Abstract
The medaka Oryzias latipes and its two sister species, O. curvinotus and O. luzonensis, possess an XX–XY sex-determination system. The medaka sex-determining gene DMY has been identified on the orthologous Y chromosome [O. latipes linkage group 1 (LG1)] of O. curvinotus. However, DMY has not been discovered in other Oryzias species. These results and molecular phylogeny suggest that DMY was generated recently [∼10 million years ago (MYA)] by gene duplication of DMRT1 in a common ancestor of O. latipes and O. curvinotus. We identified seven sex-linked markers from O. luzonensis (sister species of O. curvinotus) and constructed a sex-linkage map. Surprisingly, all seven sex-linked markers were located on an autosomal linkage group (LG12) of O. latipes. As suggested by the phylogenetic tree, the sex chromosomes of O. luzonensis should be “younger” than those of O. latipes. In the lineage leading to O. luzonensis after separation from O. curvinotus ∼5 MYA, a novel sex-determining gene may have arisen and substituted for DMY. Oryzias species should provide a useful model for evolution of the master sex-determining gene and differentiation of sex chromosomes from autosomes.
MAMMALS and birds have genetic sex determination with cytogenetically well-differentiated sex chromosomes. By contrast, various sex-determination mechanisms have evolved independently in fishes, and most species with genetic sex determination have undifferentiated sex chromosomes (Solari 1994; Devlin and Nagahama 2002). Recent studies have shown that different sex chromosomes have evolved even among closely related fishes (Woram et al. 2003; Takehana et al. 2007) or among intraspecific populations (Volff and Schartl 2001), but the mechanisms for these changes are unknown.
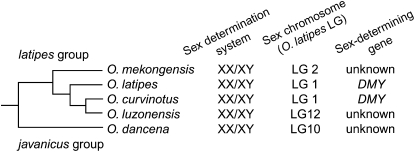
A phylogenetic tree of the medaka, Oryzias latipes, and its relatives is available in Takehana et al. (2003) and has been redrawn here in Figure 1 with the species' sex-determining system. O. latipes and a sister-species pair, O. curvinotus and O. luzonensis, have an XX–XY genetic sex-determination system (Aida 1921; Matsuda et al. 2003; Hamaguchi et al. 2004). Like other fishes, these Oryzias species have no heteromorphic sex chromosomes (Uwa and Ojima 1981; Matsuda et al. 1998), and their sex chromosomes can be regarded as being in a primitive stage of differentiation.
Figure 1.—
Phylogenetic relationship of Oryzias species based on mitochondrial DNA sequences. Using cytochrome b gene sequences from Takehana et al. (2003), we redrew a linearized neighbor-joining tree. The separation of O. luzonensis from O. curvinotus was estimated to have occurred ∼5 MYA according to a divergence rate of 2.8%/MY. DMY has been identified only in O. latipes and O. curvinotus.
A DM-domain gene, DMY, has been identified in the medaka O. latipes as the first nonmammalian sex-determining gene (Matsuda et al. 2002, 2007). DMY was conserved among other populations of O. latipes (Shinomiya et al. 2004). O. curvinotus also have DMY on the Y chromosome, which is orthologous to that of O. latipes (Matsuda et al. 2003) (see Figure 1). However, DMY has not been detected in any other fishes, such as guppy, tilapia, zebrafish, or even in the Oryzias species O. celebensis and O. mekongensis (Kondo et al. 2003).
These results suggest that DMY is not the universal primary sex-determining gene in fishes, in contrast to the mammalian SRY/Sry (Volff et al. 2003), which is well conserved among placental mammals and marsupials (Gubbay et al. 1990; Sinclair et al. 1990; Foster et al. 1992), with the exception of some species (Just et al. 1995; Soullier et al. 1998). Analysis of the Y-specific region of the O. latipes sex chromosome has demonstrated that DMY arose from a duplicated copy of the autosomal DMRT1 gene (Nanda et al. 2002; Kondo et al. 2006). This DMRT1 duplication event is estimated to have occurred ∼10 million years ago (MYA) in a common ancestor of O. latipes and O. curvinotus. However, in O. luzonensis, no functional duplicated copy of DMRT1 has been detected, although there is a pseudogene, Oludmrt1p (Kondo et al. 2004). The evolution of the sex-determining system in these closely related species, including the origin of this pseudogene, remains a mystery.
