Abstract
The effect of recurrent selective sweeps is a spatially heterogeneous reduction in neutral polymorphism throughout the genome. The pattern of reduction depends on the selective advantage and recurrence rate of the sweeps. Because many adaptive substitutions responsible for these sweeps also contribute to nonsynonymous divergence, the spatial distribution of nonsynonymous divergence also reflects the distribution of adaptive substitutions. Thus, the spatial correspondence between neutral polymorphism and nonsynonymous divergence may be especially informative about the process of adaptation. Here we study this correspondence using genomewide polymorphism data from Drosophila simulans and the divergence between D. simulans and D. melanogaster. Focusing on highly recombining portions of the autosomes, at a spatial scale appropriate to the study of selective sweeps, we find that neutral polymorphism is both lower and, as measured by a new statistic QS, less homogeneous where nonsynonymous divergence is higher and that the spatial structure of this correlation is best explained by the action of strong recurrent selective sweeps. We introduce a method to infer, from the spatial correspondence between polymorphism and divergence, the rate and selective strength of adaptation. Our results independently confirm a high rate of adaptive substitution (∼1/3000 generations) and newly suggest that many adaptations are of surprisingly great selective effect (∼1%), reducing the effective population size by ∼15% even in highly recombining regions of the genome.
PATTERNS of genetic variation within and between species arise from the interplay of several evolutionary forces, including adaptation (Lewontin 1974; Gillespie 1994). It has been argued that adaptive substitutions occur rarely and therefore should contribute negligibly to these patterns (Kimura 1983; Ohta 1992). An increasing number of studies in Drosophila, however, have found that adaptive substitution may account for a substantial fraction of the divergence between species and that adaptation may frequently influence the pattern of polymorphism within a species. In the case of divergence, applications of the McDonald–Kreitman test to multiple-gene data sets from Drosophila have concluded that up to 60% of nonsynonymous substitutions and 30% of the substitutions at regulatory sites may be driven by adaptation (Fay et al. 2002; Smith and Eyre-Walker 2002; Bierne and Eyre-Walker 2004; Andolfatto 2005; Charlesworth and Eyre-Walker 2006; Welch 2006; Shapiro et al. 2007). In the case of polymorphism, the effects of adaptation have also been seen in Drosophila: selective sweeps, which are local reductions in linked neutral polymorphism caused by adaptive substitutions, have been repeatedly detected in Drosophila populations, both in detailed studies of individual loci (e.g., Schlenke and Begun 2004; Aminetzach et al. 2005; Beisswanger et al. 2006) and in genomic scans (e.g., Glinka et al. 2003; Orengo and Aguade 2004; Ometto et al. 2005).
Outwardly, the spatial pattern of neutral polymorphism would seem to provide an ideal setting to study adaptation. Under simplifying assumptions, population genetic theory makes clear quantitative predictions about the breadth of the region that a selective sweep affects and the extent to which it reduces neutral polymorphism there (Kaplan et al. 1989; Kim and Stephan 2002). For example, in highly recombining regions of the Drosophila genome (c = 2.5 cM/Mb), a selective sweep associated with a large selection coefficient of 1% should on average depress neutral polymorphism in a region of 100 kb surrounding the adaptation to 60% of its nominal value and to 30% of its nominal value in the surrounding 50 kb (Gillespie 2004). By comparison, a selective sweep with a weaker selective coefficient of 0.1% would depress polymorphism in a narrower region, producing a 30% reduction in the nominal level of neutral polymorphism in the surrounding 10-kb region and a 60% reduction in the surrounding 5-kb region. Following a selective sweep, polymorphism is restored to its background level by mutation and genetic drift at a rate on the order of Ne generations (Przeworski 2002). The dynamic balance between sweep and restoration, extended to the genomic scale, should result in reductions in neutral polymorphism that are intermittent both in space and in time.
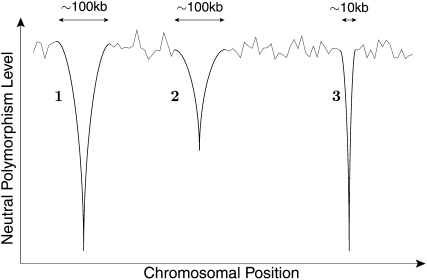
Figure 1 illustrates how the effect of recurrent selective sweeps may be recorded in the level of neutral polymorphism along a chromosomal section, at a particular instant in time. Within this section, one can observe the reductions in polymorphism caused by several selective sweeps in comparison with background levels. For example, the sweep labeled 1 has been associated with a strong selective coefficient and has occurred very recently. Sweep 2 has been associated with a similarly strong selective coefficient, but because it occurred further in the past the levels of polymorphism surrounding it have had some time to recover. Sweep 3 has occurred recently, but because it was associated with a weaker selective coefficient it reduced polymorphism in a smaller spatial region. Thus, from the spatial pattern of neutral polymorphism at a given point in time, i.e., the number and width of depressions in neutral polymorphism, one may be able to infer how frequent and how intense selected adaptations are. This approach is hindered, however, by the presence of evolutionary forces other than adaptation, perhaps most importantly demographic processes, which can also produce spatial heterogeneity in neutral polymorphism (Thornton and Jensen 2007; Thornton et al. 2007).
Figure 1.—
An illustration of the spatial effect of recurrent selective sweeps on the level of neutral polymorphism. The effects of three sweeps are shown on the background of the polymorphism level generated by mutation and random genetic drift. Sweeps 1 and 2 are of similarly strong selective advantage, but sweep 1 has taken place much more recently than sweep 2. Like sweep 1, sweep 3 has taken place recently, but is of lesser selective advantage.
The confounding effects of evolutionary processes other than adaptation might be reduced if polymorphism data were to be considered in combination with divergence data. Specifically, because a substantial number of adaptive substitutions that cause selective sweeps will appear as nonsynonymous divergences, those genomic regions that exhibit both reduced neutral polymorphism and elevated nonsynonymous divergence should more reliably indicate the presence of adaptation than would a signal of reduced polymorphism alone. Consequently, an analysis of the spatial pattern of the correspondence between neutral polymorphism and nonsynonymous divergence, as opposed to the spatial pattern of neutral polymorphism alone, might yield new insight into the adaptive process.
In this article, we explore this idea using genomewide polymorphism data from six Drosophila simulans strains and divergence data between D. simulans and D. melanogaster. Our analysis is divided into two separable parts. The first part describes how we build and verify a map, which estimates the levels of neutral polymorphism along a chromosome on the basis of polymorphism data, for the autosomes of D. simulans. The construction of such a map raises a number of statistical issues that bear on its accuracy and on its spatial resolution. Addressing these issues is important if we wish to rely on this map to study adaptation. After we develop a method for the construction of the map, we analyze its performance, eventually obtaining a map that provides an accurate representation of the variability in neutral polymorphism on the scale of ∼20 kb. Although we rely on the veracity of this map in the second part of this article, the first part is concerned solely with the problems of map building and may therefore be skipped without compromising the ability to understand the arguments that follow.
In the second part of this article we analyze the spatial correspondence between neutral polymorphism and nonsynonymous divergence in the highly recombining regions of the autosomal arms. The map of neutral polymorphism exhibits extensive spatial heterogeneity at a spatial scale of 20–200 kb, consistent with but not uniquely demonstrative of frequent, relatively strong selective sweeps. To better discern the effect of recurrent selective sweeps from that of other evolutionary forces, we examine the association between neutral polymorphism and nonsynonymous divergence in 100-kb sliding windows. Neutral polymorphism in 100-kb windows is characterized by two summary statistics: the average polymorphism in a window and a measure of the homogeneity of polymorphism within a window, QS, which is introduced for this purpose. We find that both statistics are significantly negatively correlated with the nonsynonymous divergence, where the negative correlation with QS is especially strong. On the basis of these and other analyses, we argue that these correlations are most plausibly explained as the outcome of recurrent selective sweeps.
We then proceed to show that the spatial correspondence between neutral polymorphism and nonsynonymous divergence bears information on both the strength and the rate of selective sweeps. First, we conduct a spatial randomization test that suggests that the correlations between neutral polymorphism and nonsynonymous divergence do not arise from sweeps of weak selective effect. Second, we introduce a rudimentary model that relates the two polymorphism statistics to the parameters of recurrent selective sweeps, specifically demonstrating that these statistics provide complementary information about the rate and selective coefficients of adaptive substitutions. We use an inference procedure based on this model to derive preliminary estimates of the rate and strength of adaptation from the empirical spatial correspondence between polymorphism and nonsynonymous divergence. These estimates provide independent support for the high rate of adaptation inferred in studies that used McDonald–Kreitman methodology (Fay et al. 2002; Smith and Eyre-Walker 2002; Bierne and Eyre-Walker 2004; Andolfatto 2005; Charlesworth and Eyre-Walker 2006; Welch 2006; Eyre-Walker 2006; Shapiro et al. 2007) and suggest that the effect of many of these adaptations is surprisingly strong. The inference procedure also yields an estimate of the average reduction in the effective population size caused by selective sweeps, which suggests that recurrent selective sweeps have a substantial effect on neutral polymorphism even in high-recombination regions of the Drosophila genome (Gillespie 2000, 2001).
MATERIALS AND METHODS
Data sources and initial processing:
The six-strain D. simulans alignment was obtained from the Drosophila Population Genomics Project (http://www.dpgp.org), and the release 4.0 D. melanogaster sequence, annotations, and genetic map were obtained from FlyBase (Grumbling and Strelets 2006). All base calls with phred score <35 were discarded, as was any transcript containing a stop codon in any of the seven sequences.
Starting from 7,130,159 codons in the euchromatic genome, we retained only those codons that were defined in at least four of the six D. simulans strains and were monoallelic or biallelic in the combined D. simulans and D. melanogaster sample. For a codon to be “defined” for a given strain, all three nucleotide positions in that codon were required to have a valid base call. Removing codons with more than two alleles should slightly reduce our estimates of polymorphism. Because the expected reduction in the estimate is greater where polymorphism is greater, this practice is conservative for our purposes because it can only reduce the heterogeneity observed in polymorphism. The other filtering criteria described are independent of polymorphism and divergence and introduce no bias in the estimate of polymorphism. A total of 3,059,053 codons remained, distributed among 12,146 nonoverlapping genes, which amount to 42.9% of all protein-coding DNA and 7.6% of all DNA in the euchromatic genome.
