.
EACH mammalian cell contains several hundred to thousands of copies of mitochondrial DNA (mtDNA), encoding 13 proteins (all of which are OXPHOS subunits), 22 tRNAs, and two rRNAs (Anderson et al. 1981). Point- and deletion-mutant mtDNAs have been shown to be important causes of human mitochondrial diseases, such as mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (Goto et al. 1990), Leber's hereditary optic neuropathy (Wallace et al. 1988), chronic progressive external ophthalmoplegia, and Kearns-Sayre syndrome (Holt et al. 1988).
In general, all of the mtDNA in an individual is thought to be identical (homoplasmy), because mammalian mtDNA shows strictly maternal inheritance (Kaneda et al. 1995; Shitara et al. 1998). However, in human individuals with mitochondrial disease, wild-type and mutant mtDNAs coexist at different levels (heteroplasmy), probably because some pathogenic mutant mtDNAs are lethal in the homoplasmic state. Levels of heteroplasmy (i.e., the proportions of wild-type and mutant mtDNAs) are considered to fluctuate through the random distribution of mtDNA to daughter cells during cell division (Birky 1994). Levels of heteroplasmy also depend on the specific mtDNA haplotype (Hayashi et al. 1987; Jenuth et al. 1997) and nuclear background (Dunbar et al. 1995; Battersby et al. 2003). Since the proportion of mutant mtDNA directly affects the severity of mitochondrial diseases, it is important to investigate fluctuations in the level of heteroplasmy in each tissue at various ages.
Previously, Jenuth et al. (1997) reported tissue-specific and age-related selection for different mtDNA genotypes in the same heteroplasmic mice possessing New Zealand Black (NZB) and Bagg Albino (BALB) mtDNAs. Here we used mito-mice (Inoue et al. 2000; Nakada et al. 2001), which possess wild-type and pathogenic 4696-bp deletion-mutant mtDNA (ΔmtDNA), to compare the levels of heteroplasmy of ΔmtDNA between tissues. The deletion removes six tRNAs and seven structural genes, and cells with >80% ΔmtDNA show respiratory deficiency and resultant mitochondrial diseases (Inoue et al. 2000, 2007; Nakada et al. 2001, 2004, 2006). The proportion of ΔmtDNA increased with time in cultured cells (Inoue et al. 2000) and in the tails of mito-mice (Sato et al. 2005), as has been observed in human subjects with deletion-mutant mtDNA (Larsson et al. 1990). However, the age-associated accumulation of ΔmtDNA in other tissues of mito-mice, which directly influences the expression of mitochondrial disease, has not yet been examined.
We found that no somatic tissue showed a drastic decline of ΔmtDNA with age and that each tissue had specific accumulation rates of ΔmtDNA. On the other hand, female germ cells and the resultant offspring showed a maternal age-dependent decrease in the proportions of ΔmtDNA. In terms of chromosome abnormality, it is generally accepted that the risk of having offspring with defects (such as Down syndrome in humans) increases with age. In contrast, our findings indicate that the risk of having offspring with mitochondrial defects decreases with age in mito-mice.
MATERIALS AND METHODS
Estimation of ΔmtDNA proportions by Southern blot analysis:
Southern blot analysis was performed to estimate the ΔmtDNA proportions in the tissues. A representative of Southern blot image is shown in supplemental Figure 1 at http://www.genetics.org/supplemental/. Briefly, XhoI-digested total DNA was separated on 0.6% agarose gel and transferred to a nylon membrane. Hybridization was carried out with a mtDNA probe (nucleotide positions 1859–2762) labeled by AlkPhos direct labeling and detection system (GE Healthcare, Buckinghamshire, UK). Images were obtained by exposing Hyperfilm ECL (GE Healthcare). The density of signals was measured by using the public domain National Institutes of Health (NIH) image program (developed at NIH and available at http://rsb.info.nih.gov/nih-image/). The Southern blot analysis was repeated three times for each sample and the mean of the values was represented as the proportion of ΔmtDNA.