Here, we identified seven sex-linked sequences of O. luzonensis and constructed a recombination map. The map demonstrated that the sex chromosome of O. luzonensis is orthologous to an O. latipes autosome (LG12) and, unlike in O. latipes, it does not show recombination suppression around the sex-determining region. On the basis of the draft genomic sequence of O. latipes, the sex-determining region of O. luzonensis is calculated to be <860 kbp. These results suggest that O. luzonensis has “younger” sex chromosomes than O. latipes and that the master sex-determining gene has changed at least twice in 10 million years (MY) during diversification of this species group.
MATERIALS AND METHODS
Fishes:
O. luzonensis was collected by M. J. Formacion and H. Uwa in 1982 at Solsona, Ilocos Norte, Luzon, Philippines (Formacion and Uwa 1985). O. curvinotus was collected by D. Dudgeon and H. Uwa in 1986 at Sam A. Tsuen, Plover Cove Country Park, Hong Kong (Uwa 1991). These species have been maintained as a closed colony. An inbred strain, Hd-rR, was established from the Southern population of O. latipes (Hyodo-Taguchi and Sakaizumi 1993). These fishes were supplied by a subcenter (Niigata University) of the National BioResource Project (medaka) supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan. Wild O. latipes were collected at Niitsu, Niigata Prefecture (northern population) in 2004.
Hormonal sex reversal:
Fertilized eggs of O. luzonensis were treated with either 0.025 μg/ml methyl testosterone (Sigma Chemical, St. Louis) or 0.2 μg/ml 17β-estradiol (Sigma Chemical) until hatching. They were then reared in aged tap water until sexual maturation.
Genetic crosses:
Three O. luzonensis families (Lz1–Lz3) were prepared: XX female × XY male [Lz1; number of progeny (n) = 190], XX female × XY male (Lz2; n = 93), and a sex-reversed XY female × sex reversed XX male (Lz3; n = 48). Two BC1 progeny of O. latipes were produced: (Niitsu♀ × Hd-rR♂) F1♂ × Hd-rR♀ (ND1; n = 94) and (Niitsu♀ × Hd-rR♂) F1♀ × Hd-rR♂ (ND2; n = 94). Interspecific BC1 offspring were obtained from an (O. luzonensis × O. curvinotus) F1 female crossed with an O. curvinotus male (CL1; n = 43).
Search for X–Y polymorphisms of O. luzonensis and linkage analysis:
To find polymorphisms between the X and Y of O. luzonensis, we randomly selected 250 expressed sequence tag (EST) markers from the medaka expressed sequence tag databases (http://mbase.bioweb.ne.jp/∼dclust/medaka_top.html/ and http://medaka.lab.nig.ac.jp/). ESTs were amplified using previously published primers designed for O. latipes (Naruse et al. 2004). PCR amplification was performed as follows: 33 cycles at 95° for 30 sec, 55° for 30 sec, and 72° for 3 min. Polymerase chain reaction (PCR) products were electrophoresed on polyacrylamide gels as described by Kimura et al. (2004). We adopted the PCR direct-sequencing method using an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA). An EST, OLb24.08a, was sequenced after TA cloning because its Y sequence contained a 39-bp deletion.
Linkage maps were constructed by using MAPL97 for Windows (Ukai et al. 1995). Both male and female recombination data were merged for consensus marker ordering. Each amplified marker was genotyped by restriction fragment length polymorphism (RFLP) analysis on polyacrylamide gels or single nucleotide polymorphism (SNP) analysis with the ABI PRISM 310 genetic analyzer.