Constructing a map of neutral polymorphism:
To survey polymorphism levels throughout the D. simulans genome, we built a map comprising estimates of neutral polymorphism at each genomic position. Our estimates are based on synonymous polymorphism data, which has variable sample size, n ≥ 4, and which occurs on sequence backgrounds with variable base pair composition. We obtain constant sample size throughout the data by resampling exactly four strains wherever n > 4. We denote by xp(i) the resampled polymorphism data at codon position i, where xp(i) = 1 if we observe a polymorphism at position i and xp(i) = 0 if not. We account for variation in the rate of synonymous mutation due to the base pair composition at position i by calculating the synonymous mutational opportunity, m(i), defined as the sum of the relative rates of all single-nucleotide transitions that change the codon at position i to a synonymous codon. For the purpose of this calculation we assume the codon in D. melanogaster is ancestral and that the relative rates are similar to those measured in D. melanogaster: 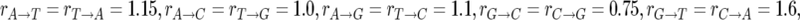 and
and 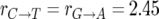 (Singh et al. 2005a; Bauer DuMont and Aquadro 2005). Note that the definition of synonymous mutational opportunity incorporates both the standard count of synonymous sites and the variation in mutation rate due to base pair composition. We thus assume that the probability of observing a synonymous polymorphism at position i is
(Singh et al. 2005a; Bauer DuMont and Aquadro 2005). Note that the definition of synonymous mutational opportunity incorporates both the standard count of synonymous sites and the variation in mutation rate due to base pair composition. We thus assume that the probability of observing a synonymous polymorphism at position i is
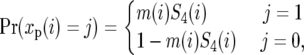 |
(1) |
where S4(i) is the probability of observing a polymorphism per unit of synonymous mutational opportunity in a sample size of 4 at position i, which is the measure we are interested in estimating.
We estimate neutral polymorphism at a given position by averaging the synonymous polymorphism in a window surrounding that position. This window, Wr(i), consists of the codons in our sample that are closest to position i, such that total synonymous mutational opportunity to the left (and to the right) of i is equal to r. Given this window our estimator of neutral polymorphism is
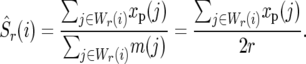 |
(2) |
The definition of the window implies that if neutral polymorphism within it is uniform, we expect to observe an equal number of synonymous polymorphisms to the right and to the left of position i.
We choose the size of the window r that best predicts the polymorphism observed in individual exons. (Strictly speaking this is half the window size, or the radius of the window. For brevity we refer to it as window size.) To find this window size we define an estimator at an exon 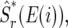 where i is the position of a codon in our sample, and E(i) is the exon in which this codon resides. The exonic estimator is defined similarly to Equation 2, with one notable distinction: the window surrounding the exon does not include codons from that exon. The exclusion of polymorphism data from a given exon in the estimation of polymorphism at that exon ensures that we do not use the same data in prediction and in its evaluation. We evaluate the predictive ability of the estimator with a given window size in terms of the likelihood of the estimator at exons given the polymorphism we observed in these exons. Namely,
where i is the position of a codon in our sample, and E(i) is the exon in which this codon resides. The exonic estimator is defined similarly to Equation 2, with one notable distinction: the window surrounding the exon does not include codons from that exon. The exclusion of polymorphism data from a given exon in the estimation of polymorphism at that exon ensures that we do not use the same data in prediction and in its evaluation. We evaluate the predictive ability of the estimator with a given window size in terms of the likelihood of the estimator at exons given the polymorphism we observed in these exons. Namely,
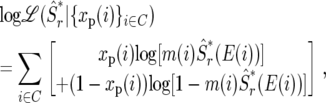 |
(3) |
where C is the set of codon positions in our sample. This likelihood is supposed to increase when the choice of window size r provides more accurate predictions. Therefore, we choose the window size that maximizes this likelihood over the polymorphism observed in a given chromosomal arm. The likelihood curve and maximum-likelihood window size for each chromosomal arm appear in supplemental Figure S1 at http://www.genetics.org/supplemental/. An analogous procedure was used to find the maximum-likelihood window sizes for the estimation of neutral divergence (supplemental Figure S2 at http://www.genetics.org/supplemental/). Once the maximum-likelihood window sizes were obtained, final maps were built using these windows based on 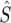 as defined in Equation 2, i.e., without removing any exons.
as defined in Equation 2, i.e., without removing any exons.
Statistics used to study the spatial correspondence between polymorphism and divergence:
We analyze the spatial correspondence between polymorphism and divergence in the highly recombining regions of the autosomes. We define regions of high recombination as those where the recombination rate c exceeded 2.5 cM/Mb. The spatial analysis is performed on a set of overlapping 100-kb windows, obtained by moving a 100-kb window across the high-recombination regions in steps of 600 bp. The number of windows obtained in this way was 78,570. To reduce noise in the measurement of polymorphism statistics arising from small sample size, we discarded any window with total synonymous mutational opportunity Ms < 1000. After this, 66,152 windows remained.
For each 100-kb window, we computed a number of statistics that are used throughout this article. We denote by Ps and Ds the number of synonymous polymorphisms and divergences observed in a window, respectively. Pn and Dn are the analogous nonsynonymous measures. We denote by ps, ds, pn, and dn the normalized versions of the above statistics, in which synonymous counts are divided by the total synonymous mutational opportunity in a window, Ms, and the nonsynonymous counts are divided by the total nonsynonymous mutational opportunity, Mn. Because of its analytical tractability, we often use Watterson's θS (Watterson 1975) instead of ps, where 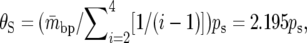 and
and 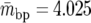 is the average mutational opportunity per base pair under the assumption of uniform base pair composition. Finally, we calculate the new statistic QS, defined as the ratio of the minimum value of our polymorphism estimate,
is the average mutational opportunity per base pair under the assumption of uniform base pair composition. Finally, we calculate the new statistic QS, defined as the ratio of the minimum value of our polymorphism estimate, 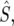 within a window to the mean of
within a window to the mean of 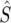 in the same window.
in the same window.
A model of the polymorphism statistics under recurrent selective sweeps:
We consider a diploid panmictic population of size N that evolves in discrete generations. We further assume a uniform model where the rate of adaptive substitutions per base pair at every site is υ, all adaptive mutations have the same selective coefficient s, and the rate of recombination per base pair c is constant.
We derive an approximation for the average heterozygosity on the basis of Gillespie's pseudohitchhiking model (Gillespie 2000), which is briefly described below. The pseudohitchhiking model in a finite population describes the dynamic of a neutral allele's frequency, under the assumption that at each time step this allele experiences either Wright–Fisher sampling or, if an adaptive substitution occurs in its vicinity, an instantaneous sweep. Gillespie considers a model where adaptive substitutions occur at a single site in the vicinity of the neutral allele. Incorporating recombination into the model implies that the swept neutral allele may not reach fixation. When an adaptive mutation destined for fixation first enters the population, it is on the same chromosome as only one copy of a neutral allele. The frequency of that copy increases to some new value, a random variable denoted y, at the expense of all other copies of that neutral allele and all other alleles, which have their frequency reduced by a fraction 1 − y. Gillespie applies the diffusion approximation to this dynamic and finds that the average heterozygosity is
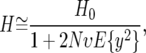 |
(4) |
where H0 is the heterozygosity without selective sweeps, i.e., the heterozygosity under neutral mutation and random genetic drift.
We consider a straightforward extension of the pseudohitchhiking model where adaptive substitutions occur at a uniform rate at every site in the vicinity of the neutral site under consideration. Under this extension the average heterozygosity becomes
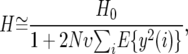 |
(5) |
where i is the distance in base pairs between the neutral site under consideration and the site of the adaptive substitution. The random variable y(i) is a function of this distance because the impact of sweeps decreases with distance. We approximate the sum in the denominator of Equation 5 on the basis of two simplifying assumptions. First, we approximate the sum by an integral, namely,
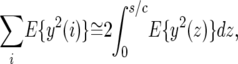 |
(6) |
where we limit the integral to s/c because sweeps beyond this distance have a negligible effect on neutral polymorphism (Kaplan et al. 1989). Second, we approximate y(z) on the basis of the deterministic hitchhiking model (Maynard Smith and Haigh 1974; Gillespie 2000), namely,
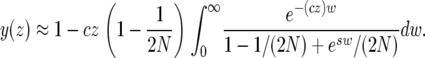 |
(7) |
Substituting Equations 6 and 7 into Equation 5, and noting that the deterministic approximation for y(z) implies the removal of the expectation from y2(z), yields the following approximation for average heterozygosity in our model,
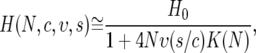 |
(8) |
where 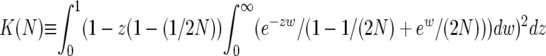 depends only on the population size. This approximation is likely an overestimate of the heterozygosity because the deterministic hitchhiking model underestimates the value of y(z) and thus the effect of recurrent selective sweeps (Gillespie 2000). Note that Wiehe and Stephan (1993) have derived an expression for the average heterozygosity similar to Equation 8, using a different modeling approach.
depends only on the population size. This approximation is likely an overestimate of the heterozygosity because the deterministic hitchhiking model underestimates the value of y(z) and thus the effect of recurrent selective sweeps (Gillespie 2000). Note that Wiehe and Stephan (1993) have derived an expression for the average heterozygosity similar to Equation 8, using a different modeling approach.