Estimation of ΔmtDNA proportions by real-time monitoring PCR:
Real-time monitoring PCR was used to estimate the proportions of ΔmtDNA in superovulated oocytes, nongrowing oocytes of newborn mito-mice, and tissues that showed no signal of ΔmtDNA by Southern blot analysis. It was performed with a TaqMan PCR reagent kit and an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA). To estimate the absolute copy number of wild-type mtDNA and ΔmtDNA, we used the standard curve method. The standard curve for the assay was calculated using a series of 10-fold dilution of titrated synthetic standard DNA. Each measurement was repeated three times and the proportions of ΔmtDNA and total mtDNA were calculated. The primer set specific for ΔmtDNA was TTTCACTATGAAGCTAAGAGCGTTAACCT and GGTGGAATCGGACCAGTAGGA. The reporter dye 6-carboxyfluorescein (FAM)-labeled TaqMan minor-groove-binder probe specific for ΔmtDNA was AACTGGTGTATGGAGATTT. The primer set specific for wild-type mtDNA was AACCTGGCACTGAGTCACCA and GGGTCTGAGTGTATATATCATGAAGAGAAT. The reporter dye FAM and the quencher dye 6-carboxy-tetramethyl-rhodamine-labeled probe was TCTGTAGCCCTTTTTGTCACATGATC.
Collection of oocytes:
Mito-mice were induced to superovulation by injection of pregnant mare serum gonadotropin and then 48 hr later by injection of human chorionic gonadotropin (hCG). Fifteen hours after the hCG injection, superovulated oocytes were collected from anesthetized mito-mice at 44 days old from the left oviduct and at 117 days old from the right oviduct. To collect nongrowing oocytes of the newborn mito-mice, ovaries were placed in PBS with 0.02% EDTA (Ca2+ free) for 10 min at 37°, and dissociated oocytes were collected using glass pipettes (Kono et al. 1996).
Ovarian grafting:
Mito-mice were euthanized and both left and right ovaries were collected. Ovaries of anesthetized host B6mtspr mice were removed from ovarian bursal cavities, and donor mito-mice ovaries were placed in ovarian bursal cavities. Two weeks after the grafting, the mice were caged with male B6 mice and offspring were obtained by natural mating. Because B6mtspr mice possess the C57BL/6J nuclear genome and Mus spretus mtDNA, we could distinguish the offspring derived from donor mito-mice (M. musculus) ovaries and host B6mtspr by analysis of mtDNA sequence polymorphisms.
Statistical analyses:
The relationship between age and proportion of ΔmtDNA in tissues was analyzed by calculating the Pearson's correlation coefficient. For comparison of the increased ΔmtDNA accumulation rate among tissues, we carried out ANCOVA and P-values of interaction were obtained. The relationships among age, proportion of ΔmtDNA, and total mtDNA content were analyzed with ANCOVA. Values with P < 0.05 were considered significant.
RESULTS
Proportions of ΔmtDNA in somatic tissues of mito-mice:
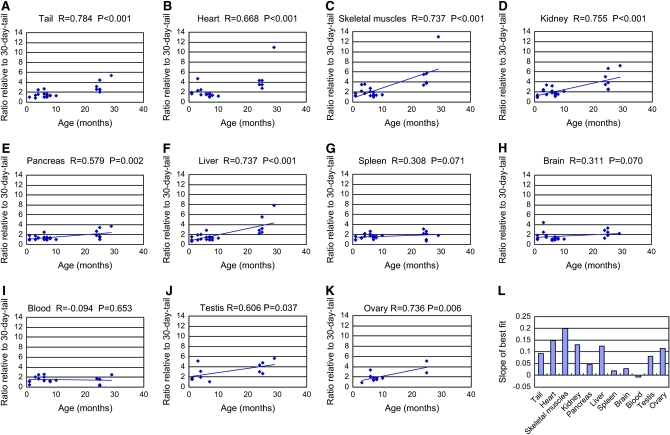
We euthanized mito-mice at various ages and examined the proportions of ΔmtDNA in their tissues. To compare the rates of accumulation of ΔmtDNA among tissues, the proportions of ΔmtDNA in each tissue of a mito-mouse were normalized with the proportion of ΔmtDNA in the tail when the same mito-mouse was 30 days old. (Figure 1, A–K). When mito-mice were young, the proportions of ΔmtDNA in tissues were almost the same as that of the tail at 30 days old. As the mito-mice aged, the proportions of ΔmtDNA became higher than that of the tail at 30 days in the tail, heart, skeletal muscles, kidney, liver, testis, and ovary. The other tissues that we examined (pancreas, spleen, brain, and blood) showed only a slight or no increase in the proportion of ΔmtDNA with age. The slopes of the best fit significantly differed when the former and the latter tissues were compared (supplemental Table 1 at http://www.genetics.org/supplemental/). The rates of accumulation of ΔmtDNA in each tissue are indicated in Figure 1L. Although we measured total mtDNA content in tissues, there was no significant correlation between age and total mtDNA content or between proportion of ΔmtDNA and total mtDNA content (supplemental Figures 2 and 3, supplemental Table 2).