Fluorescence in situ hybridization:
A bacterial artificial chromosome (BAC) genomic library, constructed from the Hd-rR strain of O. latipes (Matsuda et al. 2001), was screened, and three clones—Md0173J11 (containing SL1), Md0172B19 (containing DMRT1), and Md0171M23 (containing a body-color gene, b)—were used as probes. These BAC clones were located on the sex chromosomes (LG1) and on autosomes (LG9 and LG12) in O. latipes, respectively.
Metaphase cells from cultured caudal fins were prepared by standard cytogenetic methods (Uwa and Ojima 1981; Matsuda et al. 1998). Fluorescence in situ hybridization (FISH) was performed as described by Matsuda and Chapman (1995) and Takehana et al. (2007).
RESULTS AND DISCUSSION
O. latipes sex-determining gene DMY is absent in O. luzonensis:
Kondo et al. (2004) did not detect a sex-linked DMRT1 gene in O. luzonensis by Southern hybridization analysis. To confirm the absence of DMY, we searched O. luzonensis genomic DNA for the DMY gene by PCR with 10 primers (five forward and five reverse) designed for O. latipes DMY (see supplemental Table 1 at http://www.genetics.org/supplemental/; Figure 2A). Seven of the primer pairs (supplemental Table 2) produced a male-specific band in O. latipes and O. curvinotus (Figure 2B). In contrast, they did not amplify male-specific fragments in O. luzonensis. We also checked amplification of other genes, such as tyrosonase and b/AIM1, and obtained PCR products of the expected sizes in each of the three species (data not shown). For these genes, the synonymous substitution rate between O. latipes and O. luzonensis was similar to that between O. latipes and O. curvinotus (supplemental Table 3). Furthermore, a recent PCR survey of 47 loci, which were used for a genomewide SNP analysis (Kasahara et al. 2007), showed that 39 of these loci were successfully amplified in O. latipes, O. luzonensis, and O. curvitnotus, although the other 6 and 2 loci were not amplified in both O. luzonensis and O. curvinotus and in neither O. luzonensis nor O. curvinotus, respectively (Narita 2007). These results suggest that the lack of DMY amplification in O. luzonensis is not due to the large sequence divergence, but to the absence of DMY from the O. luzonensis genome.
Figure 2.—
Search for the DMY gene and mapping of DMY-related genes. (A) DMY structure of the O. latipes and positions of primers used in this study. Open boxes, shaded boxes, and horizontal lines indicate exons, the DM domain, and introns, respectively. (B) Agarose gel electrophoresis (1%) of PCR products using the 49S and 48U primers. Only O. latipes and O. curvinotus gave a male-specific band (DMY). (C) Polyacrylamide gel electrophoresis (9%) of PCR products using the ex4.1 and 48U primers. MboI-digested and undigested samples were loaded on the gel. The lower band of digested samples was judged as Oludmrt1, which has an Sau3AI (isochizomer of MboI) restriction site (Kondo et al. 2004). (D) Linkage analysis using CL1 cross. Map distances between markers are shown in centimorgans.
When the DMRT1 primers ex4.1 and 48U were used, two bands were obtained from both sexes in O. luzonensis. By RFLP analysis as described by Kondo et al. (2004), the lower bands were judged to represent the O. luzonensis DMRT1 (Oludmrt1) and the upper band to represent a pseudogene, Oludmrt1p (Figure 2C). Kondo et al. (2004) argued that this pseudogene might be either a degenerate version of a copy from the initial gene duplication or the result of another independent duplication of DMRT1. Our linkage analysis using an interspecific cross between O. curvinotus and O. luzonensis (CL1 cross) demonstrated that O. luzonensis DMRT1 (Oludmrt1) and Oludmrt1p are linked to markers belonging to O. latipes LG9 and LG18, respectively (Figure 2D). These data supported the second hypothesis of Kondo et al. (2004). Because no DMRT1-related genes on LG18 have been reported in O. latipes, the pseudogene may not have originated as a degenerate copy of the initial gene duplication but as an independent duplicate specific to O. luzonensis of the DMRT1(LG9). However, another possibility—that a degenerate copy of DMY on LG1 has been transposed to LG18—cannot be excluded. DMY and Oludmrt1p may thus have different origins, and DMY (LG1) may have been lost in O. luzonensis.