Next, we consider the expected minimal heterozygosity within a window of width w under recurrent selective sweeps. Under the assumptions of the uniform model, the heterozygosity is minimized at the position within the window where the last adaptive substitution took place. We can evaluate the heterozygosity at this position using the coalescent of a sample of size 2. If we assume that the adaptive substitution occurred T generations ago, a coalescence event at this neutral site can occur in two ways. Either it can occur by standard Wright–Fisher sampling or, if the lineages do not coalesce in the first T − 1 generations, they will necessarily coalesce as a result of the selective sweep. Because we are considering the position on top of the adaptive sweep, recombination can be ignored. Therefore the average depth of the coalescent L at that position is
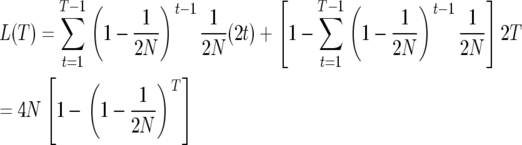 |
(9) |
When the rate of adaptive substitutions per generation in the window is small, i.e., 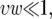 the time to the last adaptive substitution is described by the geometric distribution: T ∼ Geometric(υw). The average depth of the coalescent is therefore
the time to the last adaptive substitution is described by the geometric distribution: T ∼ Geometric(υw). The average depth of the coalescent is therefore
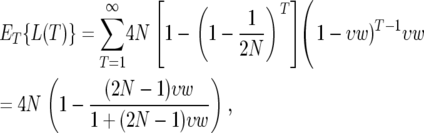 |
(10) |
and the average minimal heterozygosity is
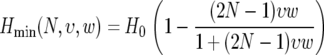 |
(11) |
Note that Hmin does not depend on the strength of adaptation. This is because the minimal polymorphism in a window appears at close proximity to the location at which the last adaptive substitution has occurred, where even a weak sweep would have driven the polymorphism to zero. At that position the level of polymorphism after the sweep depends on the time that elapsed since the completion of the sweep, in which the neutral polymorphism has partially recovered. The recovery time since the last sweep in a window depends only on the rate, and not on the strength, of adaptation. However, Hmin underestimates the minimal heterozygosity we can derive from the map of neutral polymorphism because the estimates that compose the map are always averages within a region rather than point estimates.
Equations 8 and 11 provide us with a rough model of the dependence of the θS and QS statistics on the parameters of recurrent selective sweeps. The expressions for these statistics are
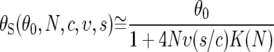 |
(12) |
and
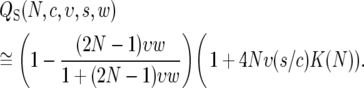 |
(13) |
Under the pseudohitchhiking approximation, the effect of recurrent selective sweeps amounts to changing the effective population size within a model of random genetic drift. Therefore, in this approximation heterozygosity and Watterson's θ are interchangeable.
Inferring genomic adaptation rate and strength from polymorphism and divergence data:
We infer the average rate υ and average selective coefficient s by fitting the above model (Equations 12 and 13) to the data. First, these data, θS, QS, and Dn, were transformed to minimize their shared dependence on variation in mutation rate and selective constraint. For that purpose, we assume that nonsynonymous divergences at window i can be expressed as a sum of two contributions. The first depends linearly on the variation in mutation and constraint, which is approximated as the deviation from the average synonymous divergence per site in a window 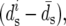 and the second, which we use as our proxy for the rate of adaptation in the window,
and the second, which we use as our proxy for the rate of adaptation in the window, 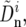 does not. Thus
does not. Thus
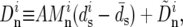 |
(14) |
where 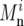 is the total nonsynonymous mutational opportunity in the window, and A is a constant computed by least squares from the data. Similarly, we transformed the average and homogeneity in polymorphism using the models
is the total nonsynonymous mutational opportunity in the window, and A is a constant computed by least squares from the data. Similarly, we transformed the average and homogeneity in polymorphism using the models 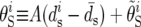 and
and 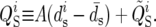
Connecting the data with the model requires us to relate our proxy of the rate of adaptation in a window, 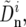 to the rate itself, υi. We assume that these are proportional to each other, namely, that
to the rate itself, υi. We assume that these are proportional to each other, namely, that
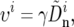 |
(15) |
where the constant of proportion is inferred from the data (Bierne and Eyre-Walker 2004). Note that in using nonsynonymous divergence as our proxy for the rate of adaptation we may be underestimating the contribution of noncoding regions. Namely, the rate of adaptation in noncoding regions would be counted only to the extent that it is correlated with nonsynonymous divergence, whereas the uncorrelated residual rate in noncoding regions would appear as noise in the estimate.
We use standard nonlinear regression analysis to fit the data to the model (e.g., Bates and Watts 1988). We define a |W| × 2 matrix of residuals as
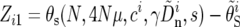 |
(16) |
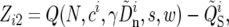 |
(17) |
where i indexes W, the set of 100-kb windows, ci is taken from the genetic map, and μ = 5.8 × 10−9 bp−1 gen−1 (Haag-Liautard et al. 2007). We find the parameters γ, s, and N by minimizing det(ZTZ). This procedure weights the noise in θS and QS equally. The average rate of selective sweeps, υ, and the average reduction in the neutral polymorphism, θ/θ0, are calculated on the basis of the inferred parameters; i.e.,
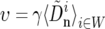 |
(18) |
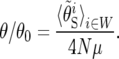 |
(19) |
Confidence intervals were found using standard tools from nonlinear regression analysis (e.g., Bates and Watts 1988).
RESULTS AND DISCUSSION
Neutral polymorphism map construction and verification:
We constructed a genomewide map of neutral polymorphism. The map was based on polymorphism data at synonymous sites from six strains of D. simulans. Among other kinds of sequence that might be used to measure neutral polymorphism, synonymous sites were chosen because they align well, abound in the euchromatic genome, and undergo weak or no selection. Initial filtration of the data involved the removal of codons present in fewer than four of the six strains and several quality checks (materials and methods). After filtering, the data set comprised just over 3 million codons distributed among 12,146 nonoverlapping genes, which amount to 42.9% of all protein-coding DNA and 7.6% of all DNA in the euchromatic genome. We considered removing synonymous polymorphism data from genes with high codon usage bias from the data set, because such sites are known to evolve under weak selection (Akashi and Eyre-Walker 1998). However, our analysis showed that the cost of the diminished sample size outweighed any benefit gained by reducing the influence of selection, so we did not remove highly codon-biased genes (appendix).
The map consists of estimates of neutral polymorphism at each position in the genome, defined as the average synonymous polymorphism across a window centered at that position, with a fixed number of synonymous sites from the sample on both sides. For these estimates, the number of synonymous sites was corrected for the difference in the mutability of different nucleotides in Drosophila; we refer to the corrected measure as synonymous mutational opportunity (materials and methods). The size of the window is a critical parameter in this procedure: if this size is too large, genuine heterogeneity along the chromosome will go unseen. Conversely, with too small a window, the true level of variation will be obscured by noise arising from the small sample size. To balance between genuine heterogeneity and sampling error, we allow the data to choose the window size for us, by comparing the performance of the estimates obtained under different window sizes at predicting observed polymorphism in exons (materials and methods). The window size that yielded the best prediction was obtained separately for each chromosome arm. For the polymorphism data, this window size was ∼1500 codons, or ∼50 kb (supplemental Figure S1). We also applied this procedure to the synonymous divergence data and obtained window sizes of ∼4500 codons (∼150 kb; supplemental Figure S2).
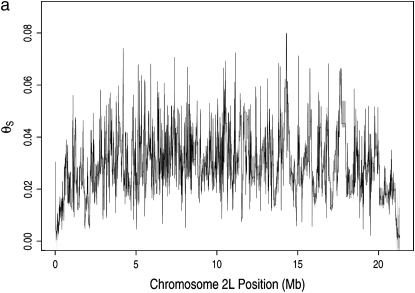
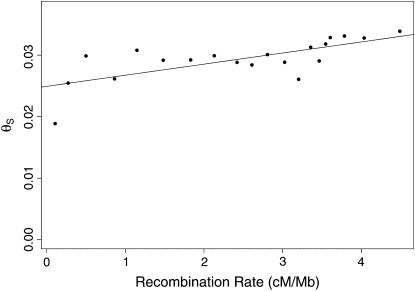
The map of neutral polymorphism exhibits heterogeneity at several spatial scales (Figure 2a, supplemental Figure S3 at http://www.genetics.org/supplemental/). By contrast, the map of neutral divergence is largely uniform (supplemental Figure S4 at http://www.genetics.org/supplemental/). For the purpose of exposition, we divide the spatial heterogeneity in neutral polymorphism into three scales: broad (>1 Mb), intermediate (20–200 kb), and fine (<10 kb). At the broad scale, ranging over the entirety of each major chromosome arm, the polymorphism estimates are greatest in the center of each arm and least at its ends. The heterogeneity at this scale likely results from the reduction of neutral polymorphism in regions of low recombination caused by recurrent selective sweeps or background selection, as previous work has suggested (Begun and Aquadro 1992; Charlesworth et al. 1993; Nordborg et al. 1996). We compared the recombination rate with the polymorphism estimates across the autosomal arms and found a significant positive correlation (Pearson's r = 0.21, P < 10−6; Figure 3), which is consistent with either of these explanations.
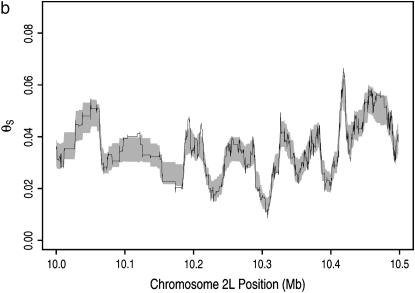
Figure 2.—
Map of neutral polymorphism estimated from data at synonymous sites. The map was calculated in units of segregating sites in a sample of size 4 per unit synonymous mutational opportunity and translated into the familiar units of θS (materials and methods). (a) Neutral polymorphism map (solid line) along chromosome arm 2L. Edges of bootstrap sleeve (shaded) are one standard deviation away from bootstrap mean; 1000 replicates are shown. (b) Same as in a, but for a 500-kb region near the center of arm 2L.
Figure 3.—
Comparison of recombination map with neutral polymorphism map across pooled autosomes. We pooled the estimates to reduce sampling error: the lists of codons were ordered by recombination rate and then grouped into pools of approximately the same total synonymous mutational opportunity. The recombination rate (x-axis) and polymorphism (y-axis) of each pool were calculated as averages across codons, where each codon was weighted according to its synonymous mutational opportunity.