Figure 1.—
Changes in proportions of ΔmtDNA in tissues with age. (A–K) Relationship between proportion of ΔmtDNA in a tissue and the age at euthanasia. Proportion of ΔmtDNA in tissues shows ratios relative to those in the tail at 30 days old. Pearson's product-moment correlation coefficient and its probability are indicated as R and P, respectively. (L) Rates of accumulation of ΔmtDNA in tissues. The slopes of best fit (ratio/month) (A–K) are indicated as bars.
One possible cause of the change of ΔmtDNA in tissues is selection against cells with high ΔmtDNA. To examine the effect of selection against cells, we removed some mito-mice, which had tissues with >80% ΔmtDNA, and examined the increase rate of ΔmtDNA. If cells with a high proportion of ΔmtDNA were selectively removed, it would affect the slope of the best fit. When we removed mice with >80% ΔmtDNA, the slopes of the best fit were not significantly different from the ones containing mice with >80% ΔmtDNA (supplemental Figure 4 at http://www.genetics.org/supplemental/). This result indicates that selection against cells within tissues is not very great in mito-mice.
Proportions of ΔmtDNA in offspring and oocytes:
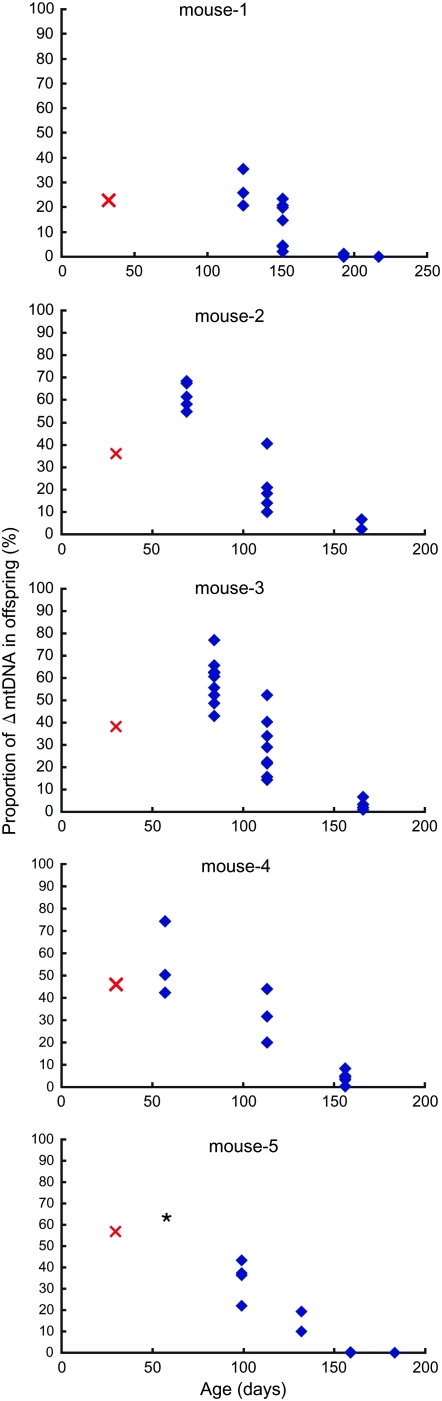
Although maternal transmission of deletion-mutant mtDNA is very rare in human cases (Larsson and Clayton 1995; Wallace 1999), it has been observed in mito-mice (Inoue et al. 2000). We examined the correlation between the proportion of ΔmtDNA in offspring and the age of maternal mito-mice. As the maternal mito-mice aged, the proportions of ΔmtDNA in their offspring declined (Figure 2; supplemental Figure 5 and supplemental Table 3 at http://www.genetics.org/supplemental/). This trend was independent of the proportion of ΔmtDNA in the maternal mito-mice. There was no correlation between maternal age and the litter size at each delivery. Some offspring delivered from mouse 1, mouse 5, and mouse S1 showed complete absence of ΔmtDNA, even with the highly sensitive PCR assay (data not shown).
Figure 2.—
Relationship between proportion of ΔmtDNA in offspring and maternal age. Proportion of ΔmtDNA in the tails of offspring at 30 days old are indicated by blue diamonds. Proportion of ΔmtDNA in the tails of maternal mito-mice at 30 days old are indicated by red crosses. Horizontal axes indicate the age of the maternal mito-mice at each delivery. At the first delivery of mouse 5, a neonate died immediately after birth; the ΔmtDNA proportion in its tail is indicated by the asterisk. In some offspring, ΔmtDNA was not detected (mouse 1: one of three in the third litter and all three in the fourth litter; mouse 5: one of two in the fourth litter and all five in the fifth litter).