The sex-linkage group of O. luzonensis is orthologous to a medaka autosome (LG12):
We found that three O. latipes ESTs (AU167284, MF01SSA025F03, and OLb24.08a) yielded male-specific banding patterns in O. luzonensis (Figure 3A). Sequencing analyses (Figure 3B) suggested that these patterns could be due to DNA heteroduplex formation (Hauser et al. 1998). These differences in electrophoretic mobility result from the heteroduplex DNA conformation of the mismatches. These heterogametic patterns were passed from father to son, confirming that O. luzonensis has an XX–XY sex-determination system, as indicated previously (Hamaguchi et al. 2004).
Figure 3.—
Sex-linked polymorphisms of O. luzonensis. (A) Polyacrylamide gel electrophoresis (9%) of PCR products for AU167284, MF01SSA025F03, and OLb24.08a. Arrowheads mark male-specific bands. (B) Sequence analysis of polymorphic PCR products. For EST AU167284, the female PCR products show seven T repeats, whereas males display both seven and eight T repeats. In EST MF01SSA025F03, males show a (C/T) SNP. In OLb24.08a, the 39-bp deletion links to maleness.
Because the markers AU167284 and OLb24.08a were already described as located on O. latipes LG12 (Naruse et al. 2004), other sequences on LG12 were examined for more sex-linked markers. Two genes, b and eyeless (Fukamachi et al. 2001; Loosli et al. 2001), two BAC end sequences (Fukamachi et al. 2001), and 22 ESTs were subjected to PCR direct sequencing. We identified four additional sex-linked SNPs (supplemental Table 4 at http://www.genetics.org/supplemental/). These seven markers were investigated for their sequence similarity and uniqueness by basic local alignment search tool (BLAST) searches against the medaka genome database (http://dolphin.lab.nig.ac.jp/medaka/). BLAST searches detected only one sequence with high similarity (sequence identity > 89%; E-value <e−45) for each (supplemental Table 4), indicating that all investigated sex-linked markers of O. luzonensis were orthologous to LG12 sequences of O. latipes.
FISH analysis (Figure 4) demonstrated that the sex chromosomes of O. luzonensis were submetacentric and not differentiated from one another cytogenetically. A BAC clone, Md0171M23, containing the tightly sex-linked gene b, hybridized on the long arms of the sex chromosomes, close to the centromere. Furthermore, it was confirmed that the sex chromosome of O. luzonensis was different from both the sex chromosome (LG1) and the DMRT1-bearing chromosome (LG9) of O. latipes.
Figure 4.—
FISH analysis of male metaphase chromosomes in O. luzonensis using O. latipes BAC clones. (A) Chromosomal location of the O. luzonensis sex-determining region (BAC Md0171M23, red) and of the O. latipes sex chromosomal marker SL1 (BAC Md0173J11, green). (B) Chromosomal location of the O. luzonensis sex-determining region (BAC Md0171M23, red) and the O. latipes DMRT1 gene (BAC Md0172B19, green).
The O. luzonensis sex chromosome appeared <5 MYA:
SRY/Sry is the only known primary sex-determining gene in higher vertebrates and is believed to have arisen 130–170 MYA (Marshall-Graves 2002). By estimating the age of DMY as ∼10 MY, Kondo et al. (2004) argued that the O. latipes sex chromosome is at an early stage of differentiation and concluded that the O. latipes Y chromosome is the youngest male-determining chromosome so far known in vertebrates.
O. latipes and O. curvinotus possess orthologous sex chromosomes (LG1) (Matsuda et al. 2003; Kondo et al. 2004); in contrast, O. luzonensis displays a different sex chromosome (LG12), suggesting that the sex chromosome shifted from LG1 to LG12 after O. luzonensis diverged from O. curvinotus. The basal species, O. mekongensis, has an LG2 sex chromosome (A. Kawaguchi, A. Shinomiya, S. Hamaguchi and M. Sakaizumi, unpublished data), indicating that the new sex chromosome of O. luzonensis (LG12) is not the result of reversion to the old sex-determination system with DMY degeneration (Figure 5). Because the separation of O. luzonensis from O. curvinotus is estimated to be ∼5 MYA on the basis of a molecular clock (Figure 1), the sex chromosome of O. luzonensis may be younger than 5 MY. O. luzonensis may have lost DMY and recruited a novel sex-determining gene on the new sex chromosome (LG12).