At the intermediate spatial scale, heterogeneity in neutral polymorphism can be seen as ∼100 large-amplitude swings along chromosome arm 2L (Figure 2a) and is further exemplified as 5–10 such swings in a typical 500-kb window from the middle of this arm (Figure 2b). At the fine scale, heterogeneity can be seen as numerous low-amplitude changes in polymorphism in Figure 2b. On both of these scales, some of the heterogeneity arises from sampling error. To measure the extent of the sampling error, we generated 1000 bootstrap replicates of the polymorphism map. Each replicate map was constructed by the same procedure used to construct the actual polymorphism map and was based on a simulated polymorphism data set in which each codon was polymorphic with the probability predicted by the actual polymorphism map at its position. The resulting bootstrap confidence intervals appear as shaded sleeves in Figure 2, a and b (and in supplemental Figures S3 and S4). Because the fluctuation on the fine scale regularly falls inside this sleeve, it probably does result primarily from sampling error. The extensive heterogeneity on the intermediate spatial scale, however, does not appear to result from sampling error because the magnitude of swings on this scale greatly exceeds the width of the bootstrap sleeve.
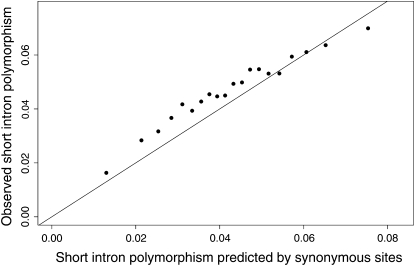
Although the intermediate-scale heterogeneity we observe does not appear to be the outcome of sampling error, it may reflect some property of synonymous sites rather than genuine heterogeneity in neutral polymorphism. To confirm that the intermediate-scale heterogeneity does reflect genuine variation in the level of neutral polymorphism, we compared the map with levels of polymorphism in short introns, an independent set of sequences thought to evolve neutrally (Halligan et al. 2004; Halligan and Keightley 2006). If the map, which is based on synonymous sites, reflects genuine changes in neutral polymorphism throughout the genome, then it should be able to predict levels of polymorphism observed in short introns well. Because broad-scale heterogeneity in polymorphism should contribute to the correspondence between synonymous and short intronic polymorphism, and because the genuineness of the intermediate-scale heterogeneity is at issue, we restricted our analysis to the high-recombination regions (c ≥ 2.5 cM/Mb) in the centers of the chromosome arms to reduce the contribution of the broad-scale phenomenon. We compiled a list of all 16,845 introns of length ≤86 bp and removed 16 and 6 bp from the 5′ and 3′ ends of each intron because these regions are under strong selective constraint (Halligan et al. 2004; Halligan and Keightley 2006). We then compared the observed level of polymorphism at each intron with the value of the polymorphism map there, after correcting for alignment error and the difference in weak selective constraint between synonymous and intronic sites (appendix). We find a very close correspondence between the levels of polymorphism from the map and those observed at short introns, which persists over the full range of polymorphism values present (Figure 4; Pearson's r = 0.210, P < 10−6; Pearson's r = 0.982, P < 10−6 when introns are pooled by predicted polymorphism). This correspondence is also observed when the entire arm is included (supplemental Figure S5 at http://www.genetics.org/supplemental/; Pearson's r = 0.250, P < 10−6; pooled Pearson's r = 0.990, P < 10−6). Taken together, the bootstrap analysis above and the close correspondence between the neutral polymorphism map and the levels of polymorphism in neutrally evolving short introns suggest strongly that much of the intermediate spatial-scale heterogeneity in polymorphism we observe is genuine.
Figure 4.—
Relationship between the level of polymorphism observed at short introns and predicted by the map based on synonymous polymorphism. To reduce sampling noise, the data were grouped by predicted polymorphism into 20 pools of similar intronic mutational opportunity.
Analysis of the spatial heterogeneity in neutral polymorphism:
We focus on the intermediate-scale heterogeneity observed in the map of neutral polymorphism for the remainder of this article. In line with previous studies (Begun and Aquadro 1992; Charlesworth et al. 1993; Nordborg et al. 1996), we also observed a pronounced broad-scale reduction in polymorphism at the low-recombination regions near the centromeric and telomeric ends of chromosomal arms (Figure 2a). To avoid conflating intermediate- with broad-scale heterogeneity, we restrict our analysis to the high-recombination regions in the center of the chromosomal arms (c ≥ 2.5 cM/Mb). We divided these high-recombination regions into overlapping 100-kb windows (materials and methods). This window size was chosen because it should capture intermediate-scale heterogeneity well. We then quantified the heterogeneity across and within the 100-kb windows, using two summary statistics. The first statistic, θS, measures average polymorphism and is defined as the ratio of the number of synonymous polymorphisms to the total synonymous mutational opportunity within a window. The second statistic, QS, is defined as the ratio of the minimum to the mean polymorphism, where both are taken from the map of neutral polymorphism within a window. QS is a measure of homogeneity in polymorphism: if neutral polymorphism varies little within the window, QS approaches its maximum, 1, but if polymorphism dips inside the window, QS takes a value close to its minimum, 0.
We compared the distribution of these statistics in the data set with the distribution expected under the simplest model of selective neutrality, namely, a model of mutation and random genetic drift in a panmictic population of constant size. The expected distribution was generated by coalescent simulations that produced a polymorphism data set, which consists of a single polymorphism observation at each location of a valid codon in the real data set. The simulations assume a constant neutral mutation rate, chosen such that the simulated mean polymorphism matched the observed mean of the combined autosomal regions, and a population size of 106 and incorporate empirical estimates of the local recombination rate from D. melanogaster (appendix). Because the expected distributions are based on the simplest model of selective neutrality, they cannot and are not used to test neutral hypotheses with more complex underlying models, such as those that result from complex demographic scenarios. We use these expected distributions only as a yardstick against which to compare the heterogeneity in polymorphism observed in the data.
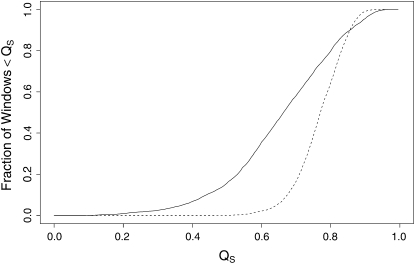
The observed average polymorphism θS is much more variable across windows than in the neutral simulations (observed σ(θS)/μ(θS) = 0.267, simulated σ(θS)/μ(θS) = 0.099, P < 10−6, Kolmogorov–Smirnov two-sample test). Furthermore, the heterogeneity within 100-kb windows, measured by QS, is much greater in the observed neutral polymorphism (Figure 5). For instance, in nearly 20% of the windows from the data, QS takes a value of ≤0.5, while so small a value is never observed in the neutral simulations. Such extensive heterogeneity on the intermediate spatial scale in regions of high recombination is expected under frequent and strong selective sweeps (Kaplan et al. 1989; Gillespie 2004). However, many other evolutionary processes, including mutation, purifying selection, and demographic processes (including population structure and variable population size) may also produce extensive heterogeneity in neutral polymorphism on the intermediate spatial scale in these regions.
Figure 5.—
Comparison of observed spatial heterogeneity in polymorphism to neutral simulations. QS was evaluated on a set of windows produced by sliding a 100-kb window along the highly recombining autosomal regions by steps of 600 bp; to reduce sampling noise, we discarded any window with total synonymous mutational opportunity <1000. This procedure was applied to both the data and the neutral simulations described in the text. For a given value of QS, say x, along the abscissa, the ordinal value is the fraction of windows in which QS exceeded x. The solid curve corresponds to the data, and the dashed curve to the neutral simulations.
Analysis of the spatial correspondence between neutral polymorphism and divergence at nonsynonymous sites:
A distinctive signature of recurrent selective sweeps may be found, if not in the pattern of polymorphism alone, in the correspondence of polymorphism with divergence. In genomic regions where adaptation is relatively frequent, we should expect both low levels of synonymous polymorphism caused by sweeps and high levels of nonsynonymous divergence caused by adaptive substitutions. Thus, recurrent sweeps should generate a negative correlation between θS and Dn, the number of nonsynonymous divergences in a 100-kb window. How might other evolutionary processes contribute to this correlation? Recent demographic events can affect polymorphism substantially (Wall et al. 2002; Haddrill et al. 2005; Jensen et al. 2005; Thornton and Jensen 2007) but have a negligible effect on divergence. Therefore, demographic processes should not generate a correlation between θS and Dn, although they may weaken an existing correlation by introducing variation in the level of θS that is not correlated with the level of Dn. Regional variation in the mutation rate or the level of selective constraint should positively associate polymorphism and divergence at both nonsynonymous and synonymous sites and in particular should generate a positive correlation between θS and Dn. Therefore, in the absence of selective sweeps, we would expect variation in mutation rate and selective constraint to generate a positive correlation between θS and Dn, and we would expect demographic processes to weaken this correlation. In the presence of selective sweeps, however, the positive correlation between θS and Dn would be reduced and could even become significantly negative were the contribution of selective sweeps to overcome the contributions from variation in mutation rate and constraint.
As a first check, we calculated the correlation between ds and dn, respectively the average synonymous and nonsynonymous divergence per site in a 100-kb window, and the correlation between θS and ds. Both correlations should mainly reflect variation in mutation rate and selective constraint, because demographic processes and selective sweeps should have little impact on ds. Therefore we expect both correlations to be positive. Both correlations are indeed significantly positive, in accordance with this reasoning [Pearson's r(ds, dn) = 0.350, P < 10−5; and Pearson's r(θS, ds) = 0.080, P < 0.015; supplemental Figure S6, a and b, at http://www.genetics.org/supplemental/). To account for the overlap between 100-kb windows in assessing the significance of these correlations, we cyclically permuted (MacLane 1999) one of the statistics from a correlation, e.g., ds, repeatedly to generate the null distribution; 105 replicates were generated. Cyclic permutation randomizes the correlation, as would a standard permutation, but additionally preserves the arrangement of each measure in space. For subsequent significance tests we proceed analogously unless otherwise noted.