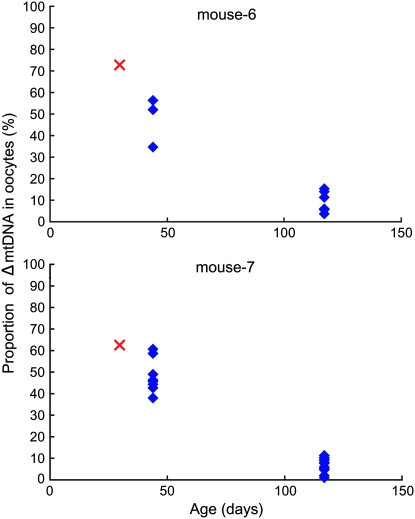
A decline in the proportion of ΔmtDNA with maternal age was also observed in oocytes collected by superovulation (Figure 3; supplemental Table 4 at http://www.genetics.org/supplemental/). This implies that oocytes with lower ΔmtDNA were ovulated as the maternal mito-mice aged and the resultant offspring had lower ΔmtDNA than their elder brothers or sisters. The decline in the proportion of ΔmtDNA in the germ cells was specific to females; the proportion of ΔmtDNA in sperm did not decrease with age (data not shown).
Figure 3.—
Proportion of ΔmtDNA in oocytes collected at different ages. Mito-mice were superovulated twice and oocytes were collected from left and right oviducts at 44 and 117 days old, respectively. Proportion of ΔmtDNA in oocytes is indicated by blue diamonds. Red crosses indicate the proportions of ΔmtDNA in the tails of oocyte-donor mito-mice at 30 days old.
Proportions of ΔmtDNA in offspring of ovary-grafted mice:
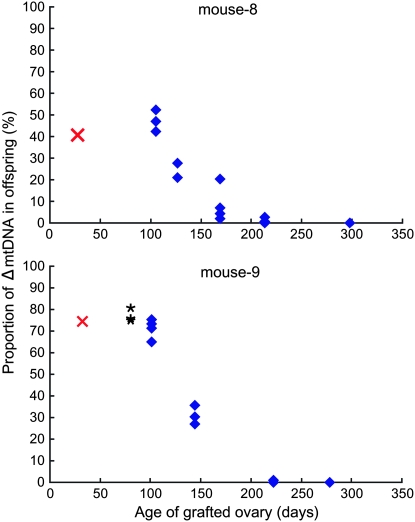
Mito-mice usually have short reproductive spans because they die early in life from renal failure caused by the presence of ΔmtDNA (Inoue et al. 2000). To obtain a prolonged reproductive span, we replaced the ovaries of B6mtspr mice with those of mito-mice. Since B6mtspr mice possess the B6 nuclear genome and M. spretus mtDNA, we could distinguish the offspring derived from the donor mito-mice (M. musculus) ovaries and the host B6mtspr ones by the analysis of mtDNA sequence polymorphisms. The ovary-grafted mice were mated with male B6 mice and offspring were obtained. The ovary-grafted mice delivered normally and most of the offspring had mito-mice-specific mtDNA genotypes (a few offspring with M. spretus mtDNA were obtained from mouse 8). The proportion of ΔmtDNA also declined in the offspring delivered from ovary-grafted mice (Figure 4; supplemental Table 5 at http://www.genetics.org/supplemental/). Some offspring delivered from both mouse 8 and mouse 9 showed complete absence of ΔmtDNA (mouse 8: two of six in the fourth litter and all three in the fifth litter; mouse 9: three of five in the fourth litter and two of three in the fifth litter).
Figure 4.—
Proportion of ΔmtDNA in offspring obtained from mice grafted with ovaries of mito-mice. Proportion of ΔmtDNA in the tails of offspring are indicated by blue diamonds. Red crosses indicate the ΔmtDNA proportions in the tails of the ovarian donor mito-mice at 30 days old. Horizontal axes indicate the age of the grafted ovaries at each delivery. From mouse 8, host-ovary-derived offspring possessing M. spretus mtDNA were obtained in the first, third, fourth, and fifth litter (not indicated in the graph). At the first delivery of mouse 9, all three neonates died immediately after birth; the ΔmtDNA proportions in their tails are indicated by asterisks. In some offspring, ΔmtDNA was not detected (mouse 8: two of six in the fourth litter and all three in the fifth litter; mouse 9: three of five in the fourth litter and two of three in the fifth litter).