Figure 5.—
Sex-determination mechanisms and sex linkage groups in Oryzias species. The phylogenetic information was taken from Takehana et al. (2005).
Comparison between the O. luzonensis sex chromosome and O. latipes LG12:
We constructed comparative linkage maps between the sex-linkage group of O. luzonensis and the autosomal linkage group (LG12) of O. latipes (Figure 6). The order of markers was completely conserved between the two maps; i.e., the sex chromosome of O. luzonensis is syntenic to O. latipes LG12. The Sex gene was tightly linked with the body-color gene b (n = 141/141) and located between eyeless and 171M23F. This region is equivalent to 859 kbp in the O. latipes genome, which includes 28 predicted genes (medaka genome sequencing project: Kasahara et al. 2007; http://dolphin.lab.nig.ac.jp/medaka/). Thus, we expect that the primary sex-determining gene of O. luzonensis lies in this interval.
Figure 6.—
Comparative recombination map between sex chromosomes of O. luzonensis and LG12 (autosome) of O. latipes. The sex-determining gene (Sex, arrowhead) of O. luzonensis is located adjacent to the b gene. Map distances between markers are shown in centimorgans and total map lengths are shown below each map.
Although the only structural difference between the O. latipes X and Y is the Y-specific region (258-kb insertion) (Kondo et al. 2006), the recombination rate in males is also strongly suppressed outside this region in a region that has a genetic map length of ∼30 cM in females (Kondo et al. 2001). This restriction of recombination is observed in O. latipes sex-reversed XX males but not in XY females (Matsuda et al. 1999), indicating that the restriction is not caused by a structural difference between X–Y but by an unknown mechanism specific to phenotypic males. In contrast to the highly restricted sex chromosome of O. latipes (Figure 2 in Kondo et al. 2001), the sex chromosome of O. luzonensis (LG12) displays only a weak reduction of the recombination rate in males and recombines well around the sex-determining region (see AU167284–MF01SSA025F03 in Figure 6). A high recombination rate between the X and Y supports the argument that the sex chromosome of O. luzonensis (LG12) is younger than that of O. latipes (LG1).
Studies of such “young” sex chromosomes are important in understanding their early evolution (Charlesworth et al. 2005). As with Oryzias species, some salmonids and sticklebacks show different sex chromosomes among closely related species (Woram et al. 2003; Peichel et al. 2004). This suggests that frequent switching between different master sex-determining genes may have occurred in many species groups that possess undifferentiated sex chromosomes. Oryzias fishes may prove to be very informative systems for studying the evolutionary processes of the early stages of sex-chromosome differentiation and of the switching mechanisms of the master sex-determining gene.
Acknowledgments
We thank Masaru Matsuda of the National Institute for Basic Biology for his helpful technical advice. Medaka genome data were provided by the National Institute of Genetics and the University of Tokyo for use in this publication/correspondence only. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to M.S. (16370094) and to K.N. (16310131).