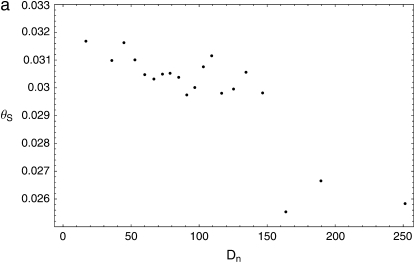
We find the correlation between θS and Dn to be significantly negative (Pearson's r = −0.115, P < 0.002; permutation test; supplemental Figure S6c at http://www.genetics.org/supplemental/); when these data are pooled by Dn to reduce sampling noise, we see that this relationship is negative and generally decreasing over the entire range of Dn (Figure 6a; pooled Pearson's r = −0.845, P < 10−5; permutation test). When we controlled for the contribution of variation in mutation and selective constraint, by computing the partial correlation of θS and Dn with respect to ds, the correlation becomes more significantly negative [Pearson's r(θS, Dn | ds) = −0.140, P < 10−5; permutation test; supplemental Figure S6d at http://www.genetics.org/supplemental/]. Because the broad-scale association of polymorphism and divergence with recombination (Begun and Aquadro 1992; Betancourt and Presgraves 2002) might contribute to the negative correlation despite the restriction to high-recombination regions, we also controlled for the recombination rate and found the correlation to be little changed [Pearson's r(θS, Dn | c) = −0.120, P < 0.001; permutation test]. Taken together, these results suggest that recurrent selective sweeps contribute substantially to the heterogeneity observed on the intermediate spatial scale.
Figure 6.—
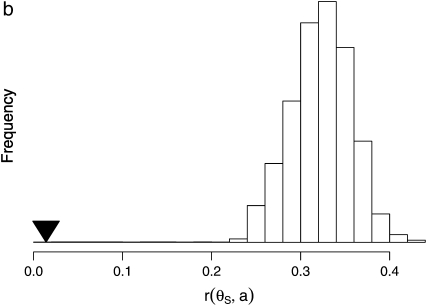
Observed relationships between proxies for the rate of adaptation and neutral polymorphism. All measures were computed over the set of 100-kb windows described in the text. (a) The relationship between the number of nonsynonymous divergences, Dn, and the average neutral polymorphism, θS. The set of windows is grouped by Dn into 20 pools of similar nonsynonymous mutational opportunity to reduce sampling noise. (b) Relationship between observed value of the correlation between the McDonald–Kreitman estimate of the number of adaptations, a = Dn − (Ds/Ps)Pn (Mcdonald and Kreitman 1991; Smith and Eyre-Walker 2002), and θS, and the null distribution of this correlation obtained by permutation, based on 1000 replicates. (c) The relationship between Dn and the homogeneity in polymorphism, QS. The set of windows has been grouped into 20 pools by Dn as in a. (d) Relationship between observed value of the correlation between Dn and θS and the null distribution of this correlation obtained by shuffling in 5-kb segments, based on 1000 replicates.
Our finding of a negative correlation between θS and Dn is likely related to that of a recent study by Shapiro et al. (2007), which found a significant negative correlation between the marginals of the McDonald–Kreitman tables in a sample of 419 genes from D. melanogaster 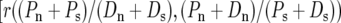 in our notation]. If lower levels of Ps(∝ θS) are associated with higher levels of Dn, we expect, everything else being equal, that lower levels of
in our notation]. If lower levels of Ps(∝ θS) are associated with higher levels of Dn, we expect, everything else being equal, that lower levels of 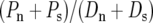 should be associated with higher levels of
should be associated with higher levels of 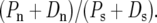 It is therefore plausible that the negative correlation between θS and Dn underlies the negative correlation observed by Shapiro et al. (2007). It is interesting to note that while Shapiro et al. (2007) suggest that these correlations may lead to a spurious detection and quantification of adaptation in a McDonald–Kreitman table pooled across many genes, we suggest these correlations are most plausibly explained as a result of positive selection. Nevertheless, these two observations do not necessarily contradict one another.
It is therefore plausible that the negative correlation between θS and Dn underlies the negative correlation observed by Shapiro et al. (2007). It is interesting to note that while Shapiro et al. (2007) suggest that these correlations may lead to a spurious detection and quantification of adaptation in a McDonald–Kreitman table pooled across many genes, we suggest these correlations are most plausibly explained as a result of positive selection. Nevertheless, these two observations do not necessarily contradict one another.
If the negative correlation between θS and Dn is indeed the result of adaptive substitutions, we should expect to see a more significant negative correlation if we replace Dn with a better proxy of the rate of adaptive substitutions. The McDonald–Kreitman estimate of the number of adaptive substitutions between D. melanogaster and D. simulans in a window, a = Dn − (Ds/Ps)Pn, is likely to be such an improved proxy (McDonald and Kreitman 1991; Smith and Eyre-Walker 2002). However, because Ps (∝ θS) is used to calculate a, θS and a will be statistically dependent across the set of windows, and this statistical dependence alone is expected to generate a strong positive correlation between them. This correlation has nothing to do with a genuine correlation between neutral polymorphism and the rate of adaptive amino acid substitution, but is rather an artifact of the estimators we use for these measures. To control for this artifactual correlation, we measured the significance of the correlation between θS and a against a null distribution that was generated from cyclic permutations, where pairs of Dn and Pn values were permuted together relative to the pairs of ds and θS. As opposed to permuting the a values themselves, this permutation procedure preserves the artifactual statistical correlation between a and θS, while dissociating the number of amino acid substitutions from the average synonymous polymorphism. Using this null distribution, we found that the correlation between a and θS is indeed significantly more negative than expected under the null distribution and considerably more significant than the correlation between θS and Dn (Pearson's r = 0.014, null mean Pearson's r = 0.322, P < 10−5; permutation test; Figure 6b).
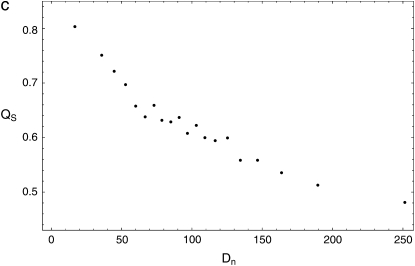
Next, we examined the relationship between QS and Dn. In regions of frequent adaptation, recurrent selective sweeps not only should reduce levels of polymorphism, but also should generate more heterogeneity in polymorphism. Specifically, recent selective sweeps produce sharp dips in levels of observed polymorphism in their vicinity, which should lead to low values of QS in regions of frequent adaptation (materials and methods). Therefore, we would expect a negative correlation between QS and Dn. We do find a strong negative correlation (Pearson's r = −0.495, P < 10−5; permutation test). How variation in mutation and selective constraint might affect QS is unclear. However, the value of the partial correlation of QS and Dn with respect to ds is similar to that of the full correlation [Pearson's r(QS, Dn | ds) = −0.480, P < 10−5; permutation test], which suggests that the negative correlation is not related to such variation. If demographic processes negligibly affect Dn, they should not have caused this correlation, although they may have weakened it substantially by introducing variation into QS that is not correlated with Dn. Figure 6c shows the strong, monotonic negative relationship between QS and Dn (pooled Pearson's r = −0.941, P < 10−5; permutation test).
It is conceivable that background selection (Charlesworth et al. 1993), like recurrent selective sweeps, could produce an association between neutral polymorphism and nonsynonymous divergence. Consider the negative correlation between θS and Dn. In regions where background selection reduces the effective population size substantially, we expect a reduced level of neutral polymorphism. If more extensive purifying selection on nonsynonymous substitutions is the cause of background selection in these regions, then we would expect such regions to show lower Dn and thus a positive correlation between θS and Dn. However, the reduction in effective population size should also elevate the rate at which slightly deleterious nonsynonymous mutations fix. If the increase in slightly deleterious substitutions in these regions outweighs the decrease due to purifying selection, this would elevate Dn and generate a negative correlation between θS and Dn. Because we do not know which of the above effects is more substantial, it is unclear whether background selection would increase or decrease the correlation between θS and Dn. Whether background selection could generate a negative correlation between QS and Dn is also not known. Thus, the current understanding of background selection, and our incomplete knowledge of the distribution of deleterious selective coefficients, does not allow conclusive predictions regarding the correlations we observe. Loewe and Charlesworth (2007) have suggested that, under a parameter regime matching that of the highly recombining autosomal regions studied here, background selection is capable of reducing neutral polymorphism at the spatial scale of a gene, i.e., our “fine” scale. When they consider multiple genes, evenly spaced at 6 kb apart, they find that the reduction in neutral polymorphism at a gene is very little changed. This suggests that in high-recombination areas background selection does not generate substantial heterogeneity in polymorphism on the intermediate spatial scale. Therefore, even if background selection were to produce a negative correlation between θS and Dn, and between QS and Dn, it would be unlikely to contribute substantially to the ∼20% reduction in θS, let alone the ∼40% reduction in QS, over the range of Dn we observe in high-recombination regions (Figure 6, a and c).
The spatial scale underlying r(θS, Dn):
The above analysis suggests that the negative correlation between θS and Dn is most plausibly explained by recurrent selective sweeps. Here we assume that recurrent selective sweeps account for this correlation and consider whether the spatial correspondence between θS and Dn contains information about the magnitude of the selective advantage associated with these sweeps. If selection were characteristically weak, this correlation would be localized. That is, because the reduction in neutral polymorphism would not extend far away from the selected site, θS would be low at the same locations that Dn is high. Alternatively, were selection characteristically strong, then neutral polymorphism would be reduced broadly around a region of high Dn.
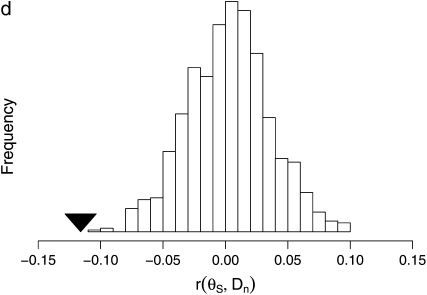
We studied the spatial scale underlying the correlation between θS and Dn by partitioning the high-recombination chromosomal regions into adjacent 5-kb segments, shuffling these segments, and then computing the correlation between θS and Dn in 100-kb windows as before. To preserve the marginal distribution of Dn, each segment was swapped with another segment with the same nonsynonymous divergence. We compared the observed correlation coefficient with the distribution of correlation coefficients based on 104 shuffled data sets. If the typical size of a swept region is <5 kb, corresponding to a relatively weak adaptation, then the association between θS and Dn should be preserved within the 100-kb windows in which they are evaluated, and thus the relationship between θS and Dn should be preserved overall. Conversely, if the typical swept region is >5 kb, corresponding to a relatively strong adaptation, then the association within the 100-kb windows should not be preserved, and the relationship between θS and Dn should be disrupted. As a control, we first determined that the positive correlation between dn and ds is not significantly changed by shuffling [Pearson's runshuffled = 0.350, mean(Pearson's rshuffled) = 0.365, P = 0.72; supplemental Figure S7 at http://www.genetics.org/supplemental/]. This is expected because much of the variation in selective constraint, which likely generates this correlation, should be present at the spatial scale of a gene (i.e., <5 kb). In contrast, the correlation between θS and Dn is significantly reduced by shuffling [Pearson's runshuffled = −0.104, mean(Pearson's rshuffled) = −0.0071, P < 10−2; Figure 6d], suggesting that adaptations of comparatively great selective advantage contribute substantially to the observed correlation between θS and Dn.