Possible mechanism of the decline of ΔmtDNA in offspring as a function of maternal age:
One possible explanation for the decline of the proportion of ΔmtDNA in oocytes with maternal age is the selection against oocytes or the post-implantation embryos. The absence of oocytes with >80% ΔmtDNA implies the presence of selection against oocytes (Sato et al. 2005). Although the efficiency of fertilization was not different between oocytes with or without ΔmtDNA (data not shown), post-implantation development was impaired when embryos contained >60% ΔmtDNA because it would reach 80% during gestation (Sato et al. 2005). These facts indicate that there is a selection against oocytes and post-implantation embryos. However, even if the selection against oocytes with a high proportion of ΔmtDNA exists in ovaries, it cannot explain the absence of oocytes or offspring with a low proportion of ΔmtDNA at a young age (Figures 2–4). It is probable that selection against oocytes or embryos does not contribute very much to the decline of the proportion of ΔmtDNA in oocytes as females age.
Another possibility for the decline of the proportion of ΔmtDNA in oocytes with maternal age is selective ovulation: oocytes with various proportions of ΔmtDNA exist in the ovary and those with higher proportions of ΔmtDNA are selectively ovulated when maternal mito-mice are young. Another possibility is that the proportions of ΔmtDNA in all oocytes are high at first, but ΔmtDNA disappears gradually in each oocyte with age. To test which is the case, we examined the proportions of ΔmtDNA in the nongrowing oocytes of newborn mito-mice. All oocytes are synchronously arrested in the diplotene stage of the first meiotic division in the newborn mouse. The result is shown in Table 1. The coefficient of variation (CV) of the proportion of ΔmtDNA in nongrowing oocytes was 0.19–0.21. This range was closer to the CV of the first litters (0.04–0.30) than to that of all litters (0.71–1.33) (supplemental Tables 3 and 5 at http://www.genetics.org/supplemental/). Moreover, nongrowing oocytes with no ΔmtDNA (as observed in Figures 2 and 4) were not observed. These results indicate that the latter hypothesis is the mechanism underlying the maternal age-associated decrease in the proportion of ΔmtDNA in the offspring.
TABLE 1.
ΔmtDNA in nongrowing oocytes from newborn mito-mice
| Nongrowing oocytes
|
|||||
|---|---|---|---|---|---|
| Tail (%) | Ovary (%) | Mean (%) | SD | CV | n |
| 57.8 | 54.6 | 60.5 | 12.8 | 0.21 | 48 |
| 55.0 | 62.4 | 66.5 | 12.9 | 0.19 | 49 |
| 69.1 | 74.7 | 73.7 | 15.8 | 0.21 | 47 |
| 55.7 | 59.1 | 68.3 | 12.6 | 0.18 | 49 |
“Tail” is the proportion of ΔmtDNA in the tail of the newborn mito-mice. “Ovary” is the proportion of ΔmtDNA in the ovary of the newborn mito-mice. “Mean” is the mean of the proportion of ΔmtDNA in nongrowing oocytes of the mito-mice. SD, standard deviation. n, number of nongrowing oocytes investigated.
DISCUSSION
Because it is smaller than wild-type mtDNA, and thus is thought to have replication advantages, deletion-mutant mtDNA tends to accumulate in the postmitotic tissues of patients with mitochondrial disease (Larsson and Clayton 1995; Wallace 1999). In cultured cells, deletion-mutant mtDNA was reported to accumulate with time (Hayashi et al. 1991) but not in another study (Tang et al. 2000). We show here that the rate of accumulation of ΔmtDNA differed among tissues. The heart, skeletal muscles, kidney, liver, testis, and ovary showed increases in the proportion of ΔmtDNA with age, but the pancreas, spleen, brain, and blood showed only a slight or no increase (Figure 1). In some of the former tissues, mito-mice show clinical phenotypes such as auriculoventricular block, renal failure, and meiotic arrest of spermatocyte (Inoue et al. 2000; Nakada et al. 2001, 2006). In the latter tissues, severe clinical phenotypes have not been observed so far. This means that the rate of accumulation of ΔmtDNA is closely related to the expression of mitochondrial diseases in mito-mice. However, the rate of accumulation of ΔmtDNA does not solely explain the expression of mitochondrial diseases, since the liver and ovary of mito-mice, which showed high rates of ΔmtDNA accumulation, have no defects. Another key to the expression of mitochondrial diseases is the tissue-specific energy requirement threshold. For example, arrest of spermatocytes occurs when the proportion of ΔmtDNA is >70% (Nakada et al. 2006), but the threshold of respiratory defects is >80% in cultured cells or in the skeletal muscles (Inoue et al. 2000; Nakada et al. 2001). Thus, to predict the phenotypic expression of mitochondrial diseases, we need to consider both the trends in ΔmtDNA accumulation and the energy requirement threshold of each tissue.