References
- Aida, T., 1921. On the inheritance of color in a fresh-water fish, Aplocheilus latipes Temmick and Schlegel, with special reference to sex-linked inheritance. Genetics 6: 554–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., B. Charlesworth and G. Marais, 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Devlin, R. H., and Y. Nagahama, 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208: 191–364. [Google Scholar]
- Formacion, M. J., and H. Uwa, 1985. Cytogenetic studies on the origin and species differentiation of the Philippine medaka, Oryzias luzonensis. J. Fish Biol. 27: 285–291. [Google Scholar]
- Foster, J. W., F. E. Brennan, G. K. Hampikian, P. N. Goodfellow, A. H. Sinclair et al., 1992. Evolution of sex determination and the Y chromosome: SRY-related sequences in marsupials. Nature 359: 531–533. [DOI] [PubMed] [Google Scholar]
- Fukamachi, S., A. Shimada and A. Shima, 2001. Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat. Genet. 28: 381–385. [DOI] [PubMed] [Google Scholar]
- Gubbay, J., J. Collignon, P. Koopman, B. Capel, A. Economou et al., 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346: 245–250. [DOI] [PubMed] [Google Scholar]
- Hamaguchi, S., Y. Toyazaki, A. Shinomiya and M. Sakaizumi, 2004. The XX-XY sex-determination system in Oryzias luzonensis and O. mekongensis revealed by the sex ratio of the progeny of sex-reversed fish. Zool. Sci. 21: 1015–1018. [DOI] [PubMed] [Google Scholar]
- Hauser, M. T., F. Adhami, M. Dorner, E. Fuchs and J. Glossl, 1998. Generation of co-dominant PCR-based markers by duplex analysis on high resolution gels. Plant J. 16: 117–125. [DOI] [PubMed] [Google Scholar]
- Hyodo-Taguchi, Y., and M. Sakaizumi, 1993. List of inbred strains of the medaka, Oryzias latipes, maintained in the Division of Biology, National Institute of Radiological Sciences. Fish Biol. J. MEDAKA 5: 29–30. [Google Scholar]
- Just, W., W. Rau, W. Vogel, M. Akhverdian, K. Fredga et al., 1995. Absence of Sry in species of the vole Ellobius. Nat. Genet. 11: 117–118. [DOI] [PubMed] [Google Scholar]
- Kasahara, M., K. Naruse, S. Sasaki, Y. Nakatani, W. Qu et al., 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719. [DOI] [PubMed] [Google Scholar]
- Kimura, T., T. Jindo, T. Narita, K. Naruse, D. Kobayashi et al., 2004. Large-scale isolation of ESTs from medaka embryos and its application to medaka developmental genetics. Mech. Dev. 121: 915–932. [DOI] [PubMed] [Google Scholar]
- Kondo, M., E. Nagao, H. Mitani and A. Shima, 2001. Differences in recombination frequencies during female and male meioses of the sex chromosomes of the medaka, Oryzias latipes. Genet. Res. 78: 23–30. [DOI] [PubMed] [Google Scholar]
- Kondo, M., I. Nanda, U. Hornung, S. Asakawa, N. Shimizu et al., 2003. Absence of the candidate male sex-determining gene dmrt1b(Y) of medaka from other fish species. Curr. Biol. 13: 416–420. [DOI] [PubMed] [Google Scholar]
- Kondo, M., I. Nanda, U. Hornung, M. Schmid and M. Schartl, 2004. Evolutionary origin of the medaka Y chromosome. Curr. Biol. 14: 1664–1669. [DOI] [PubMed] [Google Scholar]
- Kondo, M., U. Hornung, I. Nanda, S. Imai, T. Sasaki et al., 2006. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 16: 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli, F., S. Winkler, C. Burgtorf, E. Wurmbach, W. Ansorge et al., 2001. Medaka eyeless is the key factor linking retinal determination and eye growth. Development 128: 4035–4044. [DOI] [PubMed] [Google Scholar]
- Marshall-Graves, J. A., 2002. The rise and fall of SRY. Trends Genet. 18: 259–264. [DOI] [PubMed] [Google Scholar]
- Matsuda, Y., and V. M. Chapman, 1995. Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 16: 261–272. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., C. Matsuda, S. Hamaguchi and M. Sakaizumi, 1998. Identification of the sex chromosomes of the medaka, Oryzias latipes, by fluorescence in situ hybridization. Cytogenet. Cell Genet. 82: 257–262. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., S. Sotoyama, S. Hamaguchi and M. Sakaizumi, 1999. Male-specific restriction of recombination frequency in the sex chromosomes of the medaka, Oryzias latipes. Genet. Res. 73: 225–231. [Google Scholar]
- Matsuda, M., N. Kawato, S. Asakawa, N. Shimizu, Y. Nagahama et al., 2001. Construction of a BAC library derived from the inbred Hd-rR strain of the teleost fish, Oryzias latipes. Genes Genet. Syst. 76: 61–63. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., T. Sato, Y. Toyazaki, Y. Nagahama, S. Hamaguchi et al., 2003. Oryzias curvinotus has DMY, a gene that is required for male development in the medaka, O. latipes. Zool. Sci. 20: 159–161. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., A. Shinomiya, M. Kinoshita, A. Suzuki, T. Kobayashi et al., 2007. The DMY gene induces male development in genetically female (XX) medaka fish. Proc. Natl. Acad. Sci. USA 104: 3865–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler et al., 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99: 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita, T., 2007. Construction of comprehensive genomic resources for medaka, Oryzias latipes. Large-scale EST analysis and genome-wide phylogenetic study of the medaka and relatives. Ph.D. Thesis, University of Tokyo, Tokyo.