Estimating the rate, strength, and relative impact of adaptive substitution on polymorphism:
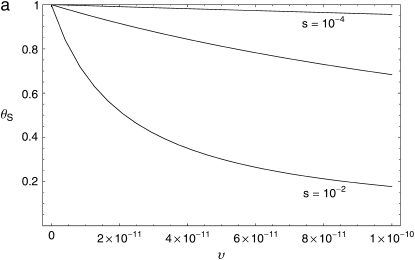
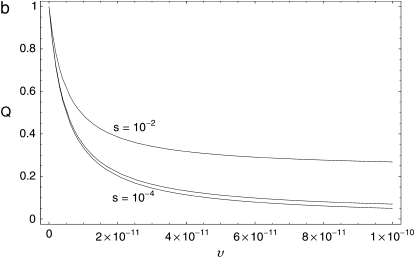
The observed associations of θS and QS with Dn plausibly reflect, and may therefore allow us to infer, the rate and strength of adaptations in D. simulans. To explore this possibility, we developed an elementary model relating θ and Q to the rate and selective advantage of adaptations (materials and methods). Figure 7 shows the predicted dependence of θ and Q on the rate υ and selective advantage s of selective sweeps. These graphs confirm that both θ and Q decrease as the rate of selective sweeps increases. However, they also show that for a given rate of adaptation, stronger selection reduces θS, but increases QS. This is because stronger selection increases the width of the region in which a sweep depresses polymorphism, which reduces θS, and also draws the mean polymorphism in a window nearer the minimum polymorphism, increasing QS.
Figure 7.—
Theoretical relationships between neutral polymorphism statistics and the rate and strength of adaptation. The range of rates, υ = 10−11–10−10 bp−1 gen−1, the recombination rate, c = 3 × 10−8 bp−1 gen−1, and the population size, N = 106, were drawn from the recent Drosophila literature (Andolfatto and Przeworski 2000; Smith and Eyre-Walker 2002; Andolfatto 2005). Curves are plotted for three values of the selective coefficient, s = 10−2, 10−3, 10−4. (a) Dependence of the average level of neutral polymorphism, θS, expressed as the ratio of its value to the neutral expectation, on the rate, υ. (b) Dependence of Qs on υ.
The fact that θS and QS respond differently to s is crucial to our ability to infer the rate and strength of adaptive substitutions. Previous attempts to infer these parameters on the basis of θS alone (e.g., Wiehe and Stephan 1993) were unable to distinguish between them because the rate and strength are confounded in their effect on the average polymorphism (Equation 12). Adding the QS statistic allows us to disentangle these parameters, because the minimal polymorphism in a window, which appears in QS, is primarily affected by the rate of adaptive sweeps and not by their strength (see derivation and intuition in materials and methods). This is because the minimal polymorphism in a window is expected to appear in close proximity to the location at which the last adaptive substitution has occurred, where even a relatively weak sweep would have driven the polymorphism to zero. Therefore, at the time the sample is taken, the minimal polymorphism in a window depends primarily on the time that elapsed since the most recent sweep, in which polymorphism partially recovers, and this time depends on the rate and not on the strength of selective sweeps.
We used nonlinear regression to fit our model to the data and infer the average rate and strength of adaptation (materials and methods). For that purpose we assume that the rate of adaptation in a window is proportional to the nonsynonymous divergence. In addition to υ and s, the model also depends on an additional unknown parameter, N, which is the effective population size in the absence of selective sweeps. On the basis of the regression we estimated the rate of adaptation to be 3.6 × 10−12 bp−1 gen−1, the selection coefficient to be 1.0 × 10−2 gen−1, and the effective population size in the absence of sweeps to be 1.5 × 106. These estimates are consistent with the compounded estimate of Wiehe and Stephan (1993), which found that (2Ns)υ > 1.3 × 10−8 compared to ∼10−7 according to our estimates.
Sweeps of weak selective strength are likely undercounted by this method because, given the limited spatial resolution of this polymorphism data set, their signatures could be masked by those of other evolutionary processes, including stronger selective sweeps and demographic processes. In other words, our method preferentially detects the impact of stronger selective sweeps. We thus expect that, compared to the true values of the rate and strength of adaptation overall, our rate estimate is an underestimate and that our estimate of the selective coefficient represents the upper end of the distribution of adaptive selective coefficients. In this light, our rate estimate of 3.6 × 10−12 bp−1 gen−1 provides independent support for the high rate of adaptation in Drosophila inferred using McDonald–Kreitman methodology, e.g., 3.6 × 10−11 bp−1 gen−1 (Andolfatto 2005) and 1.8 × 10−11 bp−1 gen−1 (Smith and Eyre-Walker 2002); see also Bierne and Eyre-Walker (2004), Charlesworth and Eyre-Walker (2006), and Welch (2006). That our rate estimate is 10–20% of the McDonald–Kreitman-based estimates suggests that 10–20% of the adaptations inferred by those methods are of a high selective coefficient of ∼1%.
The selective events preferentially registered by our inference method, namely those of greater selective strength, are of particular interest because they most influence levels of neutral polymorphism. We can measure the relative impact of selective sweeps on neutral polymorphism by θ/θ0, the ratio of observed neutral polymorphism, θ, to the level of neutral polymorphism expected in the absence of selective sweeps, θ0. We calculate θ0 as the product of 4μ and the estimate of N, where μ is the neutral mutation rate, taken to be 5.8 × 10−9 bp−1 gen−1 (Haag-Liautard et al. 2007). The point estimate of θ/θ0, 0.86, indicates that selective sweeps substantially reduce neutral polymorphism, even in high-recombination regions.
The reciprocal of this ratio, θ0/θ, is also of interest, because it is proportional to the variance in allele-frequency change per generation (Gillespie 2000, 2001) and thus to the reciprocal of twice the effective population size, 1/(2Ne). Therefore, θ0/θ reflects the combined effect of recurrent selective sweeps and other stochastic forces on the change in neutral allele frequency and the amount by which it exceeds one indicates the specific contribution of recurrent selective sweeps. The estimated value of θ0/θ, 1.16, suggests that in the high-recombination regions we studied, selective sweeps have a substantial effect on the dynamics of neutral alleles. Since this effect should be more powerful in regions of low recombination, the suggestion in Gillespie (2001) that recurrent selective sweeps are a major force in shaping neutral polymorphism appears to be likely, at least in Drosophila. This does not detract from the possible importance to neutral polymorphism of demographic processes, which have received much attention in the recent literature (Haddrill et al. 2005; Jensen et al. 2005; Thornton and Jensen 2007).
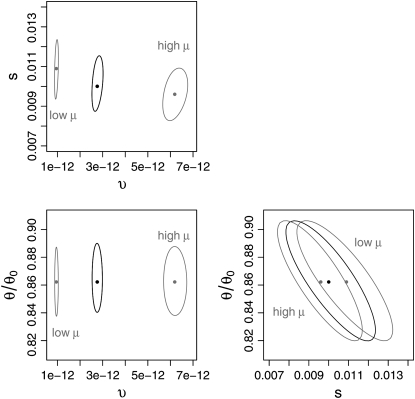
We explored the robustness of our estimates, for the moment regarding the underlying model assumptions as correct, by calculating the ∼95% confidence region about the point estimate. This confidence-interval calculation was repeated with two further values of μ, drawn from the 95% confidence interval for μ given by Haag-Liautard et al. (2007). The results are shown in Figure 8. For a given mutation rate, the point estimates are found to be relatively robust; the selection coefficient s is least well known, ranging over ∼30% of its value. This robustness with respect to the point estimate suggests that, in principle, a procedure based on the correspondence of θS and QS with divergence is capable of distinguishing adaptive rate from strength. While the mean rate of adaptation υ and selective strength s are sensitive to the value of the mutation rate, the estimated reduction in neutral polymorphism due to selective sweeps, θ/θ0, is not. This is to be expected because both θ and θ0 are proportional to the mutation rate (materials and methods).
Figure 8.—
Confidence regions about point estimates of mean adaptation rate and strength, and reduction in neutral polymorphism, for several neutral mutation rate values. The approximate 95% confidence surface, which is based on the inference procedure described in materials and methods, is displayed as three cross-sections. In each cross-section one parameter is held fixed at its inferred value while the other two are varied. The region corresponding to the mean neutral mutation rate μ = 5.8 × 10−9 bp−1 gen−1 estimated by Haag-Liautard et al. (2007) is indicated as solid lines. The lower and upper 95% neutral mutation rate estimates from the same article, respectively μ = 2.0 × 10−9 bp−1 gen−1 and μ = 1.3 × 10−8 bp−1 gen−1, are shaded and labeled accordingly. The respective point estimates are plotted as dots near the center of each respective region. The point estimates themselves, for the mean, low, and high neutral mutation rates, respectively, are (υ, s, θ/θ0) = (2.8 × 10−12, 0.010, 0.86), (0.95 × 10−12, 0.011, 0.86), and (6.2 × 10−12, 0.0096, 0.86).