The proportion of deletion-mutant mtDNA is reduced in the blood of patients with mitochondrial disease when compared with other postmitotic tissues (Lestienne and Ponsot 1988). This probably occurs because cells with reduced levels of mutant mtDNA are selected in mitotic tissue as a result of continuous cell division and mtDNA segregation. In this study, however, the tissues that showed no increase of ΔmtDNA were not confined to cells undergoing active mitosis. In addition, selection against cells within tissues seemed to make little contribution to the change in the proportion of ΔmtDNA (supplemental Figure 4 at http://www.genetics.org/supplemental/). Thus, some events occurring within cells might govern the change of the proportion of ΔmtDNA. A candidate is autophagy. The membrane-potential-dependent mitochondrial digestion by autophagy has been reported in cultured cells (Elmore et al. 2004). Another candidate is the preferential replication for specific mtDNA. The expression levels of mtDNA replication factors, such as mtTFA or PolG, might be involved in the preferential replication of specific mtDNA.
In this study, we concluded that ΔmtDNA disappears gradually from oocytes with age. However, this idea is based on the orthodox view of oogenesis: female primordial germ cells finish mitosis at an early embryonic stage and are not replenished after birth. Recently, it was reported that new oocytes and follicles are generated in the adult mouse ovary (Johnson et al. 2005a) and that germline stem cells (GSCs) in the bone marrow and peripheral blood contribute the replenishment (Johnson et al. 2005b). If GSCs were to exist and continued mitosis in adult mice, then the mtDNA population in GSCs could drift toward homoplasmy of either ΔmtDNA or wild-type mtDNA (Wallace 1981; Birky 1994). If this were the case, then the GSCs with a high proportion of ΔmtDNA would die of respiratory deficiency, and the remaining GSCs with low proportions of ΔmtDNA would survive and be ovulated. However, we observed a decline in ΔmtDNA proportions in the offspring even after ovarian grafting (Figure 4). This indicates that GSCs in the bone marrow do not contribute to the decline of ΔmtDNA proportions in oocytes. Moreover, the above hypothesis does not seem likely for two additional reasons. One is that the proportions of ΔmtDNA in rapidly dividing tissues such as the blood or spleen do not decrease with age in mito-mice (Figure 1). The other is that the mtDNA population in the offspring of mito-mice drifted only toward wild-type mtDNA (Figures 2–4). If the above hypothesis were the case, we would see a shift toward both ΔmtDNA and wild-type mtDNA. Therefore, we suppose that continued mitosis of GSCs and resultant mtDNA segregation cannot explain the gradual disappearance of ΔmtDNA.
The mechanism of decline of the proportion of ΔmtDNA in oocytes with maternal age is currently unknown. In our preliminary study, heteroplasmic mice possessing M. spretus and M. musculus mtDNA showed no segregation toward either genotype in their offspring with maternal age (data not shown). Similarly, heteroplasmic mice with NZB and BALB mtDNA showed no correlation between maternal age and segregation toward either mtDNA in their offspring (Jenuth et al. 1997). Thus, this phenomenon is specific to mito-mice, which possess pathogenic deletion-mutant mtDNA. Considering that the proportion of ΔmtDNA continued to decrease below the threshold of cellular respiratory deficiency, a reduction in the cellular energetic state does not trigger a decline in ΔmtDNA in oocytes. Like the change of ΔmtDNA accumulation rate in other tissues, the selective replication of specific mtDNA (in this case, wild-type mtDNA) or the selection against mitochondria might be involved in the decline of ΔmtDNA in oocytes.
Previously, we reported selection against male germ cells with high ΔmtDNA (Nakada et al. 2006). Female germ cells complete an entry into meiosis before birth, in contrast to male germ cells, which continue meiosis throughout their life. Therefore, meiotic selection against oocytes finishes in the early developmental stage. Females have to cope with the accumulation of ΔmtDNA while oocytes are kept in the ovary for a long time until ovulation. The decline of the proportion of ΔmtDNA in oocytes might reflect a compensatory correction mechanism that is cumulatively manifest as a function of age.