- Naruse, K., M. Tanaka, K. Mita, A. Shima, J. Postlethwait et al., 2004. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 14: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel, C. L., J. A. Ross, C. K. Matson, M. Dickson, J. Grimwood et al., 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Shinomiya, A., H. Otake, K. Togashi, S. Hamaguchi and M. Sakaizumi, 2004. Field survey of sex-reversals in the medaka, Oryzias latipes: genotypic sexing of wild populations. Zool. Sci. 21: 613–619. [DOI] [PubMed] [Google Scholar]
- Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths et al., 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346: 240–244. [DOI] [PubMed] [Google Scholar]
- Solari, A. J., 1994. Sex Chromosomes and Sex Determination in Vertebrates. CRC Press, Boca Raton, FL.
- Soullier, S., C. Hanni, F. Catzeflis, P. Berta and V. Laudet, 1998. Male sex determination in the spiny rat Tokudaia osimensis (Rodentia: Muridae) is not Sry dependent. Mamm. Genome 9: 590–592. [DOI] [PubMed] [Google Scholar]
- Takehana, Y., N. Nagai, M. Matsuda, K. Tsuchiya and M. Sakaizumi, 2003. Geographic variation and diversity of the cytochrome b gene in Japanese wild populations of medaka, Oryzias latipes. Zool. Sci. 20: 1279–1291. [DOI] [PubMed] [Google Scholar]
- Takehana, Y., K. Naruse and M. Sakaizumi, 2005. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 36: 417–428. [DOI] [PubMed] [Google Scholar]
- Takehana, Y., D. Demiyah, K. Naruse, S. Hamaguchi and M. Sakaizumi, 2007. Evolution of different Y chromosomes in two medaka species, Oryzias dancena and O. latipes. Genetics 175: 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai, Y., R. Ohsawa, A. Saito and T. Hayashi, 1995. A package of computer programs for construction of DNA polymorphism linkage maps and analysis of QTL. Breed. Sci. 45: 139–142. [Google Scholar]
- Uwa, H., 1991. Cytosystematic study of the Hainan medaka, Oryzias curvinotus, from Hong Kong (Teleostei:Oryziidae). Ichthyol. Explor. Freshw. 1: 361–367. [Google Scholar]
- Uwa, H., and Y. Ojima, 1981. Detailed and banding karyotype analyses of the Medaka, Oryzias latipes in cultured cells. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 57: 39–43. [Google Scholar]
- Volff, J. N., and M. Schartl, 2001. Variability of genetic sex determination in poeciliid fishes. Genetica. 111: 101–110. [DOI] [PubMed] [Google Scholar]
- Volff, J. N., M. Kondo and M. Schartl, 2003. Medaka dmY/dmrt1Y is not the universal primary sex-determining gene in fish. Trends Genet. 19: 196–199. [DOI] [PubMed] [Google Scholar]
- Woram, R. A., K. Gharbi, T. Sakamoto, B. Hoyheim, L. E. Holm et al., 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 13: 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]