Our model and inference procedure are limited in several respects. The deterministic hitchhiking model on which our expression for the mean polymorphism is based underestimates the effect of selective sweeps (cf. Equation 12). This should cause the inference procedure to overestimate the product υs. The coalescent derivation of the minimal polymorphism assumes we can measure the minimum at a point, whereas the limited spatial resolution dictated by the data should cause us to overestimate the true minimum. This produces an overestimate of QS and in turn an underestimate of the rate of adaptation. Using nonsynonymous divergence as a proxy for adaptation poses several problems. For example, if adaptive substitutions in noncoding regions, which appear to outnumber adaptations at nonsynonymous sites (Andolfatto 2005), do not correlate strongly in space with nonsynonymous divergence, this could cause us to underestimate the rate and strength of adaptation and the magnitude of the reduction in effective population size. Finally, the inference procedure assigned equal weights to θS and QS. Because QS is more strongly correlated with Dn than is θS, QS may be more informative about recurrent selective sweeps than θS; preliminary attempts to introduce weights to account for the relative informativeness of the two statistics produced somewhat higher estimates of υ and s and lower estimates of θ/θ0. We believe that these difficulties may be overcome by developing a more complex, maximum-likelihood inference procedure based on coalescent simulations. The rudimentary procedure introduced here, however, demonstrates that the correspondence between neutral polymorphism and divergence may be used to infer the strength and rate of adaptation and its effect on polymorphism.
Conclusion:
The spatial correspondence we document between polymorphism and divergence in Drosophila suggests strongly that selective sweeps of relatively great fitness advantage recur frequently and have a substantial effect on polymorphism, in this species. Our focus on the spatial correspondence between polymorphism and divergence as a means to study the characteristics and impact of adaptive substitutions is quite different from, though complementary to, studies of adaptation based on genomic scans of polymorphism (Glinka et al. 2003; Orengo and Aguade 2004; Ometto et al. 2005; Wright et al. 2005; Voight et al. 2006) or applications of the McDonald–Kreitman test (Fay et al. 2002; Smith and Eyre-Walker 2002; Bierne and Eyre-Walker 2004; Andolfatto 2005; Bustamante et al. 2005; Charlesworth and Eyre-Walker 2006; Eyre-Walker 2006; Welch 2006; Shapiro et al. 2007). Both genomic scans of polymorphism and the method presented here rely on the signature of selective sweeps on spatial patterns of polymorphism. However, genomic scans of polymorphism utilize additional information present in haplotype structure and allow the identification of specific regions in which adaptive substitutions may have recently taken place. On the other hand, comparing polymorphism and divergence may allow us to distinguish the effect of sweeps from that of other forces, such as demographic processes, and may therefore be more reliable for inferring the average characteristics of sweeps. Both the method presented here and methods based on McDonald–Kreitman methodology compare polymorphism with divergence and neutral with functional sites. Both methodologies also rely on strong steady-state assumptions. While the McDonald–Kreitman methodology assumes that the degree of constraint at functional sites at present has not changed since the species under consideration diverged, our method assumes that nonsynonymous divergence since the species split is a reasonable proxy for the recent rate of adaptation. Methods derived from the McDonald–Kreitman test likely capture adaptations across a large spectrum of positive selective coefficients and therefore probably provide better estimates of the overall rate of adaptation, but unlike the method presented here are not informative about the selective advantage of these adaptations. Each of these methods thus provides a complementary view of the adaptive process. In the future, more sophisticated models and inference procedures may allow for inference based on the main ideas of each and perhaps also account for the impact of other evolutionary processes, such as demography and purifying selection.
As a useful illustration of the impact of recurrent selective sweeps on the dynamics of neutral alleles, consider the trajectory of a neutral allele destined for fixation. On the basis of the mean neutral polymorphism level in D. simulans, this trajectory on average spans ∼4 × 106 generations in this population (Gillespie 2004; Andolfatto 2005). During this time, based on our estimates of the rate and selective strength of adaptation, a given site in a highly recombining region will be affected by an average of two selective sweeps in the surrounding 100 kb, and each of these sweeps will reduce the heterozygosity at the site by an average of 50% (appendix). If these estimates are close to the truth, then it is likely that genetic draft, the term for the process by which selective sweeps alter the frequencies of neutral alleles at linked sites (Gillespie 2000, 2001), is a major force in the molecular evolution of D. simulans alongside genetic drift, demographic processes, and purifying selection.
APPENDIX
Short intron data corrections:
We use data from short introns to examine the veracity of our map at predicting the levels of neutral polymorphism. To compare the levels of polymorphism at short introns with those predicted by the map, we need to correct for two complications. First, the alignment of intronic sequences contains errors that substantially distort the observations of polymorphism (result not shown). Second, different levels of weak selective constraint at synonymous and intronic sites are expected to offset the comparison.
To correct for misalignments, we took advantage of the fact that most misalignments in the data set appear as runs of nonmonomorphic sites (results not shown). We therefore removed all runs of two or more nonmonomorphic intronic sites at any coverage and applied a correction to the predicted polymorphism based on the map to account for the genuine runs of polymorphism or divergence we removed.
To correct the predicted intronic polymorphism for the removal of runs, we assume that runs are of two kinds. First, some runs will comprise spuriously observed polymorphism and divergence due to misalignments. However, as removing such runs amounts to removing random stretches of sequence, they should not affect the probability of observing a polymorphism or a divergence. Second, some runs will comprise genuine runs of polymorphism and divergence. To account for the removal of these runs we calculate the probability of observing an isolated polymorphism or divergence in a stretch of sequence in which runs were removed. The probability of observing a polymorphism after the removal of genuine runs, Pp, is
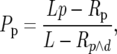 |
(A1) |
where L is the length of an intron in base pairs, p is the probability of a genuine polymorphism at an intronic site, Rp is the expected number of polymorphic sites that appear in runs, and Rp∧d is the expected overall number of intronic sites that appear in runs. Similarly, the probability of observing a divergence after the removal of genuine runs, Pd, is
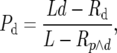 |
(A2) |
where d is the analogous probability for divergence, and Rd is the expected number of divergent sites that appear in runs. When 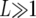 and
and 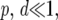 Rp, Rd, and Rp∧d can be approximated by
Rp, Rd, and Rp∧d can be approximated by
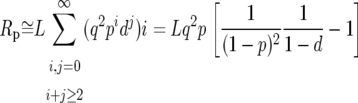 |
(A3) |
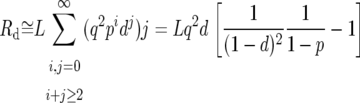 |
(A4) |
and
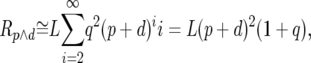 |
(A5) |
where q ≡ 1 − (p + d). We confirmed by simulation that these approximations work well for the levels of neutral polymorphism and divergence in short introns (results not shown). Substituting Equations A3–A5 into Equations A1 and A2 we find that
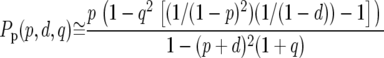 |
(A6) |
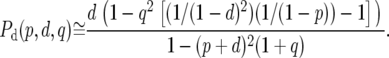 |
(A7) |
To account for the average difference in selective constraint between synonymous and intronic sites, we assume that the predicted intronic polymorphism and divergence based on the maps take the form
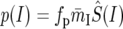 |
(A8) |
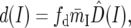 |
(A9) |
where fp and fd are parameters that account for the way the differences between intronic and synonymous selective constraint affect the differences in polymorphism and divergence, 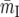 is the mean intronic mutational opportunity at intron I, and
is the mean intronic mutational opportunity at intron I, and 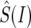 and
and 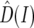 are the polymorphism and divergence map estimates at intron I. To estimate fp and fd we maximized the likelihood of the predicted intronic polymorphism with respect to these parameters as follows: we assume that the observed polymorphism and divergence, after resampling to coverage four and the filtration of runs, are multinomially distributed. Namely,
are the polymorphism and divergence map estimates at intron I. To estimate fp and fd we maximized the likelihood of the predicted intronic polymorphism with respect to these parameters as follows: we assume that the observed polymorphism and divergence, after resampling to coverage four and the filtration of runs, are multinomially distributed. Namely,
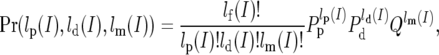 |
(A10) |
where lp(I) and ld(I) are the numbers of sites from intron I that are polymorphic and divergent among these sites, lm(I) is the number of sites that are neither, and Q ≡ 1 − (Pd + Pp). We therefore obtained the autosomal fp and fd by maximizing
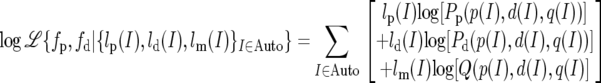 |
(A11) |
The maximum-likelihood estimates for the autosomes are fp = 1.068 and fd = 0.922.
Codon bias analysis:
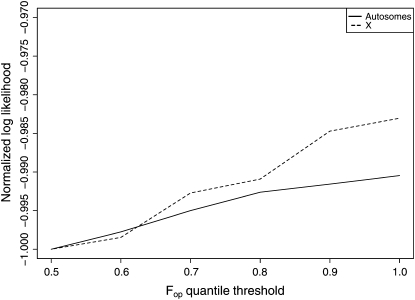
Synonymous sites in genes with high codon usage bias are known to evolve under weak selection (Akashi and Eyre-Walker 1998) in Drosophila, which could make them an unreliable proxy for neutrality. Therefore, we considered removing highly codon-biased genes from the data set used in the construction of the map. To ascertain whether removing highly codon-biased genes improves the map, we compared the performance of polymorphism maps produced from subsets of the data from which the genes in the 50th, 60th, … , 90th percentile of Fop value (Ikemura 1981) had been removed, at predicting the polymorphism in short introns. The performance of these maps at predicting the polymorphism at short introns was measured in terms of the maximum likelihood defined by Equation A11. The map built using all genes exhibited the highest likelihood (Figure A1), indicating that the cost of the diminished sample size outweighed any benefit gained by reducing the influence of selection. Therefore we did not remove highly codon-biased genes.
Figure A1.—
Log likelihood of polymorphism estimates from synonymous sites at short introns as a function of Fop quantile threshold. Curves for the autosomes (solid) and X chromosome (dashed) are shown. The X and autosomes are analyzed separately because of the different behavior of fp, fd, and codon usage bias on the X and autosomes Singh et al. (2005b). The autosomal log-likelihood values have each been divided by 49,016.8, and the X chromosomal log-likelihood values have each been divided by 5581.2 to allow the curves to appear on the same scale.