Does the phenomenon of a decrease in ΔmtDNA in offspring as maternal age increases also exist in humans? We reviewed several published reports of mitochondrial disease pedigrees showing the transmission of deletion- and point-mutant mtDNAs. In the pedigrees that we reviewed, affected subjects were not restricted to older brothers or sisters, and the proportion of mutant mtDNA was not significantly higher in older siblings (Ballinger et al. 1992; Bernes et al. 1993; Hammans et al. 1993; De Vries et al. 1994; Puoti et al. 2003). Moreover, epidemiological studies showed no correlation between maternal age and risk of inheritance of pathogenic deletion-mutant mtDNA (Brenner et al. 1998; Chinnery et al. 2004). Thus, although we cannot directly apply our findings to humans with pathogenic mutant mtDNAs, an understanding of the underlying mechanism could lead to development of a new treatment for mitochondrial diseases.
Acknowledgments
This work was supported by Grant-in-Aid 14GS0305 for Creative Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to J.-I.H.), by Grant-in-Aid for Young Scientists (A) 17689023 (to K.N.) and (B) 18790234 (to A.S.) from Japan Society for Promotion of Science, by the Research Grant 17A-10 for Nervous and Mental Disorders from the Ministry of Health, Labor and Welfare (to K.N.), and by grants for a Research Fellowship from the Japan Society for Promotion of Science for Young Scientists 09010700 (to A.K.).
References
- Anderson, S., A. T. Bankier, B. G. Barrell, M. H. de Bruijn, A. R. Coulson et al., 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. [DOI] [PubMed] [Google Scholar]
- Ballinger, S. W., J. M. Shoffner, E. V. Hedaya, I. Trounce, M. A. Polak et al., 1992. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat. Genet. 1: 11–15. [DOI] [PubMed] [Google Scholar]
- Battersby, B. J., J. C. Loredo-Osti and E. A. Shoubridge, 2003. Nuclear genetic control of mitochondrial DNA segregation. Nat. Genet. 33: 183–186. [DOI] [PubMed] [Google Scholar]
- Bernes, S. M., C. Bacino, T. R. Prezant, M. A. Pearson, T. S. Wood et al., 1993. Identical mitochondrial DNA deletion in mother with progressive external ophthalmoplegia and son with Pearson marrow-pancreas syndrome. J. Pediatr. 123: 598–602. [DOI] [PubMed] [Google Scholar]
- Birky, C. W., 1994. Relaxed and stringent genomes: why cytoplasmic genes don't obey Mendel's laws. J. Hered. 85: 355–365. [Google Scholar]
- Brenner, C. A., Y. M. Wolny, J. A. Barritt, D. W. Matt, S. Munne et al., 1998. Mitochondrial DNA deletion in human oocytes and embryos. Mol. Hum. Reprod. 4: 887–892. [DOI] [PubMed] [Google Scholar]
- Chinnery, P. F., S. DiMauro, S. Shanske, E. A. Schon, M. Zeviani et al., 2004. Risk of developing a mitochondrial DNA deletion disorder. Lancet 364: 592–596. [DOI] [PubMed] [Google Scholar]
- de Vries, D, I. de Wijs, W. Ruitenbeek, J. Begeer, P. Smit et al., 1994. Extreme variability of clinical symptoms among sibs in a MELAS family correlated with heteroplasmy for the mitochondrial A3243G mutation. J. Neurol. Sci. 124: 77–82. [DOI] [PubMed] [Google Scholar]
- Dunbar, D. R., P. A. Moonie, H. T. Jacobs and I. J. Holt, 1995. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc. Natl. Acad. Sci. USA 92: 6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore, S. P., Y. Nishimura, T. Qian, B. Herman and J. J. Lemasters, 2004. Discrimination of depolarized from polarized mitochondria by confocal fluorescence resonance energy transfer. Arch. Biochem. Biophys. 422: 145–152. [DOI] [PubMed] [Google Scholar]
- Goto, Y., I. Nonaka and S. Horai, 1990. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348: 651. [DOI] [PubMed] [Google Scholar]
- Hammans, S. R., M. G. Sweeney, M. Brockington, G. G. Lennox, N. F. Lawton et al., 1993. The mitochondrial DNA transfer RNA(Lys)A→G(8344) mutation and the syndrome of myoclonic epilepsy with ragged red fibres (MERRF). Relationship of clinical phenotype to proportion of mutant mitochondrial DNA. Brain 116: 617–632. [DOI] [PubMed] [Google Scholar]
- Hayashi, J., H. Yonekawa, J. Murakami, Y. Tagashira, O. M. Pereira-Smith et al., 1987. Mitochondrial genomes in intraspecies mammalian cell hybrids display codominant or dominant/recessive behavior. Exp. Cell Res. 172: 218–227. [DOI] [PubMed] [Google Scholar]
- Hayashi, J., S. Ohta, A. Kikuchi, M. Takemitsu, Y. Goto et al., 1991. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 88: 10614–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, I. J., A. E. Harding and J. A. Morgan-Hughes, 1988. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331: 717–719. [DOI] [PubMed] [Google Scholar]
- Inoue, K., K. Nakada, A. Ogura, K. Isobe, Y. Goto et al., 2000. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat. Genet. 26: 176–181. [DOI] [PubMed] [Google Scholar]
- Inoue, S., M. Yokota, K. Nakada, H. Miyoshi and J. Hayashi, 2007. Pathogenic mitochondrial DNA-induced respiration defects in hematopoietic cells result in anemia by suppressing erythroid differentiation. FEBS Lett. 581: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Jenuth, J. P., A. C. Peterson and E. A. Shoubridge, 1997. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 16: 93–95. [DOI] [PubMed] [Google Scholar]
- Johnson, J., J. Bagley, M. Skaznik-Wikiel, H. J. Lee, G. B. Adams et al., 2005. a Oocyte generation in adult mammalian ovaries by putative germ cells in bone marrow and peripheral blood. Cell 122: 303–315. [DOI] [PubMed] [Google Scholar]
- Johnson, J., J. Canning, T. Kaneko, J. K. Pru and J. L. Tilly, 2005. b Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature 428: 145–150. [DOI] [PubMed] [Google Scholar]
- Kaneda, H., J. Hayashi, S. Takahama, C. Taya, K. F. Lindahl et al., 1995. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc. Natl. Acad. Sci. USA 92: 4542–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono, T., Y. Obata, T. Yoshimizu, T. Nakahara and J. Carroll, 1996. Epigenetic modifications during oocyte growth correlates with extended parthenogenetic development in the mouse. Nat. Genet. 13: 91–94. [DOI] [PubMed] [Google Scholar]
- Larsson, N. G., and D. A. Clayton, 1995. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 29: 151–178. [DOI] [PubMed] [Google Scholar]
- Larsson, N. G., E. Holme, B. Kristiansson, A. Oldfors and M. Tulinius, 1990. Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome. Pediatr. Res. 28: 131–136. [DOI] [PubMed] [Google Scholar]
- Lestienne, P., and G. Ponsot, 1988. Kearns-Sayre syndrome with muscle mitochondrial DNA deletion. Lancet 1: 885. [DOI] [PubMed] [Google Scholar]
- Nakada, K., K. Inoue, T. Ono, K. Isobe, A. Ogura et al., 2001. Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med. 7: 934–940. [DOI] [PubMed] [Google Scholar]
- Nakada, K., A. Sato, H. Sone, A. Kasahara, K. Ikeda et al., 2004. Accumulation of pathogenic ΔmtDNA induced deafness but not diabetic phenotypes in mito-mice. Biochem. Biophys. Res. Commun. 323: 175–184. [DOI] [PubMed] [Google Scholar]
- Nakada, K., A. Sato, K. Yoshida, T. Morita, H. Tanaka et al., 2006. Mitochondria-related male infertility. Proc. Natl. Acad. Sci. USA 103: 15148–15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti, G., F. Carrara, S. Sampaolo, M. De Caro, C. M. Vincitorio et al., 2003. Identical large scale rearrangement of mitochondrial DNA causes Kearns-Sayre syndrome in a mother and her son. J. Med. Genet. 40: 858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, A., T. Kono, K. Nakada, K. Ishikawa, S. Inoue et al., 2005. Gene therapy for progeny of mito-mice carrying pathogenic mtDNA by nuclear transplantation. Proc. Natl. Acad. Sci. USA 102: 16765–16770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitara, H., J. Hayashi, S. Takahama, H. Kaneda and H. Yonekawa, 1998. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 148: 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y., G. Manfredi, M. Hirono and E. A. Schon, 2000. Maintenance of human rearranged mitochondrial DNAs in long-term cultured transmitochondrial cell lines. Mol. Biol. Cell 11: 2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, D. C., 1981. Mitotic segregation of mitochondrial DNAs in human cell hybrids and expression of chloramphenicol resistance. Somat. Cell Mol. Genet. 12: 41–49. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., 1999. Mitochondrial diseases in man and mouse. Science 283: 1482–1488. [DOI] [PubMed] [Google Scholar]
- Wallace, D. C., G. Singh, M. T. Lott, J. A. Hodge, T. G. Schurr et al., 1988. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242: 1427. [DOI] [PubMed] [Google Scholar]