Neutral coalescent simulations:
We generated a polymorphism map based on the simplest model of selective neutrality. The map was produced using Hudson's simulation program ms (Hudson 2002) for a panmictic population of constant size N = 106, in the following steps.
We first divided the high-recombination regions of autosomes into windows of 50 kb. The recombination rate within a window cw was estimated from the genetic map at the window's midpoint. This rate provided the recombination parameter 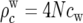 for the ms simulation of that window. ms was then used to generate the relative depth of the genealogy at each segment between recombination breakpoints within each 50-kb window, under a model with constant mutation rate, using a uniform mutational rate of ν = 10−9 to define the mutational parameter
for the ms simulation of that window. ms was then used to generate the relative depth of the genealogy at each segment between recombination breakpoints within each 50-kb window, under a model with constant mutation rate, using a uniform mutational rate of ν = 10−9 to define the mutational parameter 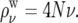 The 50-kb segments thus generated were concatenated to form simulated chromosome arms. We confirmed that dividing the chromosome arm into 50-kb windows negligibly affects the correlation between genealogy depths in neighboring segments, by examining the autocorrelation of genealogy depths. The autocorrelation fell to zero well before 50 kb (results not shown).
The 50-kb segments thus generated were concatenated to form simulated chromosome arms. We confirmed that dividing the chromosome arm into 50-kb windows negligibly affects the correlation between genealogy depths in neighboring segments, by examining the autocorrelation of genealogy depths. The autocorrelation fell to zero well before 50 kb (results not shown).
The absolute mutation rate along the chromosome arm, u, was determined by the requirement that the average polymorphism equals that observed in the data. Thus
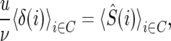 |
(A12) |
where δ(i) is the relative genealogy depth at chromosome position i, and the averages are taken over the set of chromosome positions at high-recombination regions of codons in our data C.
The genealogy depths along a chromosomal arm and the calculated mutation rate were then used to generate a polymorphism data set, 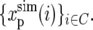 The data set was composed of polymorphism observations at each position where we possess an observation in the real polymorphism data set, where these simulated observations were randomly generated according to the probabilities
The data set was composed of polymorphism observations at each position where we possess an observation in the real polymorphism data set, where these simulated observations were randomly generated according to the probabilities
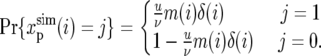 |
(A13) |
The neutral polymorphism maps based on these data were produced by the same method we used to produce the map based on real observations.
The effect of recurrent selective sweeps on a neutral allele destined for fixation:
We assume that a new neutral mutation destined for fixation takes an average of 4Ne generations to fix. If selective sweeps recur at the estimated rate of υ = 3.6 × 10−12 bp−1 gen−1, then the number of sweeps expected to occur in the 100 kb surrounding the neutral site during this time is (100 kb)4Neυ = 1.9. We simulated the change in neutral polymorphism caused by a single selective sweep using a deterministic two-locus hitchhiking model (cf. Gillespie 2004). On the basis of simulations with parameters s = 1.0 × 10−2 gen−1 as estimated, Ne = 106, c = 2.5cM/Mb, and with the distance between the neutral site and the site of adaptive substitution ranging between 0 and 50 kb, we found an average reduction of 50% in heterozygosity.
Acknowledgments
We thank Carlos Bustamante and two anonymous reviewers for many useful comments on the manuscript. We thank Molly Przeworski, Marc Feldman, Nadia Singh, Emile Zuckerkandl, Andrew Berry, and Aaron Hirsh for insights and comments on the manuscript and Yosi Rinot, Samuel Sattath, and Peter Arndt for helpful discussions. We are also grateful to Charles Langley, David Begun, Alisha Holloway, and Kristian Stevens of the Drosophila Population Genomics Project for providing assistance and access to the alignments. J.M.M. is a Howard Hughes Medical Institute Predoctoral Fellow. J.M.M. was supported in part by National Institutes of Health (NIH) grant GM 28016 to Marc Feldman. J.M.M., G.S., J.C.D., and D.A.P. were funded by NIH no. 5R01GM077368 and National Science Foundation no. 0317171 to D.A.P. G.S. was supported by a Flegg fellowship and by the Israel Science Foundation (grant no. 1435/07).
References
- Akashi, H., and A. Eyre-Walker, 1998. Translational selection and molecular evolution. Curr. Opin. Genet. Dev. 8 688–693. [DOI] [PubMed] [Google Scholar]
- Aminetzach, Y. T., J. M. Macpherson and D. A. Petrov, 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309 764–767. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437 1149–1152. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., and M. Przeworski, 2000. A genome-wide departure from the standard neutral model in natural populations of Drosophila. Genetics 156 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, D. M., and D. G. Watts, 1988. Nonlinear Regression Analysis and Its Applications. Wiley, New York.
- Bauer DuMont, V., and C. F. Aquadro, 2005. Multiple signatures of positive selection downstream of notch on the X chromosome in Drosophila melanogaster. Genetics 171 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356 519–520. [DOI] [PubMed] [Google Scholar]
- Beisswanger, S., W. Stephan and D. D. Lorenzo, 2006. Evidence for a selective sweep in the wapl region of Drosophila melanogaster. Genetics 172 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt, A. J., and D. C. Presgraves, 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99 13616–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, N., and A. Eyre-Walker, 2004. The genomic rate of adaptive amino acid substitution in Drosophila. Mol. Biol. Evol. 21 1350–1360. [DOI] [PubMed] [Google Scholar]
- Bustamante, C. D., A. Fledel-Alon, S. Williamson, R. Nielsen, M. T. Hubisz et al., 2005. Natural selection on protein-coding genes in the human genome. Nature 437 1153–1157. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, J., and A. Eyre-Walker, 2006. The rate of adaptive evolution in enteric bacteria. Mol. Biol. Evol. 23 1348–1356. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A., 2006. The genomic rate of adaptive evolution. Trends Ecol. Evol. 21 569–575. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., G. J. Wyckoff and C. I. Wu, 2002. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature 415 1024–1026. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 1994. The Causes of Molecular Evolution. Oxford University Press, London/New York/Oxford.
- Gillespie, J. H., 2000. Genetic drift in an infinite population. The pseudohitchhiking model. Genetics 155 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, J. H., 2001. Is the population size of a species relevant to its evolution? Evolution 55 2161–2169. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 2004. Population Genetics: A Concise Guide, Ed. 2. Johns Hopkins University Press, Baltimore.
- Glinka, S., L. Ometto, S. Mousset, W. Stephan and D. De Lorenzo, 2003. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multilocus approach. Genetics 165 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbling, G., and V. Strelets, 2006. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34 D484–D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard, C., M. Dorris, X. Maside, S. Macaskill, D. L. Halligan et al., 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445 82–85. [DOI] [PubMed] [Google Scholar]
- Haddrill, P. R., K. R. Thornton, B. Charlesworth and P. Andolfatto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan, D. L., and P. D. Keightley, 2006. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 16 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan, D. L., A. Eyre-Walker, P. Andolfatto and P. D. Keightley, 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18 337–338. [DOI] [PubMed] [Google Scholar]
- Ikemura, T., 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151 389–409. [DOI] [PubMed] [Google Scholar]
- Jensen, J. D., Y. Kim, V. Bauer DuMont, C. F. Aquadro and C. D. Bustamante, 2005. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, N. L., R. R. Hudson and C. H. Langley, 1989. The “hitchhiking effect” revisited. Genetics 123 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., and W. Stephan, 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK/London/New York.
- Lewontin, R. C., 1974. The Genetic Basis of Evolutionary Change. Columbia University Press, New York.
- Loewe, L., and B. Charlesworth, 2007. Background selection in single genes may explain patterns of codon bias. Genetics 175 1381–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLane, S., 1999. Algebra. American Mathematical Society, Providence, RI.
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23 23–35. [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351 652–654. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., B. Charlesworth and D. Charlesworth, 1996. The effect of recombination on background selection. Genet. Res. 67 159–174. [DOI] [PubMed] [Google Scholar]
- Ohta, T., 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23 263–286. [Google Scholar]
- Ometto, L., S. Glinka, D. De Lorenzo and W. Stephan, 2005. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 22 2119–2130. [DOI] [PubMed] [Google Scholar]
- Orengo, D. J., and M. Aguade, 2004. Detecting the footprint of positive selection in a European population of Drosophila melanogaster: multilocus pattern of variation and distance to coding regions. Genetics 167 1759–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski, M., 2002. The signature of positive selection at randomly chosen loci. Genetics 160 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke, T. A., and D. J. Begun, 2004. Strong selective sweep associated with a transposon insertion in drosophila simulans. Proc. Natl. Acad. Sci. USA 101 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, J. A., W. Huang, C. Zhang, M. J. Hubisz, J. Lu et al., 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N. D., P. F. Arndt and D. A. Petrov, 2005. a Genomic heterogeneity of background substitutional patterns in Drosophila melanogaster. Genetics 169 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N. D., J. C. Davis and D. A. Petrov, 2005. b X-linked genes evolve higher codon bias in Drosophila and Caenorhabditis. Genetics 171 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N. G., and A. Eyre-Walker, 2002. The compositional evolution of the murid genome. J. Mol. Evol. 55 197–201. [DOI] [PubMed] [Google Scholar]
- Thornton, K. R., and J. D. Jensen, 2007. Controlling the false-positive rate in multilocus genome scans for selection. Genetics 175 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K. R., J. D. Jensen, C. Becquet and P. Andolfatto, 2007. Progress and prospects in mapping recent selection in the genome. Heredity 98 340–348. [DOI] [PubMed] [Google Scholar]
- Voight, B. F., S. Kudaravalli, X. Wen and J. K. Pritchard, 2006. A map of recent positive selection in the human genome. PLoS Biol. 4 e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7 256–276. [DOI] [PubMed] [Google Scholar]
- Welch, J. J., 2006. Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics 173 821–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe, T. H. E., and W. Stephan, 1993. Analysis of a genetic hitchhiking model, and its application to DNA polymorphism data from Drosophila melanogaster. Mol. Biol. Evol. 10 842–854. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., I. V. Bi, S. G. Schroeder, M. Yamasaki, J. F. Doebley et al., 2005. The effects of artificial selection on the maize genome. Science 308 1310–1314. [DOI] [PubMed] [Google Scholar]